Abstract
Outer membrane phospholipase A (OMPLA) from Escherichia coli is an integral-membrane enzyme with a unique His–Ser–Asn catalytic triad. In serine proteases and serine esterases usually an Asp occurs in the catalytic triad; its role has been the subject of much debate. Here the role of the uncharged asparagine in the active site of OMPLA is investigated by structural characterization of the Asn156Ala mutant. Asparagine 156 is not involved in maintaining the overall active-site configuration and does not contribute significantly to the thermal stability of OMPLA. The active-site histidine retains an active conformation in the mutant notwithstanding the loss of the hydrogen bond to the asparagine side chain. Instead, stabilization of the correct tautomeric form of the histidine can account for the observed decrease in activity of the Asn156Ala mutant.
Keywords: Phospholipase, serine hydrolase, catalytic triad, active-site mutant, membrane protein, X-ray crystal structure, low-barrier hydrogen bond, histidine tautomer
Outer membrane phospholipase A (OMPLA; EC 3.1.1.32) is an integral-membrane enzyme present in the outer membrane of many Gram-negative bacteria. It catalyzes the hydrolysis of acyl ester bonds in phospholipids and lyso-phospholipids (Scandella and Kornberg 1971). The enzyme requires calcium for activity, and depletion of Ca2+ renders the enzyme inactive (Nishijima et al. 1977; Horrevoets et al. 1989; Snijder et al. 2001). The activity of OMPLA is regulated by reversible dimerization in a calcium- and substrate-dependent fashion (Dekker et al. 1997, 1999; Ubarretxena-Belandia et al. 1999). Dimerization leads to the formation of productive substrate-binding pockets and facilitates binding of calcium in the active site (Snijder et al. 1999). In Escherichia coli the enzyme has a role in colicin secretion (Pugsley and Schwartz 1984; Van Der Wal et al. 1995). In Campylobacter coli and Helicobacter pylori OMPLA has been implicated in virulence and pathogenicity (Grant et al. 1997; Dorrell et al. 1999).
E. coli OMPLA has a 12-stranded antiparallel β-barrel fold with the active site located at the exterior of the barrel (Snijder et al. 1999). The catalytic residues, His142, Ser144, and Asn156, are arranged in a serine hydrolase-like constellation (Horrevoets et al. 1991; Brok et al. 1995, 1996; Snijder et al. 1999). The asparagine residue in this catalytic triad is unique among serine hydrolases, where commonly an Asp or Glu residue is found instead. In OMPLA, substitution of the Asn for an aspartate or a glutamine resulted in a 2-fold and a 40-fold decrease in activity, respectively (Kingma et al. 2000). Removal of the asparagine's functional moiety by an Asn156Ala mutation resulted in a 20-fold rate reduction, demonstrating that although the asparagine residue contributes to catalysis, it does not have an essential role (Kingma et al. 2000). In contrast, mutation of the catalytic aspartate in dipeptidyl-peptidase IV and subtilisin resulted in much more dramatic decreases in activity, 500-fold and 104-fold, respectively (Carter and Wells 1988; David et al. 1993). A similar substitution in lipoprotein lipase completely abolished all activity (Faustinella et al. 1992).
To understand the role of the asparagine residue in the catalytic triad of OMPLA, we have determined crystal structures of the Asn156Ala mutant at various pHs and analyzed the thermal stability of wild-type and mutant protein. The structures show that no structural rearrangements or solvent molecules compensate for the loss of the stabilizing hydrogen bond between Asn156 and the catalytic histidine. Despite the loss of this hydrogen bond, the catalytic histidine retains its wild-type conformation. Instead, the catalytic importance of the asparagine can be explained by its ability to stabilize the correct tautomeric form of the histidine.
Results
3D structure of the mutant
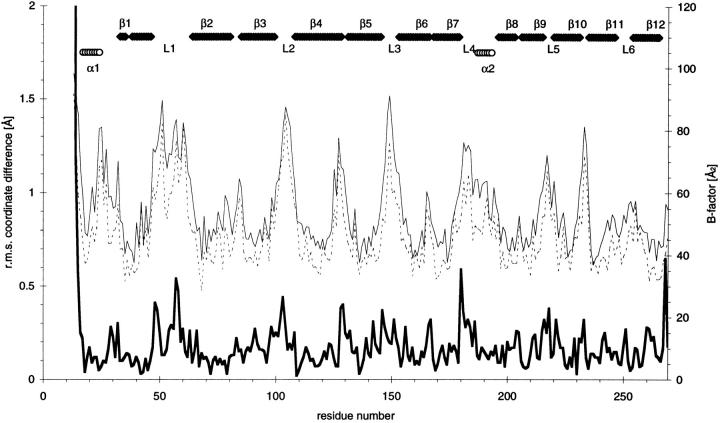
The Asn156Ala mutant (pH 6.1) has an overall fold similar to that of the native protein (Fig. 1 ▶). The r.m.s. coordinate difference between wild-type and mutant protein is as small as 0.3 Å for the common Cα atoms, and equals the estimated coordinate error in both structures (see Tables 1 and 2 for all statistics; Read 1986). The small r.m.s. coordinate difference of 0.65 Å for all 2090 common atoms stresses the overall similarity. The largest differences occur in loop regions and at the N terminus (Fig. 2 ▶) and correlate with the high crystallographic B-factors of these parts of the protein.
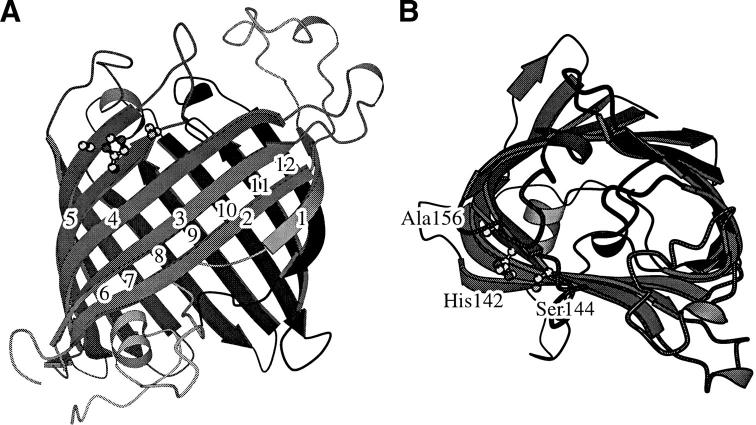
Fig. 1.
Two orthogonal views of the structure of the Asn156Ala mutant of OMPLA at pH 8.3. β-strands are shown as arrows, α-helices as spirals, and active-site residues are shown in ball-and-stick format. The β-strands are numbered in the left panel; the active-site residues are named in the right panel. The figure was prepared using MOLSCRIPT (Kraulis 1991) and Raster3D (Merritt and Bacon 1997).
Table 1.
Data collection statistics for the Asn156Ala mutant structures of E. coli OMPLA
| Data set | pH 4.6 | pH 6.1 | pH 8.3 | |
| Wavelength | (Å) | 1.5418 | 1.00 | 1.5418 |
| Resolution | (Å) | 20–2.8 | 31–2.5 | 21–2.98 |
| (2.85–2.8) | (2.54–2.5) | (3.03–2.98) | ||
| Cell dimensions | (Å) a | 78.48 | 78.17 | 78.46 |
| (Å) b | 78.48 | 78.17 | 78.46 | |
| (Å) c | 101.67 | 101.68 | 101.45 | |
| Observed reflections | 98747 | 150605 | 106289 | |
| Unique reflections | 9231 (453) | 12728 (611) | 7686 (372) | |
| Completeness | (%) | 99.3 (99.6) | 98.8 (96.1) | 99.6 (100.0) |
| Rsyma | (%) | 6.4 (34.3) | 3.7 (11.3) | 8.1 (34.0) |
| ≤I>/≤σ> | 25.4 (4.9) | 39.2 (8.3) | 24.5 (7.9) |
The values in brackets present the highest resolution bins. The crystals belong to the trigonal spacegroup P3121.
aRsym defined as ∑hkl∑i|Ii(hkl) − ≤I(hkl)>|/∑hkl∑i(hkl) * 100%.
Table 2.
Refinement statistics
| Data set | pH 4.6 | pH 6.1 | pH 8.3 |
| Resolution (Å) | 20–2.8 | 31–2.5 | 21–2.98 |
| Rcrysa (%) | 21.1 | 22.2 | 22.6 |
| Rfreeb (%) | 27.5 | 26.8 | 26.6 |
| Protein residues | 13–269 | 13–269 | 8–269 |
| Protein atoms | 2091 | 2091 | 2128 |
| Non protein atoms | 139 | 132 | 109 |
| R.m.s.d. for | |||
| Bond lengths (Å) | 0.007 | 0.006 | 0.007 |
| Bond angles (°) | 1.5 | 1.2 | 1.5 |
| Dihedral angles (°) | 25.6 | 25.7 | 25.9 |
| Average B-factor (Å2) | 49 | 53 | 46 |
aRcrys = ∑hkl,work ∥Fobs|− k|Fcalc∥/∑hkl|Fobs| * 100%.
bRfree = ∑hkl,test∥Fobs|− k|Fcalc∥/∑∼hkl|Fobs| * 100%.
Fig. 2.
Coordinate differences between Asn156Ala and wild-type OMPLA. The coordinate difference of the common Cα atoms (dark solid line); B values for mutant OMPLA (thin solid line) and wild-type OMPLA (dashed line). Secondary structure elements for residues in α-helices (open circles) and for residues in β-strands (filled diamonds).
The active site of the mutant undergoes little structural rearrangement, owing to the rigid scaffold of the β-barrel to which the active-site residues are tethered. The mutated residue 156 lacks electron density after the Cβ atom, corroborating the successful mutation of this residue from an asparagine into an alanine (Fig. 3A ▶). The mutation has not affected the position and orientation of nucleophilic serine 144. The conformation of histidine 142, however, has subtly changed. Its side chain has rotated ∼10° about χ1 and ∼40° about χ2, away from Ser144, and occupies some of the space created by the mutation (Fig. 3B ▶). As a result, the distance between the nucleophilic serine Oγ atom and the histidine Nɛ is increased from ∼3.3 Å to 3.9 Å. However, this active-site constellation is similar to that observed in the inhibited dimeric structure (Fig. 3C ▶), which we believe to be a catalytically relevant arrangement (Snijder et al. 1999).
Fig. 3.
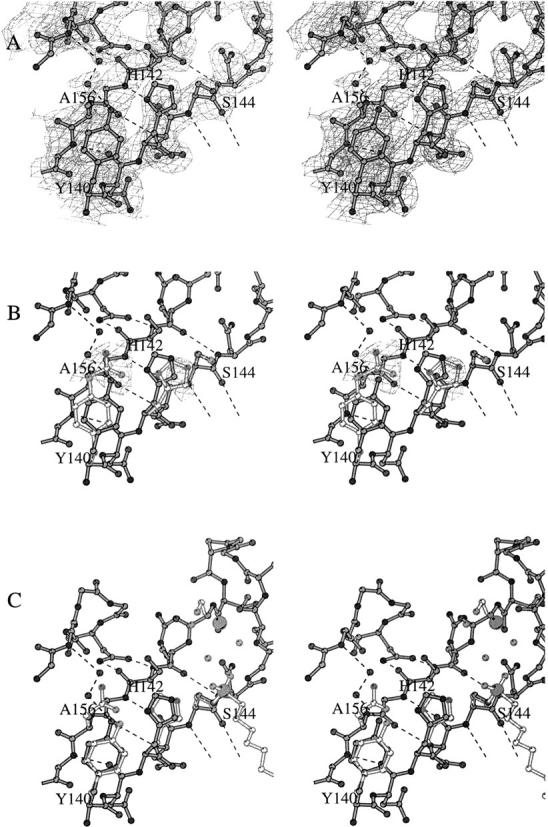
Stereo diagrams of the active site of the Asn156Ala mutant at pH 6.1. (A) Final σA-weighted electron density of the active site. (B) Superimposition of Asn156Ala and wild-type monomeric OMPLA active sites. The wild-type active-site residues are depicted in light gray. Negative (Fobs(mutant) − Fobs(native)) difference electron density is shown in light gray; the contour level is −5 σ. The two largest features in this difference density illustrate the successful mutation Asn156Ala and indicate the rotation of His142. (C) Superimposition of Asn156Ala and wild-type inhibited dimeric OMPLA. The wild-type active-site residues, the hexadecanesulfonyl inhibitor, and the catalytic calcium are depicted in light gray. Note that for some of the non-active-site residues the side chains have been omitted for clarity. This figure was constructed with BOBSCRIPT (Esnouf 1997).
Besides the movement of the imidazole ring, the space created by the mutation is partly filled by two water molecules. The first water molecule forms two hydrogen bonds, one with the hydroxyl group of tyrosine 140 and the other with the second water molecule. This second water molecule makes an additional hydrogen bond to the backbone amide of residue 181 in loop L4. None of these water molecules is at hydrogen-bonding distance from the Nδ atom of the histidine imidazole ring. Neither are other solvent molecules or protein atoms contacting His142. Although the hydroxyl group of tyrosine 140 has moved toward the imidazole side chain, it does not form a hydrogen bond as judged from the inadequate angle geometry and the large distance. Thus, as a result of the mutation of Asn156, the histidine imidazole group has lost its hydrogen bond partner.
Heat-induced unfolding of OMPLA
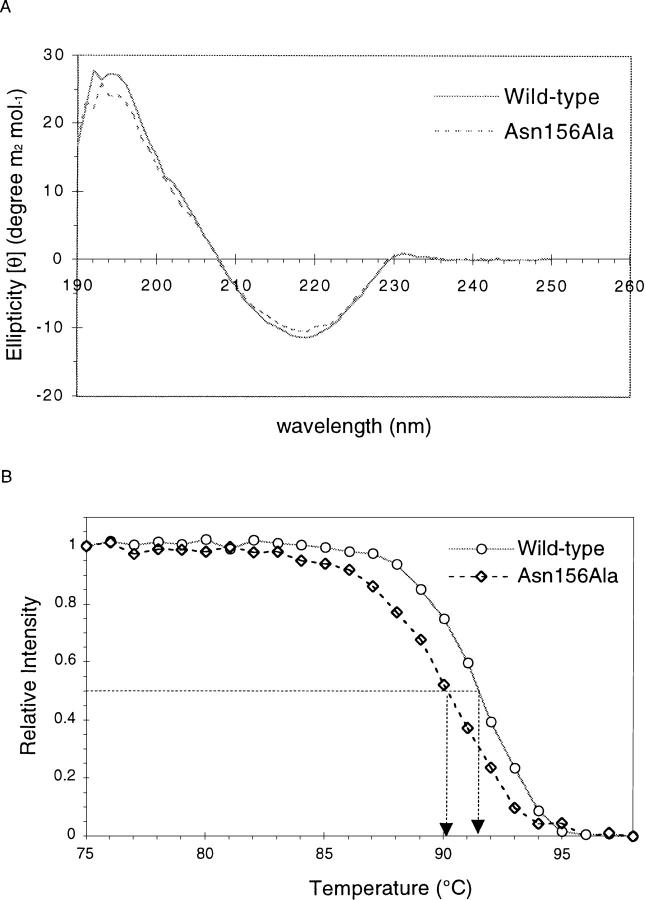
To study the importance of the His–Asn interaction for protein stability, the heat-induced unfolding of wild-type and Asn156Ala OMPLA was followed using circular dichroism (CD). The CD spectra of both species show a strong negative signal at 219 nm and a positive signal at 195 nm (Fig. 4A ▶), typical of β-sheet-containing proteins (OMPLA has 59.5% β-strand and 5.5% α-helix). Both wild-type and Asn156Ala OMPLA show an irreversible thermal transition from 83° to 95°C (Fig. 4B ▶). The melting temperatures Tm, the temperature at the midpoint of the transition curves, are 91.5°C and 90°C for wild-type and Asn156Ala, respectively.
Fig. 4.
Circular dichroism (CD) studies of OMPLA. (A) The CD spectra show a typical β-sheet spectrum with a minimum at 219 nm. The spectra are background and base-line corrected. (B) Heat-induced unfolding of OMPLA monitored by the CD signal at 219 nm. The difference in melting temperatures between mutant and wild-type protein is reproducible within 0.1°C.
Influence of the pH
The mutant has a pH-activity profile with a broad optimum at pH 8.3. At acidic pH (around pH 4) the activity is virtually zero. In the very basic pH region (pH > 10) the activity levels off to ∼75% of that at pH 8.3 (Kingma et al. 2000). We have investigated the pH dependence by examining crystal structures at three different pHs, at pH 4.6, 6.1, and 8.3, respectively. The crystal structures at pH 4.6 and pH 8.3 are very similar to the structure at pH 6.1. The r.m.s coordinate differences are all within the estimated coordinate error. Nevertheless, the decrease in distance between the Ser144 Oγ and the His142 Nɛ atoms from ∼4.0 Å at pH 4.6, via 3.9 Å at pH 6.1, to ∼3.7 Å at pH 8.3 may be related to the deprotonation of the imidazole side chain at higher pH.
Interestingly, at pH 8.3 we observed interpretable density for residues 8–12. At the lower pH used previously (pH 6.1), only clear electron density was observed starting from residue 13. At pH 8.3 a stabilizing interaction is made possible between the His9 side chain and the main-chain peptide nitrogen atom of Asn85, most likely because His9 becomes deprotonated at this pH. Residues 8–12 form a short loop at the periplasmic side of the protein surface that points toward the center of the β-barrel. In our electron density maps we found no indication where residues 1–7 could be located, but electron microscopy projection maps of OMPLA at pH 7.5 indicate that they protrude away from the barrel (Boekema et al. 1998). The orientation of these residues away from the dimerization interface of the β-barrel domain indicates that they will neither conflict with nor be involved in dimerization.
Discussion
OMPLA is a phospholipase with a serine hydrolase-like catalytic triad. Instead of the usual carboxylic acid residue, OMPLA has a neutral asparagine in the triad together with a histidine and a serine. Although serine hydrolases have been reported with a neutral main-chain carbonyl oxygen atom (Wei et al. 1995) or a histidine residue (Chen et al. 1996; Qiu et al. 1996, 1997), the occurrence of an asparagine in a serine triad has never before been observed.
In OMPLA mutation of the active-site asparagine to an alanine resulted in a modest 20-fold reduction in the reaction rate (Kingma et al. 2000). A similar substitution in the cysteine protease papain, which contains a Cys–His–Asn catalytic triad, resulted in a decrease of the activity by two orders of magnitude (Vernet et al. 1995). For dipeptidyl-peptidase IV and subtilisin, such an Asp → Ala substitution decreased the activity even 500-fold and 104-fold, respectively (Carter and Wells 1988; David et al. 1993), whereas in lipoprotein lipase it completely abolished all activity (Faustinella et al. 1992). Therefore, Asn156 in OMPLA is less strictly required for catalysis than the Asp/Asn in other hydrolases. Although the functions of the catalytic serine and histidine have been established beyond doubt as nucleophile and general base, respectively (Dodson and Wlodawer 1998), the role of the third residue in the triad is less clear. Five roles have been proposed: (1) orientation of the histidine side chain, (2) structural stabilization, (3) activation of the histidine and stabilization of the transient positive charge on the histidine, (4) formation of a low-barrier hydrogen bond (LBHB), and (5) stabilization of the catalytic competent tautomer of the histidine.
Orientation of the histidine side chain
The correct positioning of the catalytic histidine in the active sites of serine and cysteine hydrolases is crucial for enzymatic catalysis. Disruption of the spatial arrangement of the His and Ser/Cys residues leads to inactivation (Sprang et al. 1987; Mhashilkar et al. 1993; Kagawa et al. 2000). However, in OMPLA the position of the catalytic histidine is largely unaffected by the Asn156Ala mutation; the histidine keeps its favorable trans conformation. The two other favorable conformations, namely, the gauche+ and the gauche−, are effectively blocked by the bulky side chain of tyrosine 140 and by the invariant proline 116, respectively. Therefore, in OMPLA, Asn156 does not contribute to a large extent to the correct positioning of the catalytic histidine in the active site. This is in contrast to trypsin, where the active-site histidine has an unfavorable conformation (χ1 = 92°), and where mutation of the catalytic triad Asp results in the partitioning of the His between the native conformation and an alternative trans conformation (Sprang et al. 1987).
Structural stabilization
The His–Asp interaction (or His–Asn in papain) has been shown to be an important determinant for structural stability (Vernet et al. 1995; Quirk et al. 1998; Lau and Bruice 1999). Indeed, the His–Asn interaction in OMPLA is, to some extent, involved in structural stability, as illustrated by the slightly lower melting temperature of Asn156Ala OMPLA. The decrease in melting temperature is, however, much less than, for instance, for the Asp121Ala substitution in ribonuclease A (6°C; Quirk et al. 1998). The high melting temperatures of >90°C indicate the extraordinary stability of both the wild-type and Asn156Ala proteins. This exceptional thermal stability is a property of outer membrane β-barrel proteins in general (Heller 1978), wherein the many main-chain interactions between adjacent β-strands and the packing of side chains inside the β-barrel and the periplasmic turns are the principal factors for thermal stability (Koebnik 1999). As the loss of the Asn–His interaction in OMPLA has left the β-barrel and the periplasmic turns unperturbed, the paramount determinants for thermal stability are maintained, thus explaining the minor effect of the Asn156Ala mutation on the enzyme's thermostability.
Activation of the histidine and stabilization of the transient positive charge on the histidine
The acidic group in classical serine hydrolases functions in increasing the pKa of the histidine, thereby increasing the nucleophilicity of the serine (Dodson and Wlodawer 1998). An Ala or Asn residue instead of an Asp may therefore compromise catalytic efficiency through a decrease in nucleophilicity of the serine. Conversely, the Asn156Asp mutant of OMPLA is significantly more active (although only at elevated pH; Kingma et al. 2000). Nevertheless, as wild-type OMPLA efficiently hydrolyzes phospholipids (with a specific activity of ∼40–50 sec−1; Brok et al. 1996; Dekker et al. 1997), other factors must make up for a lowered pKa of the active-site histidine and the accordingly less nucleophilic serine. We suggest that the nearby calcium ion in the active site of dimeric OMPLA contributes to competent catalysis (Snijder et al. 1999). The positive charge of the calcium, which is not compensated by its ligands, may directly increase the nucleophilicity of the active-site serine. Furthermore, mediated by water molecules, the calcium ion enhances the polarization of the carbonyl bond of the ester substrate, thus creating a more electrophilic substrate center. In addition, in concert with three hydrogen-bond donors, the calcium ion stabilizes the oxyanion intermediates formed during the reaction (oxyanion hole). The increased polarization of the carbonyl function reduces the need for a strongly nucleophilic serine, and this may explain why OMPLA tolerates an asparagine and even an alanine residue in its catalytic triad. The same considerations are valid for the breakdown of the enzyme–acyl intermediate by the hydrolytic water molecule.
Formation of a low-barrier hydrogen bond (LBHB)
A much debated role of the Asp–His couple in serine proteases is the possible formation of a short and exceptionally strong hydrogen bond (low-barrier hydrogen bond, LBHB) during the transition state. It was proposed that such an LBHB could lower the energy of the transition state by as much as 20 kcal/mole, thereby facilitating efficient catalysis (Cleland and Kreevoy 1994; Frey et al. 1994). However, the existence and involvement of LBHBs in enzyme catalysis have been seriously questioned (Warshel et al. 1995; Ash et al. 1997). In the solvent-exposed active site of OMPLA, the presence of an LBHB between Asn156 and His142 is impossible, because the formation of an LBHB requires the absence of H-bonding solvents and matching of the pKa values for the donor and acceptor (Cleland and Kreevoy 1994; Frey et al. 1994). Neither criterion is met, and OMPLA therefore presents an example of a serine hydrolase that functions efficiently without the need of an LBHB. Because a peptide bond is intrinsically less reactive than an ester bond, the activity of serine proteases might still require a strong hydrogen bond in the active site.
Stabilization of the catalytic competent tautomer of the histidine
Finally, the third residue of the triad selects the correct histidine tautomer to accept the serine hydroxyl proton during catalysis. In basic aqueous solutions, the Nɛ—H tautomer dominates the imidazole form in free L-histidine, histidine derivatives, oligopeptides and polypeptides (Reynolds et al. 1973; Deslauriers et al. 1974), and proteins (Wilbur and Allerhand 1977; Bhattacharya et al. 1997). In Asn156Ala OMPLA, the His142 side chain is solvent-exposed and lacks specific protein and solvent interactions. Although the exact tautomeric state of His142 cannot be determined at the current resolution (2.5–3.0 Å), it is likely that in this mutant the His142 imidazole is substantially protonated on its Nɛ atom, and cannot accept a proton from the nucleophilic serine. In wild-type enzyme the hydrogen-bond interaction of His142 with the carboxamide oxygen of Asn156 may shift the tautomeric equilibrium toward the catalytic competent form with the Nɛ atom unprotonated. This is similar to what has been observed for other serine esterases (Sprang et al. 1987; Dodson and Wlodawer 1998). From the five roles proposed for the Asn/Asp residue in the classical catalytic triad, four functions (positioning of the histidine, contributing to structural stability, activation of the histidine, and formation of a LBHB) are hardly significant for OMPLA. Therefore, the catalytic importance of Asn156 in OMPLA seems mainly to stem from its role in stabilization of the correct tautomeric form of the active-site histidine. In α-lytic protease the Asp–His couple increases the amount of the correct histidine tautomer approximately 24-fold (Bachovchin and Roberts 1978). This change is of the same order as the decrease in activity observed for the Asn156Ala mutant, showing that, indeed, the lack of tautomer stabilization may account for the observed decrease in activity.
Materials and methods
The overexpression, refolding, and purification of the Asn156Ala mutant has been described earlier (Kingma et al. 2000). The mutant OMPLA was crystallized following a procedure similar to that of wild-type OMPLA (Blaauw et al. 1995). Hanging drops consisting of 3 μL of protein solution and 2 μL of reservoir solution were suspended over a 1-mL reservoir containing 27% (v/v) 2-methyl-2,4-pentanediol (MPD), 0.4–1.0 mM CaCl2, and 0.1 M Bis-Tris buffer at pH 6.0–6.3. The initial protein solution contained 10 mg/mL of Asn156Ala OMPLA, 10 mM KCl, 1% (w/v) 1-O-n-octyl-β-D-glucopyranoside (β-OG), and 0.2 mM Tris-HCl at pH 6.6. In these setups, one to six crystals grew to typical sizes of 0.5 × 0.25 × 0.15 mm3 after 1 wk. Crystals were brought to pH 8.3 and pH 4.6 by 20-min soaks in four stabilizing mother liquors of increasing and decreasing pH, respectively.
Data were collected from cryocooled crystals at either the protein crystallography beamline 5.2 R of the ELETTRA synchrotron in Triëste (data set pH 6.1) using a 345 mm MarResearch image plate detector, or in-house with a Mac Science DIP2000 image plate detector (Nonius) mounted on a Nonius FR591 rotating anode generator providing Cu Kα radiation. All data were processed using DENZO, SCALEPACK (Otwinowski and Minor 1997), and TRUNCATE (French and Wilson 1978; Collaborative Computational Project Number 4 1994).
The structure of monomeric native OMPLA (pdb entry 1QD5; Snijder et al. 1999) without detergent and water molecules was used as a starting model for refinement. The active-site residues His142, Ser144, and Asn156 were replaced by alanine residues. The coordinates of all remaining atoms were given a random shift of maximally 0.5 Å using MOLEMAN (Kleywegt and Jones 1997) to reduce model bias. Refinement was performed with the CNS software suite (Brünger et al. 1998). A random set of 10% of the unique reflections was set apart to calculate a free R-factor (Brünger 1992). Care was taken to include all reflections in the free R-set that previously had been used for cross-validation of the refinement of native monomeric OMPLA. After rigid-body refinement, several rounds of positional and individual restrained B-factor refinement were performed, using maximum likelihood functions applied to amplitudes. Bulk solvent correction and anisotropic B-factor scaling were applied throughout the refinement. No σ cutoffs were used. After every refinement round, σA-weighted 2Fobs − Fcalc and Fobs − Fcalc electron density maps (Read 1986) were subjected to visual inspection using O (Jones et al. 1991). Water molecules were placed in spherical density, where there was substantial positive difference density. The side chains of the active-site residues His142 and Ser144 were added back to the model in the course of refinement, both showing clear electron density. Various ordered detergent and MPD molecules were ideied and included in the models.
Heat-induced unfolding of wild-type and Asn156Ala OMPLA was studied by far-UV circular dichroism (CD). For these experiments 0.4 mg/mL protein was dialyzed for 6 d against a solution of 10 mM Tris-HCl at pH 6.6 and 1.1 % (w/v) β-OG. For the measurements the samples were diluted to a concentration of 0.1 mg/mL. All solutions were degassed prior to data collection. The CD experiments were performed on an Aviv 62A DS spectrometer equipped with a thermoelectric sample holder. Data were recorded in a 1-mm path length cell. Temperature scans between 25° and 98° C were performed in 1°C temperature steps with 30 sec of equilibration time per step. The unfolding of OMPLA was monitored by measuring the average CD signal at 219 nm for 20 sec.
The structures and experimental data of the Asn156Ala mutant of OMPLA have been deposited with the Protein Data Bank with accession codes 1ILD (pH 4.6), 1ILZ (pH 6.1), and 1IM0 (pH 8.3).
Acknowledgments
We thank the staff of the protein crystallography beamline at the ELETTRA synchrotron, Trieste, for assistance during data collection. We also thank M. de Vocht for assistance with the CD experiments. The investigations were supported by the Netherlands Foundation for Chemical Research (CW) with financial aid from the Netherlands Organisation for Scieic Research (NWO).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
Bis-Tris, bis-(2-hydroxyethyl)imino-tris hydroxymethylmethane
MPD, 2-methyl-2,4-pentanediol
OMPLA, outer membrane phospholipase A
LBHB, low-barrier hydrogen bond
β-OG, 1-O-n-octyl-β-D-glucopyranoside
Tris, tris(hydroxy-methyl)aminomethane
CD, circular dichroism
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/ps.17701.
References
- Ash, E.L., Sudmeier, J.L., De Fabo, E.C., and Bachovchin, W.W. 1997. A low-barrier hydrogen bond in the catalytic triad of serine proteases? Theory versus experiment. Science 278 1128–1132. [DOI] [PubMed] [Google Scholar]
- Bachovchin, W.W. and Roberts, J.D. 1978. Nitrogen-15 nuclear magnetic resonance spectroscopy. The state of histidine in the catalytic triad of α-lytic protease. Implications for the charge-relay mechanism of peptide-bond cleavage by serine proteases. J. Am. Chem. Soc. 100 8041–8047. [Google Scholar]
- Bhattacharya, S., Sukits, S.F., MacLaughlin, K.L., and Lecomte, J.T. 1997. The tautomeric state of histidines in myoglobin. Biophys. J. 73 3230–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaauw, M., Dekker, N., Verheij, H.M., Kalk, K.H., and Dijkstra, B.W. 1995. Crystallization and preliminary X-ray analysis of outer membrane phospholipase A from Escherichia coli. FEBS Lett. 373 10–12. [DOI] [PubMed] [Google Scholar]
- Boekema, E.J., Stuart, M., Koning, R.I., Keegstra, W., Brisson, A., Verheij, H.M., and Dekker, N. 1998. A 7.4-angstrom projection structure of outer membrane phospholipase A from Escherichia coli by electron crystallography. J. Struct. Biol. 123 67–71. [DOI] [PubMed] [Google Scholar]
- Brok, R.G.P.M., Dekker, N., Gerrits, N., Verheij, H.M., and Tommassen, J. 1995. A conserved histidine residue of Escherichia coli outer-membrane phospholipase A is important for activity. Eur. J. Biochem. 234 934–938. [DOI] [PubMed] [Google Scholar]
- Brok, R.G.P.M., Ubarretxena-Belandia, I., Dekker, N., Tommassen, J., and Verheij, H.M. 1996. Escherichia coli outer membrane phospholipase A: Role of two serines in enzymatic activity. Biochemistry 35 7787–7793. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T. 1992. Free R value: A novel statistical quantity for assessing the accuracy of structures. Nature 355 472–475. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54 905–921. [DOI] [PubMed] [Google Scholar]
- Carter, P. and Wells, J.A. 1988. Dissecting the catalytic triad of a serine protease. Nature 332 564–568. [DOI] [PubMed] [Google Scholar]
- Chen, P., Tsuge, H., Almassy, R.J., Gribskov, C.L., Katoh, S., Vanderpool, D.L., Margosiak, S.A., Pinko, C., Matthews, D.A., and Kan, C.C. 1996. Structure of the human cytomegalovirus protease catalytic domain reveals a novel serine protease fold and catalytic triad. Cell 86 835–843. [DOI] [PubMed] [Google Scholar]
- Cleland, W.W. and Kreevoy, M.M. 1994. Low-barrier hydrogen bonds and enzymic catalysis. Science 264 1887–1890. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4. 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D 50 760–763. [DOI] [PubMed] [Google Scholar]
- David, F., Bernard, A.M., Pierres, M., and Marguet, D. 1993. Ideication of serine 624, aspartic acid 702, and histidine 734 as the catalytic triad residues of mouse dipeptidyl-peptidase IV (CD26). A member of a novel family of nonclassical serine hydrolases. J. Biol. Chem. 268 17247–17252. [PubMed] [Google Scholar]
- Dekker, N., Tommassen, J., Lustig, A., Rosenbusch, J.P., and Verheij, H.M. 1997. Dimerization regulates the enzymatic activity of Escherichia coli outer membrane phospholipase A. J. Biol. Chem. 272 3179–3184. [DOI] [PubMed] [Google Scholar]
- Dekker, N., Tommassen, J., and Verheij, H.M. 1999. Bacteriocin release protein triggers dimerization of outer membrane phospholipase A in vivo. J. Bacteriol. 181 3281–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslauriers, R., McGregor, W.H., Sarantakis, D., and Smith, I.C. 1974. Carbon-13 nuclear magnetic resonance studies of structure and function in thyrotropin-releasing factor. Determination of the tautomeric form of histidine and relationship to biology activity. Biochemistry 13 3443–3448. [DOI] [PubMed] [Google Scholar]
- Dodson, G. and Wlodawer, A. 1998. Catalytic triads and their relatives. Trends Biochem. Sci. 23 347–352. [DOI] [PubMed] [Google Scholar]
- Dorrell, N., Celeste Martino, M., Stabler, R.A., Ward, S.J., Zhang, Z.W., McColm, A.A., Farthing, M.J.G., and Wren, B.W. 1999. Characterization of Helicobacter pylori PldA, a phospholipase with a role in colonization of the gastric mucosa. Gastroenterology 117 1098–1104. [DOI] [PubMed] [Google Scholar]
- Esnouf, R.M. 1997. An extensively modified version of Molscript that includes greatly enhanced coloring capabilities. J. Mol. Graphics 15 132–134. [DOI] [PubMed] [Google Scholar]
- Faustinella, F., Smith, L.C., and Chan, L. 1992. Functional topology of a surface loop shielding the catalytic center in lipoprotein lipase. Biochemistry 31 7219–7223. [DOI] [PubMed] [Google Scholar]
- French, G.S. and Wilson, K.S. 1978. On the treatment of negative intensity observations. Acta Crystallogr. A 34 517–525. [Google Scholar]
- Frey, P.A., Whitt, S.A., and Tobin, J.B. 1994. A low-barrier hydrogen bond in the catalytic triad of serine proteases. Science 264 1927–1930. [DOI] [PubMed] [Google Scholar]
- Grant, K.A., Ubarretxena-Belandia, I., Dekker, N., Richardson, P.T., and Park, S.F. 1997. Molecular characterization of pldA, the structural gene for a phospholipase A from Campylobacter coli, and its contribution to cell-associated hemolysis. Infect. Immun. 65 1172–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller, K.B. 1978. Apparent molecular weights of a heat-modifiable protein from the outer membrane of Escherichia coli in gels with different acrylamide concentrations. J. Bacteriol. 134 1181–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrevoets, A.J.G., Hackeng, T.M., Verheij, H.M., Dijkman, R., and De Haas, G.H. 1989. Kinetic characterization of Escherichia coli outer membrane phospholipase A using mixed detergent–lipid micelles. Biochemistry 28 1139–1147. [DOI] [PubMed] [Google Scholar]
- Horrevoets, A.J., Verheij, H.M., and de Haas, G.H. 1991. Inactivation of Escherichia coli outer-membrane phospholipase A by the affinity label hexadecanesulfonyl fluoride. Evidence for an active-site serine. Eur. J. Biochem. 198 247–253. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.-Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47 110–119. [DOI] [PubMed] [Google Scholar]
- Kagawa, T.F., Cooney, J.C., Baker, H.M., McSweeney, S., Liu, M., Gubba, S., Musser, J.M., and Baker, E.N. 2000. Crystal structure of the zymogen form of the group A Streptococcus virulence factor SpeB: An integrin-binding cysteine protease. Proc. Natl. Acad. Sci. USA 97 2235–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma, R.L., Fragiathaki, M., Snijder, H.J., Dijkstra, B.W., Verheij, H.M., Dekker, N., and Egmond, M.R. 2000. Unusual catalytic triad of Escherichia coli outer membrane phospholipase A. Biochemistry 39 10017–10022. [DOI] [PubMed] [Google Scholar]
- Kleywegt, G.J. and Jones, T.A. 1997. Model building and refinement practice. Methods Enzymol. 277 208–230. [DOI] [PubMed] [Google Scholar]
- Koebnik, R. 1999. Structural and functional roles of the surface-exposed loops of the β-barrel membrane protein OmpA from Escherichia coli. J. Bacteriol. 181 3688–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis, P.J. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Lau, E.Y. and Bruice, T.C. 1999. Consequences of breaking the Asp–His hydrogen bond of the catalytic triad: Effects on the structure and dynamics of the serine esterase cutinase. Biophys. J. 77 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt, E.A. and Bacon, J.A. 1997. Raster3D: Photorealistic molecular graphics. Methods Enzymol. 277 505–524. [DOI] [PubMed] [Google Scholar]
- Mhashilkar, A.M., Viswanatha, T., Chibber, B.A., and Castellino, F.J. 1993. Breaching the conformational integrity of the catalytic triad of the serine protease plasmin: Localized disruption of a side chain of His-603 strongly inhibits the amidolytic activity of human plasmin. Proc. Natl. Acad. Sci. USA 90 5374–5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishijima, M., Nakaike, S., Tamori, Y., and Nojima, S. 1977. Detergent-resistant phospholipase A of Escherichia coli K-12. Eur. J. Biochem. 73 115–124. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Pugsley, A.P. and Schwartz, M. 1984. Colicin E2 release: Lysis, leakage or secretion? Possible role of a phospholipase. EMBO J. 3 2393–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, X., Culp, J.S., DiLella, A.G., Hellmig, B., Hoog, S.S., Janson, C.A., Smith, W.W., and Abdel-Meguid, S.S. 1996. Unique fold and active site in cytomegalovirus protease. Nature 383 275–279. [DOI] [PubMed] [Google Scholar]
- Qiu, X., Janson, C.A., Culp, J.S., Richardson, S.B., Debouck, C., Smith, W.W., and Abdel-Meguid, S.S. 1997. Crystal structure of varicella-zoster virus protease. Proc. Natl. Acad. Sci. USA 94 2874–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirk, D.J., Park, C., Thompson, J.E., and Raines, R.T. 1998. His–Asp catalytic dyad of ribonuclease A: Conformational stability of the wild-type, D121N, D121A, and H119A enzymes. Biochemistry 37 17958–17964. [DOI] [PubMed] [Google Scholar]
- Read, R.J. 1986. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A 42 140–149. [Google Scholar]
- Reynolds, W.F., Peat, I.R., Freedman, M.H., and Lyerla, J.R. 1973. Determination of the tautomeric form of the imidazole ring of L-histidine in basic solution by carbon-13 magnetic resonance spectroscopy. J. Am. Chem. Soc. 95 328–331. [DOI] [PubMed] [Google Scholar]
- Scandella, C.J. and Kornberg, A. 1971. A membrane-bound phospholipase A1 purified from Escherichia coli. Biochemistry 10 4447–4456. [DOI] [PubMed] [Google Scholar]
- Snijder, H.J., Ubarretxena-Belandia, I., Blaauw, M., Kalk, K.H., Verheij, H.M., Egmond, M.R., Dekker, N., and Dijkstra, B.W. 1999. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature 401 717–721. [DOI] [PubMed] [Google Scholar]
- Snijder, H.J., Kingma, R.L., Kalk, K.H., Dekker, N., Egmond, M.R., and Dijkstra, B.W. 2001. Structural investigations of calcium binding and its role in activity and activation of outer membrane phospholipase A from Escherichia coli. J. Mol. Biol. 309 477–489. [DOI] [PubMed] [Google Scholar]
- Sprang, S., Standing, T., Fletterick, R.J., Stroud, R.M., Finer Moore, J., Xuong, N.H., Hamlin, R., Rutter, W.J., and Craik, C.S. 1987. The three-dimensional structure of Asn102 mutant of trypsin: Role of Asp102 in serine protease catalysis. Science 237 905–909. [DOI] [PubMed] [Google Scholar]
- Ubarretxena-Belandia, I., Hozeman, L., van der Brink-van der Laan, E., Pap, E.H.M., Egmond, M.R., Verheij, H.M., and Dekker, N. 1999. Outer membrane phospholipase A is dimeric in phospholipid bilayers: A cross-linking and fluorescence resonance energy transfer study. Biochemistry 38 7398–7405. [DOI] [PubMed] [Google Scholar]
- Van der Wal, F.J., Luirink, J., and Oudega, B. 1995. Bacteriocin release proteins: Mode of action, structure, and biotechnological application. FEMS Microb. Rev. 17 381–399. [DOI] [PubMed] [Google Scholar]
- Vernet, T., Tessier, D.C., Chatellier, J., Plouffe, C., Sing Lee, T., Thomas, D.Y., Storer, A.C., and Ménard, R. 1995. Structural and functional roles of asparagine 175 in the cysteine protease papain. J. Biol. Chem. 270 16645–16652. [DOI] [PubMed] [Google Scholar]
- Warshel, A., Papazyan, A., and Kollman, P.A. 1995. On low-barrier hydrogen bonds and enzyme catalysis. Science 269 102–103. [DOI] [PubMed] [Google Scholar]
- Wei, Y., Schottel, J.L., Derewenda, U., Swenson, L., Patkar, S., and Derewenda, Z.S. 1995. A novel variant of the catalytic triad in the Streptomyces scabies esterase. Nat. Struct. Biol. 2 218–223. [DOI] [PubMed] [Google Scholar]
- Wilbur, D.J. and Allerhand, A. 1977. Titration behavior and tautomeric states of individual histidine residues of myoglobins. Application of natural abundance carbon 13 nuclear magnetic resonance spectroscopy. J. Biol. Chem. 252 4968–4975. [PubMed] [Google Scholar]