Abstract
Eukaryotic histone proteins condense DNA into compact structures called nucleosomes. Nucleosomes were viewed as a distinguishing feature of eukaryotes prior to ideication of histone orthologs in methanogens. Although evolutionarily distinct from methanogens, the methane-producing hyperthermophile Methanopyrus kandleri produces a novel, 154-residue histone (HMk). Amino acid sequence comparisons show that HMk differs from both methanogenic and eukaryotic histones, in that it contains two histone-fold ms within a single chain. The two HMk histone-fold ms, N and C terminal, are 28% identical in amino acid sequence to each other and ∼21% identical in amino acid sequence to other histone proteins. Here we present the 1.37-Å-resolution crystal structure of HMk and report that the HMk monomer structure is homologous to the eukaryotic histone heterodimers. In the crystal, HMk forms a dimer homologous to [H3–H4]2 in the eukaryotic nucleosome. Based on the spatial similarities to structural ms found in the eukaryotic nucleosome that are important for DNA-binding, we infer that the Methanopyrus histone binds DNA in a manner similar to the eukaryotic histone tetramer [H3–H4]2.
Keywords: Hyperthermophiles, DNA-binding proteins, high resolution, histone, MAD phasing, methanogen, molecular models, nucleosome, recombinant proteins, selenomethionine
Eukaryotic histone proteins condense DNA into compact structures called nucleosomes. This process was long viewed as a distinguishing feature of eukaryotes until histone orthologs were ideied in methanogens (Sandman et al. 1990). Recently an unusual histone has been ideied in Methanopyrus kandleri (Slesarev et al. 1998), an organism that is among the most heat-tolerant of hyperthermophilic prokaryotes, with a maximum growth temperature of 112°C. Although it produces methane, Methanopyrus is phylogenetically unrelated to other methanogens (Burggraf et al. 1991).
When the Methanopyrus histone protein gene was isolated (Slesarev et al. 1998), its sequence was found to contain unique features as well as features that were intermediate between those found in methanogens (Sandman et al. 1990; Reeve et al. 1997) and in eukaryotes (Thomas and Kornberg 1975; Klug et al. 1980; Arents and Moudrianakis 1995; Luger et al. 1997). Initial amino acid sequence comparisons showed that the Methanopyrus histone (HMk) is twice the length, at 154 residues, of methanogen histones (Reeve et al. 1997). HMk differs from all other histones, both methanogenic and eukaryotic, in containing two histone-fold ms within a single HMk chain. The two HMk histone-fold ms, N and C terminal, are 28% identical in amino acid sequence to each other and ∼21% identical in amino acid sequence to other histones (Slesarev et al. 1998).
Here we present the 1.37-Å-resolution crystal structure of HMk. Consisting of a tandem repeat of the histone fold, the HMk monomer structure is similar to a eukaryotic histone heterodimer. In the crystal, HMk forms a dimer homologous to [H3–H4]2 in the eukaryotic nucleosome. We infer that the Methanopyrus histone binds DNA in a manner similar to the eukaryotic histone tetramer [H3–H4]2, based on the spatial similarities to structural ms found in the eukaryotic nucleosome that are important for DNA-binding.
Results and Discussion
Structure of HMk
The Methanopyrus histone crystal structure was determined using multiwavelength anomalous dispersion (MAD) phasing and refined to 1.37-Å resolution using SHELXL to a final R factor of 16.3% (Table 1). HMk crystals belong to space group P41212 (a = 57.4, c = 97.0) and contain 1 HMk monomer per asymmetric unit. The HMk monomer structure contains two histone-fold domains (Fig. 1A ▶), an N-terminal and a C-terminal domain, which are assembled like a eukaryotic histone heterodimer (Figs. 1A, 2A ▶ ▶). Three helices in a short–long–short arrangement comprise each histone-fold domain. The two domains of HMk are related by a pseudo-2-fold axis along the long molecular axis. These domains are connected by a 13-residue coil at the center of the molecule, colored orange in Figure 1A ▶. The interdomain interface is composed of apolar contacts, mostly alanine, leucine, and isoleucine side chains, which bury 1750 Å2 of surface area in each of the two domains (Fig. 3 ▶). Because the N- and C-terminal domains are oriented antiparallel to one another, helices α1N and α1C are on one molecular face, and helices α3N and α3C are on the other (Fig. 1A ▶). Short β-strands from each domain, found at both ends of the molecule, form two short parallel β-sheets.
Table 1.
Summary of crystallographic data and atomic refinement for HMk
| Crystallographic data | |
| Space group | P41212 |
| Cell dimensions (Å) | a = 57.4 c = 97.0 |
| Data collection statistics | |
| Data set | SSRL |
| Wavelength (Å) | λ = 0.77 |
| Resolution range (Å) | 30–1.37 |
| Completeness (%); Overall (last shell) | 99.7 (99.9) |
| Multiplicity | 5 |
| Rmergea; Overall (last shell) | 6.4 (47.5) |
| Phasing statisticsb | |
| Phasing power | |
| Acentricc | 1.5 |
| Centric | 1.2 |
| Rcullisd centric | 0.52 |
| Figure of merit before density modification | 0.63 |
| Figure of merit after density modification | 0.72 |
| Refinement statistics | |
| Resolution range (Å) | 30.0–1.37 |
| Number of protein atoms | 1254 |
| Number of solvent atoms and ions | 170 |
| Rcryste (Rfree) (%) | 16.3 (20.8) |
| Average temperature factor (protein) (Å2) | 16.2 |
| Average temperature factor (solvent) (Å2) | 31.7 |
| RMSD from ideal geometry | |
| Bond lengths | 0.016 Å |
| Bond angles | 2.88° |
aRmerge = ∑HKL|I-〈I〉|/∑I.
b Phasing statistics are from the combined NSLS X-12C, 3-wavelength MAD data set; high remote (λ3 = 0.98), peak (λ2 = 0.979) and inflection point (λ1 = 0.992).
c Phasing power and Rcullis statistics for acentric reflections are presented as isomorphous/anomalous.
d Rcullis = ∑HKL∥Fph ± FP| − Fh,calc∥/∑HKL|Fph − FP|.
e Rcryst = ∑HKL∥ Fobs|−|Fcalc∥/∑HKL|Fobs|.
Fig. 1.
(A) Ribbon diagram of the HMk monomer structure. α-Helices are depicted in cyan, β-sheets in green, and coils in orange. Helices are labeled at their N termini as follows: the helices in domain 1, the N-terminal domain, are denoted α1N, α2N, and α3N; the helices in domain 2, the C-terminal domain, are denoted α1C, α2C, and α3C. Note the orange loop connecting the two domains that wraps behind the molecule at about the center of the molecule. Also note the short, parallel β-sheets located at both ends of the long molecular axis. (B) Ribbon diagram of the HMk dimer. α-Helices are depicted in cyan, β-sheets in green, and coils in orange. The four-helix bundle formed by the dimer interface is visible at the top center of the molecule.
Fig. 2.
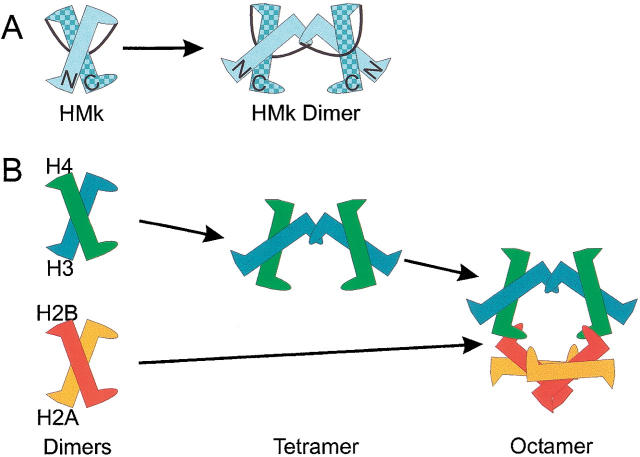
Nomenclature and schematic representation of assemblies observed for several histones. The histone fold is stylized here as a pointed N terminus representing the short helix 1, a long central region representing the longer helix 2, and a rounded C terminus representing the short helix 3. A eukaryotic nucleosome is comprised of 145–147 bp of DNA and two copies each of four histone proteins (H2A, H2B, H3, and H4) (Thomas and Kornberg 1975; Arents and Moudrianakis 1995; Luger et al. 1997). A complete nucleosome histone octomer may be viewed as a left-handed spiral protein assembly constructed from three subassemblies. (A) (Left) An HMk monomer contains two histone-fold ms, the N- and C-terminal domains, tethered by a 13-residue loop. (Right) An HMk dimer formed through crystallographic contacts associates through C-terminal helices of the N-terminal domain. (B) (Left) The eight histone proteins assemble as two copies each of two different heterodimers (H2A–H2B and H3–H4) (Thomas and Kornberg 1975; Luger et al. 1997). (Center) [H3–H4] assembles as [H3–H4]2. This complex initiates DNA-binding, positions the nucleosome, and forms stable nucleosomelike structures in complex with DNA (Dong and van Holde 1991; Hayes et al. 1991). (Right) The nucleosome is completed by adding [H2A–H2B] to each end of the [H3–H4]2 tetramer.
Fig. 3.
Sequence alignments based upon structure alignments for various histones. Only the histone-fold segments are shown. Residues buried in the dimerization interfaces are colored HMk N- and C-terminal domains (cyan), HMfB homodimer (purple), H2A–H2B (yellow), and H3–H4 (orange). Residues buried in the four-helix-bundle interface are colored HMk (dark blue); H2B, H3, and H4 (red). Some charged residues important in DNA interactions in the eukaryotic histone α1α1 m are indicated in aqua, and the region is denoted by α1 at the top of the alignments. The corresponding residues for methanogen histones are also indicated in aqua. The Arg–Thr pairs that form the L1L2 ms are colored in aqua and are indicated by an arrow at the bottom of the alignments. Helical regions are underlined. Loop 2 salt bridges are illustrated by schematic side chains. This figure is similar in style to a figure that appeared in Luger et al. 1997.
The HMk dimer is formed by a crystallographic 2-fold rotation about an axis tilted ∼30° with respect to the long molecular axis (Figs. 1B, 2A ▶ ▶ right). The dimer interface is a four-helix bundle, formed between the N-terminal domain helices α2N, α3N, and their symmetry mates. Twenty-seven residues, mostly small, apolar side chains, bury 1150 Å2 of surface area per monomer in this dimer interface (Fig. 3 ▶). Two salt bridges formed between Lys 55 and Glu 61 and their symmetry mates span the dimer interface at the four-helix bundle (Fig. 1B ▶).
Comparison of HMk to other histones
Whereas amino acid sequence alignments reveal low, but significant, sequence identity among HMk, methanogen, and eukaryotic histones (Fig. 3 ▶), structural alignments strongly idey common secondary, tertiary, and quaternary structures among HMk, methanogen, and eukaryotic histones (Fig. 4 ▶) that align, yielding low root mean square deviations (RMSDs). The N- and C-terminal domains of HMk align with an RMSD of 2.0 Å, and the HMk monomer aligns to other histone dimers with RMSDs between 1.5 and 2.7 Å (Satow et al. 1986; Starich et al. 1996; Luger et al. 1997). The histone folds are similar among HMk, methanogen, and eukaryotic histones, although eukaryotic histone H3 has an elongated helix α1. Like the methanogen histones, HMk lacks the N- or C-terminal extensions found in eukaryotic histones, where they are essential for down-regulating nucleosome assembly and play a role in higher nucleosome assembly (Luger et al. 1997; de la Barre et al. 2000).
Fig. 4.
Ribbon diagrams of HMk (cyan) aligned to various histone proteins. (A) HMk aligned to H2A–H2B (Luger et al. 1997). (B) HMk aligned to H3–H4 (Luger et al. 1997). (C) C-Terminal domain of HMk aligned to HMfB (Starich et al. 1996). (D) Superposition of HMk dimer created from the crystallographic 2-fold axis (cyan) and (H3–H4)2 tetramer. Note the structural similarity between HMk and the other histones. Also note the similar arrangement of domains and interfaces between HMk and [H3–H4]. However, in the HMk structure, the C termini contact one another, whereas in the nucleosome structure the [H3–H4]2 has a gap. Structure alignments were performed using ALIGN (Satow et al. 1986).
The HMk dimer aligns well with the eukaryotic [H3–H4]2 tetramer with an RMSD of ∼2.0 Å (Fig. 4D ▶; Satow et al. 1986; Luger et al. 1997), but differs in certain aspects. The HMk dimer assembly has a |Lp-shaped appearance wherein the four-helix bundle is at the point of the |Lp and an HMk monomer lies along each arm of the |Lp. In the HMk dimer, the two arms of the |Lp are drawn together compared to [H3–H4]2 (Fig. 4D ▶). Closing these arms has the consequence in the HMk dimer that there is more surface area buried per monomer relative to the eukaryotic histones (1153 Å2 in HMk vs. 628 Å2 in H3–H4).
The HMk monomer is homologous to both eukaryotic heterodimers [H2A–H2B] and [H3–H4]. When H3–H4 and H2A–H2B dimers in the eukaryotic structure are replaced with their HMk homolog, four-helix-bundle interactions are observed at four locations in the model. Two of these occur between HMk α3N helices and two between HMk α3C helices. This contrasts with the HMk crystal structure, where the terminal ends of the two α3C helices are used to form a |Lp -arm closure structure, thereby effectively preventing formation of two of the four-helix bundles that were found in the modeled eukaryotic nucleosome. This suggests that the |Lp -arm closure structure may have a role in preventing formation of higher-order structures.
Function of HMk
HMk associates with and condenses DNA similarly to the eukaryotic histones, suggesting a physiological role for HMk in DNA condensation (Musgrave et al. 2000). Similarities between regions in the eukaryotic histones that are important for protein–DNA interactions in the eukaryotic nucleosome structure and the corresponding regions in HMk indicate that HMk binds DNA in a manner similar to that of the eukaryotic histones. In the eukaryotic nucleosome structure there are three regions responsible for DNA-binding: an α1α1 site, formed by the N-terminal α1 helices; and two L1L2 binding sites, formed by an L1-Arg and an L2-Thr from different histone monomers, located at both ends of the long molecular axis of the histone dimers (Fig. 3 ▶; Luger et al. 1997). The α1α1 m in HMk formed by α1N and α1C is about equivalent in charge and charged residue positioning to the [H3–H4] α1α1 m (+7 charge in HMk vs. +8 in [H3–H4]; Fig. 3 ▶). L1L2 ms are found at both ends of the HMk molecule formed between R22–T134 and R99–T57 (Fig. 3 ▶). A structural alignment between the HMk dimer and [H3–H4]2 + DNA (Luger et al. 1997) shows similarly placed histone structural elements between the HMk dimer and [H3–H4]2 relative to the DNA. This alignment indicates that HMk would bind DNA, forming a left-handed spiral assembly, and is in agreement with biochemical data that indicate that HMk binds DNA, forming a left-handed spiral assembly (Musgrave et al. 2000).
HMk as an intermediate
Nucleosomes are present in all eukaryotes. Therefore, it can be assumed that the last common ancestor of eukaryotes possessed nucleosomes; their origins would appear to be even earlier. Histones are also found in methanogens. Methanogen histones have a histone fold, and they compact DNA into regular nucleosomelike structures. They are therefore orthologous to eukaryotic histones (Starich et al. 1996; Reeve et al. 1997; Slesarev et al. 1998; Zhu et al. 1998; Musgrave et al. 2000). Methanopyrus kandleri is phylogenetically distinct from methanogens and, consistent with this, its histone, which contains a tandem repeat of the histone fold in a single chain, differs from those found in methanogens. The structural similarities between the Methanopyrus HMk dimer and its [H3–H4]2 ortholog in eukaryotes indicate that the HMk structure is intermediate between that of methanogens and eukaryotes, and provides a glimpse into a step in the evolution of the eukaryotic nucleosome.
Materials and methods
Protein preparation
Chemically competent Escherichia coli methionine auxotrophic strain BL21 (Novagen BL21-DE3 ) cells were prepared and transformed with a pet32 plasmid encoding for HMk. Selenomethionyl HMk was produced using the previously published growth protocol (Slesarev et al. 1998) and an established selenomethionine-incorporation protocol (Hendrickson et al. 1990; Yang et al. 1990). Electrospray mass spectrometry demonstrated the protein produced was full-length, fully incorporated selenomethionyl protein (data not shown).
Crystallization and data collection
Selenomethionyl-HMk (SeHMk) was concentrated to 20 mg/mL in 1M NaCl, 50 mM Tris at pH 8.0, and 1 mM βME. Optimal HMk crystals were obtained by suspending a 1-μL hanging drop of the protein solution over a well containing 50 mM Tris at pH 8.5 and 400 mM NaCl. SeHMk crystallized as tetragonal bipyramids in space group P41212 (a = 57.4 Å, c = 97.0 Å) with 1 molecule per asymmetric unit (Vm = 2.35 Å3/D). Crystals were cryoprotected using a solution of 40% MPD, 400 mM NaCl, and 50 mM Tris at pH 8.5. No increase in mosaicity and little change in unit cell dimensions were noted upon freezing the crystals. Selenomethionyl-HMk data usable to 1.37 Å were collected at Stanford Synchrotron Radiation Laboratory (SSRL) beamline A-2.
MAD data collection and phasing
Multiwavelength anomalous dispersion (MAD) data were collected at beamline X12C at National Synchrotron Light Source (NSLS) using a Brandeis 2X2 CCD detector. MAD data for phasing were collected using a single cryoprotected SeHMk crystal. A 3-wavelength MAD experiment was performed. These data were collected to 1.47-Å resolution. Data from the 3-wavelength MAD experiment were indexed, integrated, and reduced using DENZO and SCALEPACK (Otwinowski and Minor 1997). Phases were obtained using SOLVE (Terwilliger and Berendzen 1999).
Model building and refinement
The initial 1.50-Å electron density map was obtained from phasing the MAD experiment, low-remote wavelength (λ = 0.95 Å). The map quality was improved through solvent flattening and histogram matching using DM (CCP4) (Cowtan 1994), which increased the figure of merit from 0.63 to 0.72. In the initial building round, 151 residues of 154 were manually placed into the modified electron density using the baton-build and lego-side-chain algorithms in O (Jones et al. 1991). Real-space refinement in O was then used to better fit the model to electron density. The 1.5-Å model was refined through a single refinement round using phase-restrained positional, then phase-restrained, simulated annealing refinements in CNS Version 0.4 (Brunger et al. 1998). The resulting gap between R and Rfree was small (31.4% vs. 33.0%). This model was then used to phase directly the 1.37-Å resolution Se-Met dataset from SSRL, and refinement proceeded using SHELXL (Sheldrick and Schneider 1997). Riding hydrogen atoms, added using the automatic hydrogen-building routines in SHELXL, and anisotropic U values were refined in the last few refinement rounds. The final round of refinement included all data in a full-matrix, least-squares refinement protocol that included anisotropic temperature-factor refinement. The structure was evaluated after almost every refinement round using several programs; PROCHECK (Laskowski et al. 1996), WHATCHECK (Hooft et al. 1996), ERRAT (Colovos and Yeates 1993), 3D-1D profiles (Bowie et al. 1991), and SFCHECK (Vaguine et al. 1999). The HMk model contains residues 4–154 (26 residues with multiple conformations), 182 water atoms (4 with multiple conformations), and 1 Cl− ion. Surface area buried in the various histone assemblies was calculated using CNS Version 0.4 (Lee and Richards 1971; Brunger et al. 1998).
Coordinates
Coordinates have been deposited in the Protein Data Bank (accession code 1F1E).
Acknowledgments
We are indebted to D. Eisenberg for assistance, guidance, and insight with this project and manuscript. We thank T.C. Terwilliger and J. Feigon for discussions as well as M. Soltis (SSRL) and R. Sweet (NSLS beamline X12C) for their assistance in data collection. This work was supported by grants from NIH and DOE to D. Eisenberg, by grants from NSF and DOE to J.A.L., and by grants from HHMI and the Russian Foundation for Basic Research to A.S. Diffraction data for this study were collected at Brookhaven National Laboratory in the Biology Department single-crystal diffraction facility at beamline X12-C in the NSLS and at SSRL. NSLS is supported by the United States Department of Energy Offices of Health and Environmental Research and of Basic Energy Sciences, by the National Science Foundation, and by the National Institutes of Health. SSRL is funded by the Department of Energy and the National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
H2A, eukaryotic histone 2A
H2B, eukaryotic histone 2B
H3, eukaryotic histone 3
H4, eukaryotic histone 4
HMk, histone from Methanopyrus kandleri
HMfB, histone from Methanothermus fervidus type B
MAD, multiwavelength anomalous dispersion
RMSD, root mean square deviation
Se-Met, selenomethionine
SeHMk, selenomethionyl histone from Methanopyrus kandleri
NSLS, National Synchrotron Light Source
SSRL, Stanford Synchrotron Radiation Laboratory
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/ps.10901.
References
- Arents, G. and Moudrianakis, E.N. 1995. The histone fold: A ubiquitous architectural m utilized in DNA compaction and protein dimerization. Proc. Natl. Acad. Sci. USA 92 11170–11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie, J.U., Luthy, R., and Eisenberg, D. 1991. A method to idey protein sequences that fold into a known three-dimensional structure. Science 253 164–170. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 905–921. [DOI] [PubMed] [Google Scholar]
- Burggraf, S., Stetter, K.O., Rouviere, P., and Woese, C.R. 1991. Methanopyrus kandleri: An archael methanogen unrelated to all other known methanogens. Systemic Appl. Microbiol. 14 346–351. [DOI] [PubMed] [Google Scholar]
- Colovos, C. and Yeates, T.O. 1993. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 2 1511–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan, K. 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D 50 760–763. [DOI] [PubMed] [Google Scholar]
- de la Barre, A.E., Gerson V., Gout, S., Creaven, M., Allis, C.D., and Dimitrov, S. 2000. Core histone N-termini play an essential role in mitotic chromosome condensation. EMBO J. 19 379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, F. and van Holde, K.E. 1991. Nucleosome positioning is determined by the (H3-H4) 2 tetramer. Proc. Natl. Acad. Sci. USA 88 10596–10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, J.J., Clark, D.J., and Wolffe, A.P. 1991. Histone contributions to the structure of DNA in the nucleosome. Proc. Natl. Acad. Sci. USA 88 6829–6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson, W.A., Horton, J.R., and LeMaster, D.M. 1990. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction MAD: A vehicle for direct determination of three-dimensional structure. EMBO J. 9 1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooft, R.W., Sander, C., Scharf, M., and Vriend, G. 1996. The PDBFINDER database: A summary of PDB, DSSP and HSSP information with added value. Comput. Appl. Biosci. 12 525–529. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47 110–119. [DOI] [PubMed] [Google Scholar]
- Klug, A., Rhodes, D., Smith, J., Finch, J.T., and Thomas, J.O. 1980. A low resolution structure for the histone core of the nucleosome. Nature 287 509–516. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., Rullmannn, J.A., MacArthur, M.W., Kaptein, R., and Thornton, J.M. 1996. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8 477–486. [DOI] [PubMed] [Google Scholar]
- Lee, B. and Richards, F.M. 1971. The interpretation of protein structures: Estimation of static accessibility. J. Mol. Biol. 55 379–400. [DOI] [PubMed] [Google Scholar]
- Luger, K., Mader, A.W., Richmond, R.K., Sargent, D.F., and Richmond, T.J. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389 251–260. [DOI] [PubMed] [Google Scholar]
- Musgrave, D., Forterre, P., and Slesarev, A. 2000. Negative constrained DNA supercoiling in archaeal nucleosomes. Mol. Microbiol. 35 341–349. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymology 276 307–326. [DOI] [PubMed] [Google Scholar]
- Reeve, J.N., Sandman, K., and Daniels, C.J. 1997. Archaeal histones, nucleosomes, and transcription initiation. Cell 89 999–1002. [DOI] [PubMed] [Google Scholar]
- Sandman, K., Krzycki, J.A., Dobrinski, B., Lurz, R., and Reeve, J.N. 1990. HMf, a DNA-binding protein isolated from the hyperthermophilic archaeon Methanothermus fervidus, is most closely related to histones. Proc. Natl. Acad. Sci. USA 87 5788–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satow, Y., Cohen, G.H., Padlan, E.A., and Davies, D.R. 1986. Phosphocholine binding immunoglobulin Fab McPC603. An X-ray diffraction study at 2.7 Å. J. Mol. Biol. 190 593–604. [DOI] [PubMed] [Google Scholar]
- Sheldrick, G.M. and Schneider, T.R. 1997. SHELXL: High resolution refinement. Methods Enzymology 277 319–343. [PubMed] [Google Scholar]
- Slesarev, A.I., Belova, G.I., Kozyavkin, S.A., and Lake, J.A. 1998. Evidence for an early prokaryotic origin of histones H2A and H4 prior to the emergence of eukaryotes. Nucleic Acids Res. 26 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starich, M.R., Sandman, K., Reeve, J.N., and Summers, M.F. 1996. NMR structure of HMfB from the hyperthermophile, Methanothermus fervidus, confirms that this archaeal protein is a histone. J. Mol. Biol. 255 187–203. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C. and Berendzen, J. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J.O. and Kornberg, R.D. 1975. An octamer of histones in chromatin and free in solution. Proc. Natl. Acad. Sci. USA 72 2626–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaguine, A.A., Richelle, J., and Wodak, S.J. 1999. SFCHECK: A unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D Biol. Crystallogr. 55 191–205. [DOI] [PubMed] [Google Scholar]
- Yang, W., Hendrickson, W.A., Kalman, E.T., and Crouch, R.J. 1990. Expression, purification, and crystallization of natural and selenomethionyl recombinant ribonuclease H from Escherichia coli. J. Biol. Chem. 265 13553–13559. [PubMed] [Google Scholar]
- Zhu, W., Sandman, K., Lee, G.E., Reeve, J.N., and Summers, M.F. 1998. NMR structure and comparison of the archaeal histone HFoB from the mesophile Methanobacterium formicicum with HMfB from the hyperthermophile Methanothermus fervidus. Biochemistry 37 10573–10580. [DOI] [PubMed] [Google Scholar]