Abstract
We present a novel and efficient approach for assessing protein–protein complex formation, which combines ab initio docking calculations performed with the protein docking algorithm BiGGER and chemical shift perturbation data collected with heteronuclear single quantum coherence (HSQC) or TROSY nuclear magnetic resonance (NMR) spectroscopy. This method, termed "restrained soft-docking," is validated for several known protein complexes. These data demonstrate that restrained soft-docking extends the size limitations of NMR spectroscopy and provides an alternative method for investigating macromolecular protein complexes that requires less experimental time, effort, and resources. The potential utility of this novel NMR and simulated docking approach in current structural genomic initiatives is discussed.
Keywords: NMR, BiGGER, soft docking, protein complex, structure
There are currently two methods for high-resolution three-dimensional structure determination of proteins and protein complexes: nuclear magnetic resonance (NMR) spectroscopy and X-ray crystallography. An analysis of the Protein Data Bank (http://www.rcsb.org) ideied that during 1999, approximately 290 such structures were determined by NMR spectroscopy whereas 1528 structures were solved by X-ray crystallography. Of these, less than 5% are protein–protein complexes. It is well recognized that macromolecular structure elucidation by NMR is limited by the size of the protein or protein complex, and although complex formation often stabilizes flexible loops or domains, structure determination of these complexes by X-ray crystallography is frequently impeded by difficulties in the cocrystallization process. These experimental impediments have hindered structural investigations of large physiological complexes (>100 kD) that suffer from these aforementioned properties and thus necessitate an alternative strategy to accurately idey the recognition interface within these macromolecular complexes.
NMR is an established method for three-dimensional structure determination of small proteins (<20 kD). The molecular mass range amenable to structure determination by NMR has increased significantly in recent years with the development of triple resonance pulse sequence technology and increased magnetic field strengths (Pervushin et al. 1997). Combining triple resonance experiments with recombinant expression methods, which provide heteronuclear (15N and 13C) and deuterium (2H)-labeled proteins has significantly reduced these size limitations (Clore and Gronenborn 1998; Gardner and Kay 1998). In addition, TROSY (transverse relaxation optimized spectroscopy) experiments with their constructive transverse relaxation interference have permitted high-resolution data collection and resonance assignment for several large proteins, including the 110 kD homo-octameric protein 7,8-dihydroneopterin aldolase (DHNA), and facilitated the ideication of the protein–protein interaction interface in the FimC-adhesin FimH complex (Pellecchia et al. 1999; Guntert et al. 2000). Merging these labeling strategies and experimental techniques with the dynamic range of NMR spectroscopy provides structural biologists with a powerful tool ideally suited to investigate larger protein structures and probe protein–ligand (Bolon et al. 1999) and protein–protein interaction interfaces (Morelli et al. 2000a).
The noncovalent assembly of proteins into macromolecular complexes is a critical paradigm in many biological systems. Many groups have investigated protein–protein interactions using NMR spectroscopy or molecular docking approaches to better understand the interplay of these interactions in biochemical processes (Kuntz et al. 1982; Janin and Wodak 1985; Jiang and Kim 1991; Abagyan and Totrov 1994; Norel et al. 1994; Jackson and Sternberg 1995; Vakser 1995; Pellecchia et al. 1999; Takahashi et al. 2000). NMR investigations of these interactions have typically used intermolecular nuclear Overhauser enhancement data (NOEs) with or without residual dipolar couplings, TROSY spectroscopy, and TROSY spectroscopy combined with hydrogen–deuterium (H-D) exchange of backbone amide groups following radio frequency cross-saturation transfer (Garrett et al. 1997; Clore 2000; Takahashi et al. 2000). Alternatively, molecular docking approaches employ an interaction function to evaluate the probability of each putative binding mode. Scoring functions have been described which use geometric complementarity between the two molecules (Katchalski-Katzir et al. 1992), electrostatic interaction potentials (Bacon and Moult 1992), free energy of complex formation (Jackson and Sternberg 1995), and statistically observed information about amino acid interactions or combinations of the above terms (Cherfils et al. 1991; Duncan and Olson 1993). The real challenge facing structural biologists using a molecular docking algorithm is the ability to unequivocally idey the correct structure or family of structures among those calculated, due to the inherent limitations of the individual scoring functions used. The idea of using NMR data for assessing protein–ligand interactions by molecular docking was originally proposed by Maurer and colleagues to investigate the binding of a fibrinogen A-α-like peptide to thrombin (S195A). They used NOE distance constraints and the flexible docking program ECEPP/3 (Maurer et al. 1999). However, all attempts to extend this approach to automate the study of protein–protein recognition and macromolecular complex assembly have been unsuccessful until recently (Morelli et al. 2000b).
The near completion of several genome sequencing projects including the human genome and those of various microbial pathogens emphasizes the need for new tools and strategies for predicting and characterizing how proteins recognize and bind endogenous ligands and/or protein partners. The analysis of chemical shift (resonance frequency) perturbations upon complex formation is a significant source of structural information that has been extensively employed to map protein–drug, protein–peptide, and protein–protein interaction interfaces and has helped clarify biological function and mutagenesis studies (Foster et al. 1998; McKay et al. 1998; Zhou et al. 1999; Li et al. 2000). The coupling of this analysis with TROSY spectroscopy or cross-saturation transfer methods has extended the size limitation and sensitivity of the complexes available to these investigations (Pervushin et al. 1997; Salzmann et al. 1998; Takahashi et al. 2000). In the present study, we validated a novel approach for the characterization of protein–protein complexes which couples NMR chemical shift perturbation analysis with computational ab initio calculations, using the protein docking algorithm BiGGER (Biomolecular complex Generation with Global Evaluation and Ranking) (Palma et al. 2000). This method assumes that the structure of each protein in its unbound conformation is known and that upon complex formation the individual proteins undergo minor conformational perturbations that are restricted to the side chains of surface exposed amino acids. This approximation holds true for the majority of physiological macromolecular complexes (Lo Conte et al. 1999). In addition, the docking algorithm BiGGER takes into account the flexibility of individual amino acid side chains at the protein surface; hence the designation "soft-docking."
We validated this "restrained soft-docking" approach using several known macromolecular complexes, including the N-terminal domain of enzyme 1 (EIN) in complex with histidine phosphocarrier protein (HPr) (Garrett et al. 1999), the complex between barnase and barstar (Buckle et al. 1994), a protein–peptide complex involving Tom20 and a presequence peptide (Abe et al. 2000), and yeast cytochrome c complexed to the cytochrome c peroxidase (Pelletier and Kraut 1992). In addition, we compared our theoretically calculated structure of the EIN/HPr complex with the structure that was recently calculated using an alternative approach that required the inclusion of intermolecular NOEs and residual dipolar coupling (RDC) information (Clore 2000). We demonstrate here that our method is ideally suited to investigate large protein–protein complexes, while requiring significantly less experimental time and data analysis. We also propose a broader application for this method for the investigation of binary and ternary complexes, a prerequisite for extending the utility of NMR for use in structural genomics.
Results and Discussion
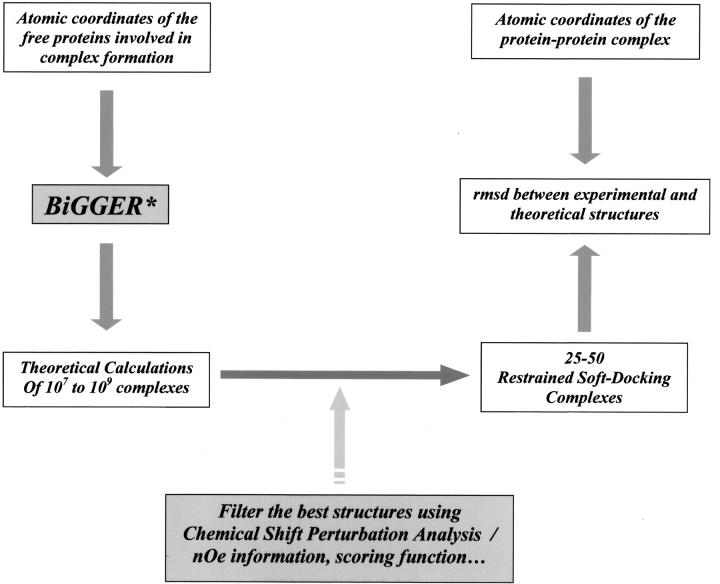
Obtaining the structure of a protein–protein complex by NMR spectroscopy or X-ray crystallography is both difficult and time-consuming. To validate our "restrained soft-docking" method (Morelli et al. 2000b) (as outlined in Fig. 1 ▶), we ideied known protein complexes that satisfied our stringent selection criteria, including availability of the atomic coordinates of the free proteins involved in the complex, coordinates of the protein–protein complex, the observation that the backbone conformation of neither protein is significantly perturbed upon complex formation, and NMR data assessing this complex formation. The NMR data, or experimental filters, are chemical shift perturbations of one (provides a single filter) or both proteins (a double filter) and/or amide proton-deuterium (NH-ND) exchange experiments (Jones et al. 1993; Jeng et al. 1994; Garrett et al. 1997; Abe et al. 2000). Following ab initio molecular docking calculations and the use of our NMR experimental filters, the theoretically selected complexes are compared with the known structures, determined previously using NMR or X-ray crystallography (Table 1).
Fig. 1.
Validation of the "restrained soft-docking" approach. The atomic coordinates of the free proteins are entered into the protein docking algorithm BiGGER* (Palma et al. 2000Palma et al. 2000), with the larger and smaller proteins of the complex representing the "target" and "probe," respectively. Initial ab initio calculations by BiGGER produce 107–109 binding configurations, which are filtered and scored on the basis of geometric surface complementarity, pairwise amino acid interaction propensities, interaction electrostatic potential, and solvation energy upon complex formation. These alternatively docked geometric configurations are then filtered and scored using NMR chemical shift perturbation analysis data, NOE information, or H-D exchange data to idey the 1000 best theoretical solutions. These solutions are scored and ranked according to their fit to these NMR data. The family of structures which is ranked highest is superimposed with the known complex structure, and the RMS deviation (RMSD) was measured.
Table 1.
Comparison of experimental and theoretical complexes investigated
| Dissociation constant (KD) | Experimental interface SASA (Å2) | Theoretical interface SASA (Å2) | rmsd (Å) | |
| EIN/HPr | 6 μMa | 1948 | 2203 | 1.6 |
| Barnase/Barstar | 0.2 pMb | 1556 | 1368 | 0.79 |
| Tom20/Presequence | 20 μMc | 1000 | 1000 | 0.54 |
| Cyt c/Ccp | ≈μMd | 1140 | 1670 | 2.2 |
The experimental protein–protein interaction interface observed in the X-ray or NMR structure of each complex is compared with those obtained using our restrained soft-docking approach. The theoretical interface calculations for each complex were determined using the protein–protein interaction server developed by Prof. Janet Thornton and Dr. S. Jones (http://www.biochem.ucl.ac.uk). The rmsd of the backbone trace between the experimental structure and our best solution for each complex is included.a Garrett et al. 1999; bBuckle et al. 1994; cAbe et al. 2000; dPelletier and Kraut 1992.
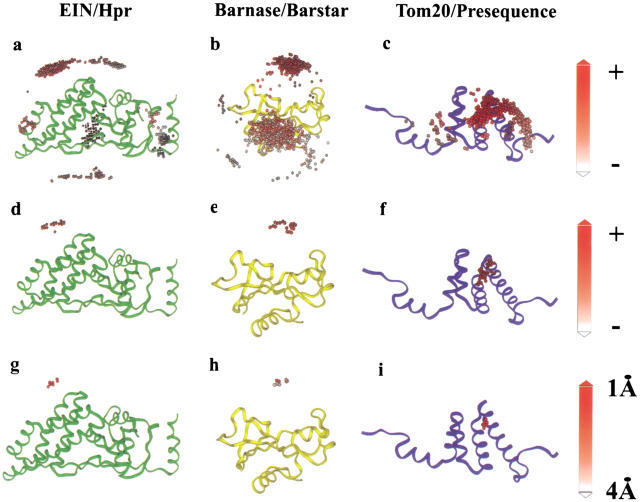
The conformational changes observed upon complex formation are not correlated with the size of the complex but rather depend on the size of the interface. The largest interaction interfaces, which bury 2000–4660 Å2, often expose a small surface before complex formation and thus require extensive conformational perturbation (Lo Conte et al. 1999). Our approach tolerates relatively minor conformational changes upon complex formation and therefore should be limited to those complexes presenting small (<1200 Å2) or standard (1600 ± 400 Å2) interaction interfaces, which are common in physiological complexes (Jones and Thornton 1997a; Lo Conte et al. 1999). The results of the docking calculations are presented in Figure 2 ▶. Panels a, b, and c of Figure 2 ▶ idey the 1000 alternatively docked geometric configurations for each complex, illustrating the backbone trace of EIN (Fig. 2a ▶), Barnase (Fig. 2b ▶), or Tom20 (Fig. 2c ▶) superimposed with the putative positions of their partner molecules, which are represented by a solid sphere denoting their center of mass. Each docked solution is color-coded according to the level of compliance with the observed chemical shift perturbation restraints (higher scoring solutions represented in red). Figure 2d ▶, e, and f illustrate the highest ranked structures according to the above NMR-restrained docking calculations, and Figure 2g ▶, h, and i idey the 10 solutions that are closest to the known structure of each complex.
Fig. 2.
Docking of the complexes assessed following ab initio calculations and NMR chemical shift perturbation analysis and/or H-D exchange filtering. Panels a, b, and c illustrate the backbone trace (N, Cα, C`) of EIN, Barnase, and Tom20, respectively, superimposed with the 1000 putative docking configurations of their complex partners, which are represented by small solid spheres that denote the center of mass for each docking position, color-coded according to the NMR filtering interaction score (higher scoring solutions, which are scored positive, are represented in red). Panels d, e, and f: the highest ranked cluster of structures obtained from these calculations is shown for each complex (illustrated as red spheres). Panels g, h, and i idey the best solutions for each complex, which are scored from red to white (1–4Å) according to the RMS deviation (calculated by BiGGER) of the calculated backbone trace with the previously determined structure.
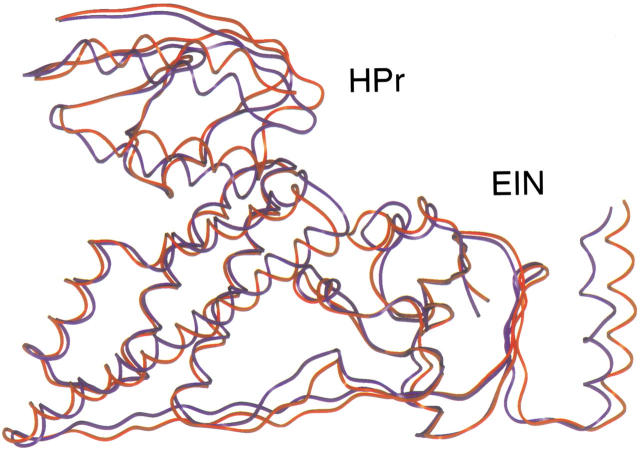
An ideal complex for demonstrating the feasibility and accuracy of this approach was the EIN of the phosphoenolpyruvate:sugar phosphotransferase system in complex with HPr. The structure of this complex was solved using NMR spectroscopy and possesses an interaction interface of approximately 1913 Å2, which is within the standard interface range (Garrett et al. 1999). The structures of EIN and HPr in their unbound conformation and in complex were previously determined (Jia et al. 1993; Liao et al. 1996). In addition, 15N chemical shift perturbation analysis has been performed with 15N-labeled EIN in the presence of unlabeled HPr and 15N-labeled HPr in the presence of unlabeled EIN, thus providing us the mapping of the interaction site for each protein (Garrett et al. 1997). The 1000 best solutions of the EIN (backbone trace) / HPr complex following BiGGER ab initio calculations are illustrated in Figure 2a ▶. Each sphere represents the center of mass for each docking position. After the application of the NMR filter, the 1000 best solutions from the ab initio calculation can be further ranked according to their compliance with the NMR restraints. The spheres representing the best score with the NMR data are shown in red, and those with the worst fit are white. Figure 2d ▶ ideies a subset of 24 structures extracted from Figure 2a ▶ that optimally satisfy the experimentally determined NMR restraints. These data demonstrate that the inclusion of the chemical shift perturbation analysis with the initial ab initio scoring procedure dramatically improves the capacity to idey a single family of structures. The 10 top-ranked docking solutions of HPr (several of the HPr docking positions are degenerate as a result of identical translational and orientational information) are illustrated in Figure 2g ▶. Each of these solutions has a root mean square (RMS) deviation of less than 2 Å for the backbone trace (N, Cα, C`) (Fig. 2g ▶) when compared with the average structure of the EIN/ HPr complex solved by NMR spectroscopy. The spectroscopically determined EIN/HPr structure itself has an inherent RMS deviation of approximately 1.3 Å (Garrett et al. 1999). Moreover, the best calculated complex structure has an RMS deviation of 1.6 Å, when compared with the experimentally determined average structure (Fig. 3 ▶). Taken together these data suggest that the structure obtained using our "restrained soft-docking" approach is equivalent to the structure determined by NMR or X-ray crystallography.
Fig. 3.
Superimpostion of the EIN/ HPr complex determined using "restrained soft-docking" (illustrated in blue) with the previously determined average NMR structure (presented in red) (Garrett et al. 1999Garrett et al. 1999). These structures were superimposed and the RMSD determined using the BiGGER software package. The EIN and HPr proteins of this complex are labeled.
We then performed an analogous study using the same approach to investigate the complex between the extracellular ribonuclease from Bacillus amyloliquifaciens, barnase, and its inhibitor barstar, which was originally solved by X-ray crystallography (Buckle et al. 1994). The interaction interface has been investigated using both chemical shift perturbation analysis of 15N-labeled barnase in complex with unlabeled barstar and hydrogen-deuterium (H-D) exchange of the backbone amide groups of barnase upon complex formation with barstar (Jones et al. 1993). However, complementary data demonstrating the effect of barnase on barstar are not available. Ab initio calculations employing BiGGER ideied two significantly different interaction sites (Fig. 2b ▶). The inclusion of barnase chemical shift perturbation analysis and H-D exchange data in the ab initio calculations again permitted ranking of the original set of structures. After we filtered out solutions that violated the observed NMR restraints, only 30 complexes were ideied; these represent a single family of structures (colored red in Fig. 2e ▶). Each of the 10 highest-ranked structures (Fig. 2h ▶) within this family possessed an RMS deviation of less than 4 Å and the best solution has an RMS deviation of 0.79 Å when compared with the structure solved by x-ray crystallography.
Many mitochondrial proteins are synthesized in the cytosol as precursor proteins and imported into the mitochondria via a presequence signal peptide. NMR spectroscopy was recently used to determine the complex between the translocase outer mitochondrial membrane receptor protein (Tom20) and a presequence peptide derived from the rat aldehyde dehydrogenase (Abe et al. 2000). To demonstrate the flexibility of our approach (Fig. 1 ▶), we calculated the complex of Tom20 with the presequence peptide using the structure of the complex and chemical shift perturbation analysis information. However, the absence of molecular coordinates for the free molecules prevented us from performing true ab initio calculations, because we separated both molecules by editing the atomic coordinate file (pdb). The 1000 highest-scored solutions are illustrated in Figure 2c ▶, color-coded according to the NMR filtering score from the best (red) to the worst (white). The 94 best solutions are represented by a single cluster of orientations (Fig. 2f ▶), and the 10 best structures within this family all possess RMS deviations of less than 1.5 Å when compared with the NMR structure of the complex (Fig. 2i ▶).
The last complex investigated was that of cytochrome c (cc) with cytochrome c peroxidase (ccp), an electron-transfer complex, which further supported our approach (Pelletier and Kraut 1992). This complex was amenable to analysis by our algorithm because both of these proteins have been thoroughly characterized in both the free and bound states, and their three-dimensional structures are known (Louie and Brayer 1990; Wang et al. 1990). However, without chemical shift perturbation data available to filter our theoretical complexes, we used an H-D exchange experiment in which a protection factor for cc had been determined following complex formation with ccp (Jeng et al. 1994). This information provided us with a single filter for cc upon complex formation. The initial solutions were filtered with the H-D exchange data, and these results ideied 310 solutions that possessed a Haem–Haem distance of less than 30 Å (the known distance between the iron centers is 26 Å) (Pelletier and Kraut 1992). The second highest-ranking structure within these solutions has an RMS deviation of 2.87 Å when compared with the structure previously determined by X-ray crystallography. The highest-ranked complex was oriented in an alternative-binding site which did not converge (RMS deviation of 21.85 Å) with the known structure (data not shown). However, other groups have proposed that an alternative binding site for cc upon complex formation with ccp exists in solution, which might explain our results; this remains a topic of discussion. These data suggest that the use of additional NMR data for ccp (chemical shift perturbation analysis, H-D exchange or other NMR constraints), which would provide us with a double filter, would enhance the accuracy of the theoretical solutions, resulting in the ideication of a single family of structures.
In a recent publication by Clore (2000), an alternative approach to investigate the EIN/HPr complex was presented that required intermolecular NOE restraints, RDC measurements, and rigid body minimization. This approach is both expensive, necessitating a double labeling strategy for the proteins involved, and considerably more time-consuming, requiring significantly more data collection and analysis. The RDC measurements provided additional orientational parameters (one protein with respect to its partner) to supplement the translational and orientational information provided by intermolecular NOEs (Garrett et al. 1999). However, this method predicted the correct EIN/ HPr complex (NMR structure) only when both experimental filters (NOEs and RDC) were included. While this is an excellent alternative strategy, our restrained soft-docking approach does not require additional sample preparation and/or data collection (RDC measurements), laborious and time-consuming assignment of intermolecular NOEs, or additional energy minimization. In addition, the enhanced sensitivity of the HSQC and TROSY methods permit chemical shift perturbation analysis of protein–protein complexes at very low protein concentrations (micromolar), which could facilitate automated screening of potential protein–protein interactions and structural studies of less soluble complexes.
Conclusion
Lo Conte and others have extensively analyzed recognition sites of complexes with known structures and determined that these "patches" differ in charge distribution, solvation potential, planarity, and accessible surface area (Jones and Thornton 1997a,b; Lo Conte et al. 1999). The heterogeneity in the nature of these sites makes ideication of protein–protein interfaces by theoretical criteria difficult. Our current analyses investigate protein–protein recognition sites using a novel approach for probing these interfaces combining the power of ab initio molecular docking with chemical shift perturbation analysis. We believe that the ability to screen complex formation rapidly using HSQC experiments (requiring 1 to 2 hours) with ab initio calculations performed by BiGGER (performed in a few hours on a personal computer) provides a powerful tool for studying protein–protein complexes that will facilitate the ideication of lead compounds for rational drug design and ultimately become integrated within current structural genomic applications.
Materials and methods
The structural coordinates of the bound and free forms of the proteins involved in complex formation were taken from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB). The accession codes for the Hpr/EIN complex solved by NMR (Garrett et al. 1999) and the X-ray crystallography structures of free EIN (Liao et al. 1996) and Hpr (Jia et al. 1993) are 3EZA, 1ZYM, and 1POH, respectively. The barnase/barstar complex accession code is 1brs (Buckle et al. 1994); the accession codes for the unbound barnase and barstar are 1a2p (Mauguen et al. 1982) and 1a19 (Ratnaparkhi et al. 1998), respectively. The Tom20 presequence complex accession code is 1om2. No structures are available for the unbound forms (Abe et al. 2000). The PDB accession code for the cytochrome c/cytochrome c peroxidase complex is 2pcc (Pelletier and Kraut 1992). The accession codes for the free cytochrome c and cytochrome c peroxidase are 1ccp (Wang et al. 1990) and 1ycc (Louie and Brayer 1990), respectively.
Protein docking
Molecular interaction simulations between each of the protein pairs were performed using the protein docking algorithm BiGGER (Palma et al. 2000). This algorithm performs a comprehensive and systematic search of the complete 6th-dimension binding space of both molecules without the use of supplemental binding site information. Approximately 107–109 binding configurations are initially assessed and subsequently edited with a sequence of filters and scoring functions based on the evaluation of geometric surface complementarity, the pairwise amino acid propensities to contact across the interface, the interaction electrostatic potential, and the solvation energy change upon complex formation (Wang et al. 1995).
In the present work, a set of 1000 alternative binding solutions were generated by BiGGER and subsequently filtered using data experimentally derived from NMR chemical shift perturbation analysis and/or other NMR restraints.
Acknowledgments
We thank Prof. Barbara Furie for reading the manuscript. The project is supported by the French Embassy and by the ICCTI (project 316C2).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/ps.07501.
References
- Abagyan, R. and Totrov, M. 1994. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J. Mol. Biol. 235 983–1002. [DOI] [PubMed] [Google Scholar]
- Abe, Y., Shodai, T., Muto, T., Mihara, K., Torii, H., Nishikawa, S., Endo, T., and Kohda, D. 2000. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell 100 551–560. [DOI] [PubMed] [Google Scholar]
- Bacon, D.J. and Moult, J. 1992. Docking by least-squares fitting of molecular surface patterns. J. Mol. Biol. 225 849–858. [DOI] [PubMed] [Google Scholar]
- Bolon, P.J., Al-Hashimi, H.M., and Prestegard, J.H. 1999. Residual dipolar coupling derived orientational constraints on ligand geometry in a 53 kDa protein-ligand complex. J. Mol. Biol. 293 107–115. [DOI] [PubMed] [Google Scholar]
- Buckle, A.M., Schreiber, G., and Fersht, A.R. 1994. Protein–protein recognition: Crystal structural analysis of a barnase–barstar complex at 2.0-A resolution. Biochemistry 33 8878–8889. [DOI] [PubMed] [Google Scholar]
- Cherfils, J., Duquerroy, S., and Janin. J. 1991. Protein–protein recognition analyzed by docking simulation. Proteins 11 271–280. [DOI] [PubMed] [Google Scholar]
- Clore, G.M. 2000. Accurate and rapid docking of protein–protein complexes on the basis of intermolecular nuclear overhauser enhancement data and dipolar couplings by rigid body minimization. Proc. Natl. Acad. Sci. 97 9021–9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore, G.M. and Gronenborn, A.M. 1998. Determining the structures of large proteins and protein complexes by NMR. Trends Biotechnol. 16 22–34. [DOI] [PubMed] [Google Scholar]
- Duncan, B.S. and Olson, A.J. 1993. Shape analysis of molecular surfaces. Biopolymers 33 231–238. [DOI] [PubMed] [Google Scholar]
- Foster, M.P., Wuttke, D.S., Clemens, K.R., Jahnke, W., Radhakrishnan, I., Tennant, L., Reymond, M., Chung, J., and Wright, P.E. 1998. Chemical shift as a probe of molecular interfaces: NMR studies of DNA binding by the three amino-terminal zinc finger domains from transcription factor IIIA. J. Biomol. NMR. 12 51–71. [DOI] [PubMed] [Google Scholar]
- Gardner, K.H. and Kay, L.E. 1998. The use of 2H, 13C, 15N multidimensional NMR to study the structure and dynamics of proteins. Annu. Rev. Biophys. Biomol. Struct. 27 357–406. [DOI] [PubMed] [Google Scholar]
- Garrett, D.S., Seok, Y.J., Peterkofsky, A., Clore, G.M., and Gronenborn. A.M. 1997. Ideication by NMR of the binding surface for the histidine-containing phosphocarrier protein HPr on the N-terminal domain of enzyme I of the Escherichia coli phosphotransferase system. Biochemistry 36 4393–4398. [DOI] [PubMed] [Google Scholar]
- Garrett, D.S., Seok, Y.J., Peterkofsky, A., Gronenborn, A.M., and Clore, G.M. 1999. Solution structure of the 40,000 Mr phosphoryl transfer complex between the N-terminal domain of enzyme I and HPr. Nat. Struct. Biol. 6 166–173. [DOI] [PubMed] [Google Scholar]
- Guntert, P., Salzmann, M., Braun, D., and Wuthrich, K. 2000. Sequence-specific NMR assignment of proteins by global fragment mapping with the program MAPPER [In Process Citation]. J. Biomol. NMR 18 129–137. [DOI] [PubMed] [Google Scholar]
- Jackson, R.M. and Sternberg, M.J. 1995. A continuum model for protein–protein interactions: Application to the docking problem. J. Mol. Biol. 250 258–275. [DOI] [PubMed] [Google Scholar]
- Janin, J. and Wodak, S.J. 1985. Reaction pathway for the quaternary structure change in hemoglobin. Biopolymers 24 509–526. [DOI] [PubMed] [Google Scholar]
- Jeng, M.F., Englander, S.W., Pardue, K., Rogalskyj, J.S., and McLendon, G. 1994. Structural dynamics in an electron-transfer complex. Nat. Struct. Biol. 1 234–238. [DOI] [PubMed] [Google Scholar]
- Jia, Z., Quail, J.W., Waygood, E.B., and Delbaere, L.T. 1993. The 2.0-A resolution structure of Escherichia coli histidine-containing phosphocarrier protein HPr. A redetermination. J. Biol. Chem. 268 22490–22501. [DOI] [PubMed] [Google Scholar]
- Jiang, F. and Kim, S.H. 1991. "Soft docking": Matching of molecular surface cubes. J. Mol. Biol. 219 79–102. [DOI] [PubMed] [Google Scholar]
- Jones, D.N., Bycroft, M., Lubienski, M.J., and Fersht, A.R. 1993. Ideication of the barstar binding site of barnase by NMR spectroscopy and hydrogen-deuterium exchange. FEBS Lett. 331 165–172. [DOI] [PubMed] [Google Scholar]
- Jones, S. and Thornton, J.M. 1997a. Analysis of protein–protein interaction sites using surface patches. J. Mol. Biol. 272 121–132. [DOI] [PubMed] [Google Scholar]
- Jones, S. and Thornton, J.M. 1997b. Prediction of protein–protein interaction sites using patch analysis. J. Mol. Biol. 272 133–143. [DOI] [PubMed] [Google Scholar]
- Katchalski-Katzir, E., Shariv, I., Eisenstein, M., Friesem, A.A., Aflalo, C., and Vakser, I.A. 1992. Molecular surface recognition: Determination of geometric fit between proteins and their ligands by correlation techniques. Proc. Natl. Acad. Sci. 89 2195–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz, I.D., Blaney, J.M., Oatley, S.J., Langridge, R., and Ferrin, T.E. 1982. A geometric approach to macromolecule-ligand interactions. J. Mol. Biol. 161 269–288. [DOI] [PubMed] [Google Scholar]
- Li, M.X., Spyracopoulos, L., Beier, N., Putkey, J.A., and Sykes, B.D. 2000. Interaction of cardiac troponin C with Ca(2+) sensitizer EMD 57033 and cardiac troponin I inhibitory peptide. Biochemistry 39 8782–8790. [DOI] [PubMed] [Google Scholar]
- Liao, D.I., Silverton, E., Seok, Y.J., Lee, B.R., Peterkofsky, A., and Davies, D.R. 1996. The first step in sugar transport: Crystal structure of the amino terminal domain of enzyme I of the E. coli PEP: Sugar phosphotransferase system and a model of the phosphotransfer complex with HPr. Structure 4 861–872. [DOI] [PubMed] [Google Scholar]
- Lo Conte, L., Chothia, C., and Janin, J. 1999. The atomic structure of protein–protein recognition sites. J. Mol. Biol. 285 2177–2198. [DOI] [PubMed] [Google Scholar]
- Louie, G.V. and Brayer, G.D. 1990. High-resolution refinement of yeast iso-1-cytochrome c and comparisons with other eukaryotic cytochromes c. J. Mol. Biol. 214 527–555. [DOI] [PubMed] [Google Scholar]
- Mauguen, Y., Hartley, R.W., Dodson, E.J., Dodson, G.G., Bricogne, G., Chothia, C., and Jack, A. 1982. Molecular structure of a new family of ribonucleases. Nature 297 162–164. [DOI] [PubMed] [Google Scholar]
- Maurer, M.C., Trosset, J.Y., Lester, C.C., DiBella, E.E., and Scheraga, H.A. 1999. New general approach for determining the solution structure of a ligand bound weakly to a receptor: Structure of a fibrinogen A-α-like peptide bound to thrombin (S195A) obtained using NOE distance constraints and an ECEPP/3 flexible docking program. Proteins 34 29–48. [DOI] [PubMed] [Google Scholar]
- McKay, R.T., Pearlstone, J.R., Corson, D.C., Gagne, S.M., Smillie, L.B., and Sykes, B.D. 1998. Structure and interaction site of the regulatory domain of troponin-C when complexed with the 96–148 region of troponin-I. Biochemistry 37 12419–12430. [DOI] [PubMed] [Google Scholar]
- Morelli, X., Czjzek, M., Hatchikian, C.E., Bornet, O., Fontecilla-Camps, J.C., Palma, N.P., Moura, J.J., and Guerlesquin, F. 2000a. Structural model of the Fe-hydrogenase/cytochrome c553 complex combining transverse relaxation-optimized spectroscopy experiments and soft docking calculations. J. Biol. Chem. 275 23204–23210. [DOI] [PubMed] [Google Scholar]
- Morelli, X., Dolla, A., Czjzek, M., Palma, P.N., Blasco, F., Krippahl, L., Moura, J.J., and Guerlesquin, F. 2000b. Heteronuclear NMR and soft docking: An experimental approach for a structural model of the cytochrome c553-ferredoxin complex. Biochemistry 39 2530–2537. [DOI] [PubMed] [Google Scholar]
- Norel, R., Lin, S.L., Wolfson, H.J., and Nussinov, R. 1994. Shape complementarity at protein–protein interfaces. Biopolymers 34 933–940. [DOI] [PubMed] [Google Scholar]
- Palma, P.N., Krippahl, L., Wampler, J.E., and Moura, J.J. 2000. BiGGER: A new (soft) docking algorithm for predicting protein interactions. Proteins 39 372–384. [PubMed] [Google Scholar]
- Pellecchia, M., Sebbel, P., Hermanns, U., Wuthrich, K., and Glockshuber, R. 1999. Pilus chaperone FimC-adhesin FimH interactions mapped by TROSY-NMR. Nat. Struct. Biol. 6 336–339. [DOI] [PubMed] [Google Scholar]
- Pelletier, H. and Kraut, J. 1992. Crystal structure of a complex between electron transfer partners, cytochrome c peroxidase and cytochrome c [see comments]. Science 258 1748–1755. [DOI] [PubMed] [Google Scholar]
- Pervushin, K., Riek, R., Wider, G., and Wuthrich, K. 1997. Attenuated T2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. 94 12366– 12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnaparkhi, G.S., Ramachandran, S., Udgaonkar, J.B., and Varadarajan, R. 1998. Discrepancies between the NMR and X-ray structures of uncomplexed barstar: Analysis suggests that packing densities of protein structures determined by NMR are unreliable. Biochemistry 37 6958–6966. [DOI] [PubMed] [Google Scholar]
- Salzmann, M., Pervushin, K., Wider, G., Senn, H., and Wuthrich, K. 1998. TROSY in triple-resonance experiments: New perspectives for sequential NMR assignment of large proteins. Proc. Natl. Acad. Sci. 95 13585–13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, H., Nakanishi, T., Kami, K., Arata, Y., and Shimada, I. 2000. A novel NMR method for determining the interfaces of large protein–protein complexes [see comments]. Nat. Struct. Biol. 7 220–223. [DOI] [PubMed] [Google Scholar]
- Vakser, I.A. 1995. Protein docking for low-resolution structures. Protein Eng. 8 371–377. [DOI] [PubMed] [Google Scholar]
- Wang, J.M., Mauro, M., Edwards, S.L., Oatley, S.J., Fishel, L.A., Ashford, V.A., Xuong, N.H., and Kraut, J. 1990. X-ray structures of recombinant yeast cytochrome c peroxidase and three heme-cleft mutants prepared by site-directed mutagenesis. Biochemistry 29 7160–7173. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Zhang, H., and Scott, R.A. 1995. A new computational model for protein folding based on atomic solvation. Protein Sci. 4 1402–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P., Chou, J., Olea, R.S., Yuan, J., and Wagner, G. 1999. Solution structure of Apaf-1 CARD and its interaction with caspase-9 CARD: A structural basis for specific adaptor/caspase interaction. Proc. Natl. Acad. Sci. 96 11265–11270. [DOI] [PMC free article] [PubMed] [Google Scholar]