Abstract
Analysis of complete genome sequences has made it clear that fibronectin type II (FN2) modules are present only in the vertebrate lineage, raising intriguing questions about the origin of this module type. Kringle domains display many similarities to FN2 domains; therefore it was suggested previously that they are highly divergent descendants of the same ancestral protein-fold. Since kringles are present in arthropodes, nematodes, and invertebrate chordates as well as in vertebrates, it is suggested that the FN2 domain arose in the vertebrate lineage through major structural modification of the more ancestral kringle fold. To explore this structural transition, in the present work we compare key structural features of two highly divergent kringle domains (the kringle of Caenorhabditis elegans Ror receptor tyrosine kinase and the kringle of rat neurotrypsin) with those of plasminogen kringles and FN2 domains. Our NMR conformation fingerprinting analysis indicates that characteristic 1H-NMR markers of kringle or FN2 native folding, such as the dispersion of Trp aromatic connectivities and shifts of the Leu46/Thr16 methyl signals, both decrease in the order kringles > neurotrypsin kringle > FN2 domains. These results suggest that the neurotrypsin kringle may represent an intermediate form between typical kringles and FN2 domains.
Keywords: Fibronectin type II domain, kringle domain, neurotrypsin, NMR spectroscopy, evolution of protein folds
Fibronectin type II modules (FN2 modules) are small, compact two-disulphide-bond domains of about 40 amino acid residues first ideied in the extracellular matrix protein, fibronectin, and in some seminal fluid proteins (Esch et al. 1983; Skorstengaard et al. 1986; Seidah et al. 1987;). Related domains are found in the extracytoplasmic parts of membrane-associated proteins, such as members of the mannose receptor–phospholipase A2 receptor family (Taylor et al. 1990; Ishizaki et al. 1994; Jiang and Nussenzweig 1995), mannose-6-phosphate receptors (Morgan et al. 1987), and the pancreas-specific sel-1 proteins of vertebrates (Harada et al. 1999; Biunno et al. 2000). FN2 modules are also present in matrix metalloproteinases MMP-2 and MMP-9 (Collier et al. 1988; Wilhelm et al. 1989) as well as in the serine proteases, factor XII, and hepatocyte growth factor activator (McMullen and Fujikawa 1985; Miyazawa et al. 1993).
NMR and/or X-ray crystallographic structures are known for the second FN2 module of bovine seminal plasma protein PDC-109 (Constantine et al. 1992), the two FN2 domains of fibronectin (Pickford et al. 1997; Sticht et al. 1998), and the three FN2 domains of gelatinase A/MMP-2 (Briknarová et al. 1999; Morgunova et al. 1999).
An interesting aspect of the evolutionary history of FN2 domains is that, despite their widespread occurrence in diverse proteins of vertebrates, FN2 modules are absent from invertebrates, including the completely sequenced genomes of Caenorhabditis elegans and Drosophila melanogaster (cf. the SMART and Pfam databases; Sonnhammer et al. 1997; Schultz et al. 1998,2000). It is interesting to point out that whereas SEL-1 genes of vertebrates contain an FN2 domain, this domain is missing from the invertebrate orthologs of sel-1 (Harada et al. 1999). The fact that FN2 is restricted to the chordate lineage seems to suggest that this domain type has arisen in this lineage through major structural modification of a more ancestral domain type.
In an earlier study we have noted a distant sequence similarity between FN2 modules and kringles of proteases and suggested that they are divergent members of the same fold family (Patthy et al. 1984). In harmony with this proposal, FN2 domains revealed a fold with many similarities to the protein-fold of protease kringles. FN2 domains are similar to kringles inasmuch as they are also characterized by two short antiparallel β-sheets and an exposed aromatic-rich ligand binding site (Briknarová et al. 1999; Tordai and Patthy 1999), as well as two cystine bridges in close, quasi-orthogonal juxtaposition (Constantine et al. 1992)..
In view of the distant relation of FN2 domains and kringles it seems possible that FN2 modules have evolved from kringles. Kringles do indeed have a longer evolutionary history than FN2 modules since they are present both in vertebrates and in invertebrates such as C. elegans and D. melanogaster (cf. the SMART and Pfam databases; Sonnhammer et al. 1997; Schultz et al. 1998,2000).
Kringles are usually 80 amino acid residue-long, and were first found in members of the trypsin-family: prothrombin (Magnusson et al. 1975), plasminogen (Sottrup-Jensen et al. 1978), urokinase (Günzler et al. 1982), tissue-plasminogen activator (Pennica et al. 1983), hepatocyte growth factor (Nakamura et al. 1989), macrophage-stimulating protein (Han et al. 1991), coagulation factor XII (McMullen and Fujikawa 1985), hepatocyte growth factor activator (Miyazawa et al. 1993), hyaluronan-binding protein (Choi-Miura et al. 1996), a novel serine protease of the ascidian Herdmania momus (Arnold et al. 1997), and the brain-specific serine protease, neurotrypsin/motopsin (Gschwend et al. 1997; Yamamura et al. 1997; Proba et al. 1998; Iijima et al. 1999).
More recently, kringles were also found in the extracellular regions of diverse members of the Ror-type receptor tyrosine kinase family (Masiakowski and Carroll 1992; Jennings et al. 1993; Wilson et al. 1993; Oishi et al. 1997; Forrester et al. 1999; Fu et al. 1999).
Association of kringles with receptor tyrosine kinases is more ancient than with serine proteases. C. elegans and D. melanogaster have kringle-containing homologs of vertebrate Ror-type receptor tyrosine kinases (Wilson et al. 1993; Oishi et al. 1997; Forrester et al. 1999), whereas serine proteases with kringles seem to be restricted to the chordate lineage. We may therefore assume that kringles have been joined to protease domains (cf. Patthy 1985) only in the chordate lineage and that the conquest of this novel functional environment may have been accompanied by structural readjustments in the kringle domains.
Although more than two dozen kringle structures are deposited in the Protein Data Bank (PDB) (http://www.rcsb.org), these structures represent a rather closely related group of protease kringles (kringles of prothrombin, plasminogens, plasminogen-related proteins, plasminogen activators). In order to understand the structural rearrangements that have accompanied the use of kringles in proteolytic systems, it is important to define the structures of kringles representing the more ancestral Ror receptor tyrosine kinase family as well as kringles of more divergent members of the protease family.
In this paper we report the expression, purification, and refolding of the kringle domain of the C. elegans Ror receptor tyrosine kinase and the kringle of rat neurotrypsin, as well as their conformational fingerprinting via NMR spectroscopy. Structural comparison of these divergent members of the kringle family with structurally well-characterized kringle and FN2 domains suggests that the kringle domain of neurotrypsin represents an intermediate form in the transition from kringles to FN2 domains.
Results
In the present work we have determined the key structural features of the kringle of C. elegans Ror-type receptor tyrosine kinase and the kringle of rat neurotrypsin by NMR spectroscopy. The basis of our conformational fingerprinting approach is that the unique structural organization of the hydrophobic core of kringles is reflected in some common characteristics of the NMR spectra of kringles (Trexler et al. 1983; Atkinson and Williams 1990; Byeon and Llinás 1991; Cox et al. 1994; Hansen et al. 1994; Li et al. 1994; Rejante and Llinás 1994b; Byeon et al. 1995). Accordingly, conservation or structural modification of the core structure in novel kringle modules may be assessed by NMR spectroscopic fingerprinting of the solution structure of the kringles.
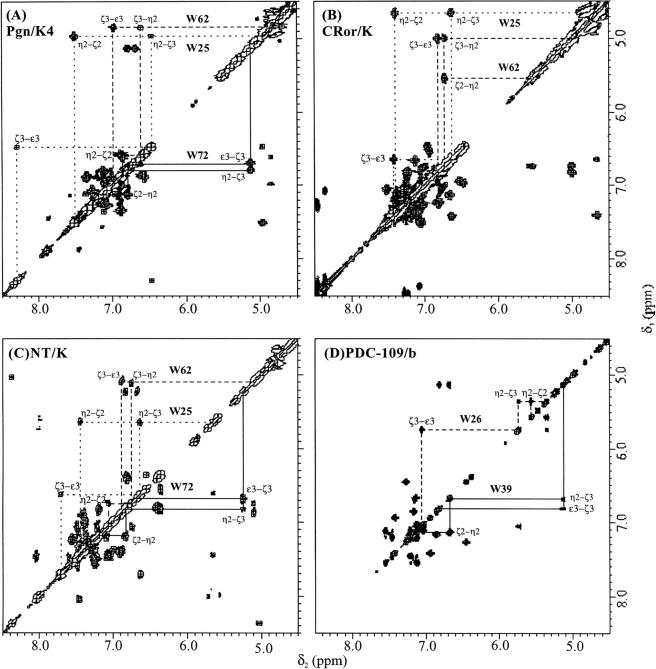
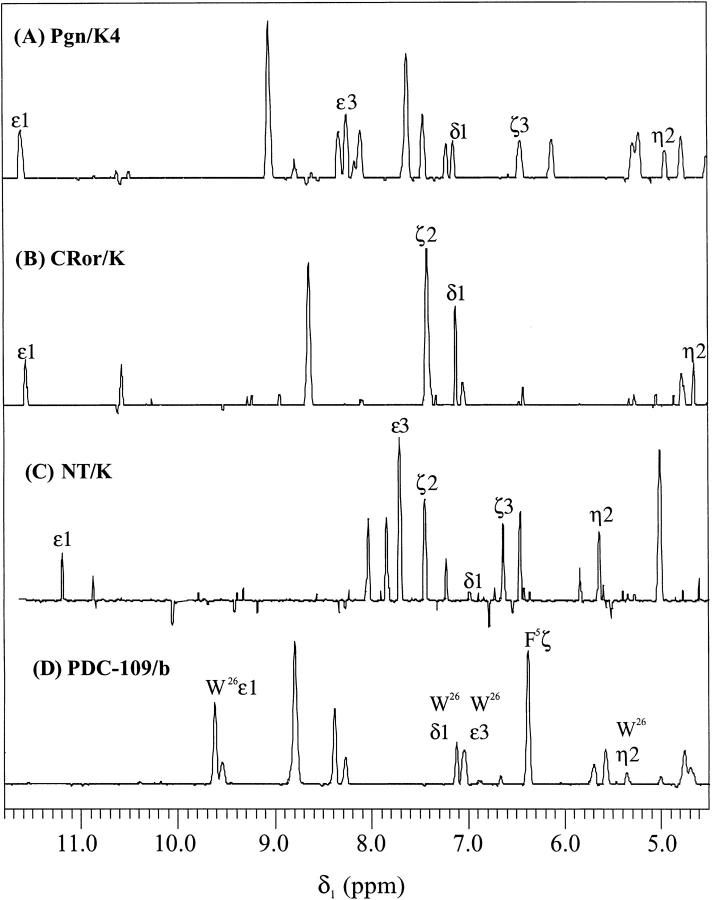
A recurrent feature of the NMR spectra of kringles is the dispersion of Trp aromatic signals arising from the strictly conserved tryptophans of the conserved hydrophobic core (W25, W62 of kringles in Fig.1). Some kringles (e.g., kringle 4 of plasminogen in Fig.1) also contain a third tryptophan, W72, that forms part of a lysine-binding site (Hochschwender and Laursen 1981).
In the NMR spectrum of protease kringles (as exemplified by plasminogen kringle 4, Pgn/K4 in Table 1), the corresponding Trp sidechain indole 1H resonances span a characteristic region, between ∼ 4.5 ppm and ∼ 11.6 ppm, thus affording a reliable signature of kringle folding. The 1H-NMR dispersion of the Trp aromatic signals reflect ring-current shifts stemming from mutually interacting aromatic side-chains which contribute to the buildup of the hydrophobic core, a structural feature conserved in all five of the plasminogen kringles (Thewes et al. 1988; Rejante and Llinás 1994a). As is apparent from inspection of Figure 2 ▶ A, B, and C, the spectra of both CRor/K and NT/K exhibit similar sets of dispersed resonances for their Trp25, Trp62 (and in the latter for its Trp72) as Pgn/K4.
Table 1.
Assigned conserved tryptophan aromatic spin systems and shifted methyl resonances in the 1H-NMR spectra of kringle-related homologs
| Chemical shiftsa (ppm) | ||||||||
| Spin system | Module | ɛ1 (NH1) | δ1 (CH2) | ɛ3 (CH4) | ζ3 (CH5) | |gn2 (CH6) | ζ2 (CH7) | CH3 |
| Trp25 | Pgn/K4 | 11.61 | 7.13 | 8.30 | 6.47 | 4.97 | 7.51 | |
| CRor/K | 11.56 | 7.12 | 7.42 | 6.64 | 4.66 | 7.41 | ||
| NT/K | 11.20 | 6.99 | 7.71 | 6.64 | 5.65 | 7.45 | ||
| Trp62 | Pgn/K4 | 10.95 | 7.40 | 6.98 | 4.85 | 6.61 | 6.86 | |
| CRor/K | 10.35 | 7.53 | 6.83 | 5.01 | 6.73 | 5.55 | ||
| NT/K | 9.58 | 7.56 | 6.89 | 5.10 | 6.75 | 7.06 | ||
| (Trp26) | PDC-109/b | 9.63 | 7.07 | 7.05 | 5.74 | 5.36 | 5.57 | |
| Trp72 | Pgn/K4 | 9.95 | 6.86 | 6.69 | 5.13 | 6.79 | 7.13 | |
| NT/K | 8.97 | 6.85 | 6.68 | 5.25 | 6.83 | 7.19 | ||
| (Trp39) | PDC-109/b | 9.80 | 7.22 | 6.82 | 5.13 | 6.69 | 7.12 | |
| Leu46 | Pgn/K4 | −1.07 | ||||||
| CRor/K | −0.67 | |||||||
| NT/K | −0.39 | |||||||
| (Thr16) | PDC-109/b | 0.09 | ||||||
a NMR data recorded on 1–2 mM 1H2O/2H2O (90/10%) solutions, pH 5.12, 300 K.
Fig. 2.
500 MHz 1H-NMR COSY spectra of homologous kringle and FN2 domains: aromatic connectivities of conserved tryptophan residues. (A) The fourth kringle of human plasminogen (Pgn/K4). (B) The kringle of Ror-type receptor tyrosine kinase of C. elegans (CRor/K). (C) The kringle of rat neurotrypsin (NT/K). (D) The second FN2 domain of bovine seminal fluid protein PDC-109. C. elegans Ror kringle and rat neurotrypsin kringle resonances were ideied from 2D COSY, NOESY, and TOCSY experiments by reference to the human Pgn/K4 spectrum. The assignments of the human Pgn/K4 and PDC-109/b spectra have been reported (Atkinson and Williams 1990; Constantine et al. 1991, 1992. Spectra recorded at 300 K, on 1 mM protein samples dissolved in 2H2O, pH* 4.8.
The Trp72 indole group is known to be exposed at the ligand-binding site of K4 (Hochschwender and Laursen 1981; De Marco et al. 1989) where it contributes hydrophobic component to the kringle–ligand interaction. The striking similarity between Trp72 connectivities in the Pgn/K4 and NT/K COSY spectra (Fig. 2 ▶) suggests that Trp72 exists in a similarly exposed environment in NT/K. On the other hand, the lesser dispersion (Table 1) of the Trp25 and Trp62 aromatic signals in the NT/K (δ = 5.55 ppm and 4.48 ppm, respectively) relative to both the Pgn/K4 (δ = 6.64 ppm and 6.10 ppm, respectively) and CRor/K (δ = 6.93 ppm and 5.34 ppm, respectively) reveals an altered packing of the corresponding, conserved side chain groups in the NT/K.
In the case of PDC-109/b, the alignment with kringles suggests that Trp26 of FN2-domains corresponds to Trp62 of kringles, Trp39 of FN2 domains corresponds to Trp72 of kringles, whereas Trp25 of kringles corresponds to Phe5 of FN2 domains (cf. Fig.1). It is revealing that PDC-109/b exhibits a qualitatively similar, kringle-like pattern for the Trp aromatic proton resonances, with the PDC-109/b Trp39 COSY connectivities closely mimicking those of the corresponding Trp72 of kringles (Fig. 2 ▶). The fact that Trp39 of FN2 domains are also involved in ligand binding (Briknarova et al. 1999) is consistent with a close structural and functional homology between exposed hydrophobic binding site components in kringles and the FN2 domains.
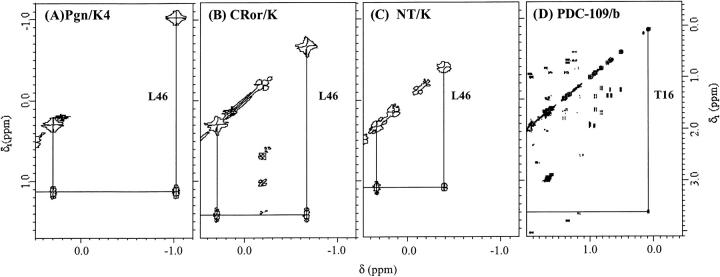
Another characteristic 1H-NMR marker of the native structure of kringles is the pair of high-field shifted methyl doublet signals which have been assigned to the Leu46 δ,δ`CH3 protons (Llinás et al. 1983; Bokman et al. 1993), with Leu46 being a highly conserved residue among all known homologous kringle sequences. These shifted signals afford a fingerprint of native kringle folding, readily ideiable in the spectra of CRor/K (Fig. 3B ▶) and NT/K (Fig. 3C ▶) by reference to the Pgn/K4 spectrum (Fig. 3A ▶). As is apparent from inspection of Figures 3A ▶, B, and C, the most shifted Leu46 δCH3 resonance, which appears at ∼ −1 ppm in the Pgn/K4 spectrum, uniformly shifts to ∼ −0.7 ppm in the CRor/K and ∼ −0.4 ppm in the NT/K spectra. By comparison, the PDC/109-b 1H-NMR spectrum shows a methyl resonance at ∼ 0.09 ppm, assigned to the Thr16>CH3 (Constantine et al. 1991). This observation suggests that this residue might fulfill a structural role similar to that of the conserved Leu46 in the kringles, where the magnetic shielding of the Leu46 δ,δ`CH3 protons arises from anisotropic ring-current effects resulting from side-chain aromatic groups at the kringle hydrophobic core (De Marco et al. 1985).
Fig. 3.
High-field shifted Leu46 CH3δ,δ` resonances of kringle-domains and Thr16 CH3> resonances of FN2 domain: COSY connectivities. Experimental conditions as for Figure 2 ▶.
Most noteworthy, the Leu46 δCH3 is in close contact with the aromatic ring of Trp25, a residue strictly conserved among all kringle homologs. Figure 4A ▶ shows a slice along δ1 dimension of the 2D NOESY spectrum (τmix 100 ms) of the Pgn/K4, at δ2 corresponding to the ∼ −1 ppm Leu46 δCH3 resonance. As is apparent from inspection of Figure 4A ▶, a significant proton–proton cross-relaxation occurs between the Leu46 δCH3 and the Trp25 indole ring Hδ1 (CH2), Hɛ1 (NH1), Hɛ3 (CH4), Hη2 (CH6), and Hζ3 (CH5). In the cases of CRor/K and NT/K the NOESY experiments also reveal Leu46 proximity to the Trp25 Hδ1, Hɛ1, Hη2, and Hζ2 (CH7) (Fig. 4B ▶) and Hδ1, Hɛ1, Hɛ3, Hζ2, and Hζ3 (Fig. 4C ▶), respectively, the relative intensity of the NOEs indicating that the packing of the methyl group against the Trp25 ring varies among the three kringles. By analogy, the PDC-109/b Thr16 >CH3 doublet at ∼ 0.09 ppm (Fig. 3D ▶) happens to be in close contact with both the Phe5 ring Hζ (CH4) and the Hδ1, Hɛ1, Hɛ3, Hη2, and Hζ2 of Trp26 (Fig. 4D ▶) that aligns with the kringles' Trp62 (Fig. 1 ▶). Hence, although the kringle Trp25 is not conserved in the PDC-109/b FN2 (Fig. 1 ▶), the Phe5, jointly with Trp26 (aligned with Trp62 in the kringle sequence), define the aromatic environment of the Thr16 >CH3 group. It is thus suggested that in FN2 domains, the strictly conserved Thr16 residue could play a structural role similar to that of Leu46 in the kringle homologs, namely that of nucleating the module's hydrophobic core. Indeed, in Pgn/K4, the Trp62 ring is ∼ 7.5 Å from the Leu46 methyl (Wu et al. 1991), and an NOE between the two groups is readily detectable (Ramesh et al. 1987). Thus, only a small displacement of the kringle Trp62 toward the Leu46 methyl would be required in order to generate the proximity observed between Trp26 and Thr16 in the PDC-109/b FN2 domain. Interestingly, jointly with Thr16, both Phe5 and Trp26 are strictly conserved in FN2 modules. This suggests—as proposed for the conserved Trp25, Leu46, and Trp62 in kringles (Trexler and Patthy1983)—that these three residues are structural determinants of FN2 domains.
Fig. 4.
1H-NMR NOESY connectivity analogies between kringle and FN2 domains: hydrophobic core. (A–C) 1D slice along the indirect dimension δ1 from the Leu46 CH3δ doublet at −107 ppm (Pgn/K4), −0.669 ppm (CRor/K), and −0.385 ppm (NTK/K) showing NOE connectivities to Trp25. (D) PDC-109/b: slice along the indirect dimension δ1 from the Thr16 CH3> doublet at 0.085 ppm showing NOE connectivities to Phe5 and Trp26. Data collected at 300 K, on 1 mM protein solutions in 90/10% 1H2O/2H2O (v/v), pH 5.2, mixing time 100 ms.
Fig. 1.
Multiple alignment of the amino acid sequences of representative members of the FN2- and kringle-module families. Abbreviations: PDC-109/b, the second FN2 domain of bovine seminal fluid protein PDC-109; NT/K, the kringle domain of the rat neurotrypsin; CRor/K, the kringle domain of C. elegans Ror-type receptor tyrosine kinase; Pgn/K4, the fourth kringle domain of human plasminogen; coK, consensus sequence of kringles. The numbers at the top refer to the residue numbering of FN2 domains, those at the bottom refer to the residue numbering of kringles. Cysteine residues are highlighted in bold.
Discussion
Kringle structures are constrained by three intramolecular disulfide bonds in a 1–6, 2–4, 3–5 pattern that generates a characteristic three-loop structure. The similarity of key spectral characteristics of NT/K and CRor/K to those of typical protease kringles indicates that the overall topology of the protein fold is maintained by these distantly related and functionally dissimilar domains. On the other hand, the structure of NT/K differs quite significantly from typical protease kringles such as those of plasminogen. As has been noted earlier (Yamamura et al. 1997), the segments between the first two cysteines (Cys1 and Cys22) and the last two cysteines of kringles (Cys75 and Cys80) are significantly shorter in the kringle of neurotrypsin than in all other kringles (cf. Fig. 1 ▶). The concomitant shorter lengths of both the N- and C-terminal stretches in NT/K may well represent a structural requirement to preserve the Cys1–Cys80 disulfide pairing that brings C and N termini together. Thus, it is suggested that in order to maintain the pretzel-like kringle structure in NT/K, the shortening of the Cys75–Cys80 stretch has been accommodated by a concomitant shortening of the Cys1–Cys22 segment. It is noteworthy in this respect that a major difference between kringles and FN2 domains is that the segments corresponding to the N- and C-terminal stretches of kringles (and the Cys1–Cys80 disulphide bond) are missing from FN2 domains (cf. Fig. 1 ▶). It seems likely that the drastic shortening of both segments in NT/K forebodes their truncation in typical FN2 domains.
In summary, as revealed by the strikingly uniform patterns of key 1H-NMR COSY/NOESY connectivities (Figs. 2–4 ▶ ▶), the Pgn, Cror, and NT kringles are endowed with similar folding characteristics. Nevertheless, a number of 1H-NMR spectroscopic signatures exhibited by the NT/K are also shared by FN2 domains, suggesting that NT/K may represent an intermediate form between typical kringles and FN2 domains. In particular, typical 1H-NMR markers of kringle or FN2 native folding, such as the dispersion of similarly patterned Trp aromatic connectivities and shifts of the magnetically shielded Leu46 (K)/ Thr16(FN2) methyl signals (Table 1, Figs. 2, 3, 4 ▶ ▶ ▶), are revealing in that they both show a decrease in the order Pgn/K4 > CRor/K > NT/K > PDC-109/b. This relates to the packing of the hydrophobic core which, as the NOESY experiments reveal (Fig. 3 ▶), is analogous for the four modules in that it clusters aromatic rings in close interaction with the kringle Leu46, or PDC-109b Thr16, sidechain methyl group. We are thus led to conclude that the NT/K domain may be viewed as filling a gap in the structural transition from a "typical" kringle to FN2 domains.
Materials and methods
Restriction enzymes, PCR primers, vectors, bacterial strains
Restriction enzymes were purchased from Promega and New England Biolabs. The M13 sequencing reagents used for dideoxy sequencing of cloned DNA fragments were from Amersham Pharmacia Biotech. PCR primers were obtained from Integrated DNA Technologies and from Pharmacia Biotech.
Plasmid pmed23 (Lukacsovich et al. 1987) was from Dr. P. Venetianer (Biological Research Center, Szeged, Hungary). Escherichia coli strain JM-109 was used to propagate and amplify expression plasmids.
The kringle domain of the C. elegans Ror receptor tyrosine kinase
The recombinant kringle domain of the C. elegans Ror receptor tyrosine kinase was expressed in E. coli. The plasmid expressing the kringle domain of this receptor tyrosine kinase was constructed as follows: PCR-primers (sense: 5 ATATGGCCATACCCATTG GTGTTATGTGAACAGT, antisense: 5 TCGAAGCTTAAT CACTTGGACATTGTGGAACATCAC) were designed to amplify the segment corresponding to the kringle domain from a nematode genomic DNA library (Stratagene). The sequence of the PCR product was verified by cloning it into the Sma I site of the M13mp18 sequencing vector followed by dideoxy sequencing (Sanger et al. 1977). The insert of the M13mp18 was excised by cleaving it with MscI and HindIII and the insert was cloned into PvuII/HindIII-digested expression vector pmed 23 (Lukacsovich et al. 1987). The resulting construct (pmed23Cror/K) encodes a fusion protein (βgalCRor/K) containing the N-terminal 36 residues of β-galactosidase plus the kringle domain of the nematode receptor tyrosine kinase and has the sequence: MTMITDSLAVVLQR R D W E N P G V T QLNRLAAHPPFASHTHWCYVNSGTQYEGT VAQTSSGKQCAPWIDSTSRDFNVHRFPELMNSKNYSRNPG GKKSRPWCYSKPNGQEEYCDVPQCPSD*
E. coli cells carrying recombinant pmed23CrorK plasmids were grown, expression of β-galactosidase fusion proteins was induced with 100 μmole IPTG (Serva), and inclusion bodies containing recombinant protein were isolated as described previously (Bányai and Patthy 1991). The inclusion bodies were dissolved in 60 mL of 0.1 M Tris-HCl, 8 M urea, 10 mM EDTA, 0.1 M dithiothreitol (Sigma) at pH 8.0, and the solution was incubated for 60 min with constant stirring at 25°C.. Insoluble cellular debris were removed by centrifugation and the solubilized proteins were chromatographed on a Sephacryl S-300 column equilibrated with 100 mM Tris-HCl, 8 M urea, 10 mM EDTA, 0.1% 2-mercaptoethanol. The fractions containing the fusion proteins were ideied by SDS-PAGE, pooled, and dialysed against 0.1 M Tris-HCl, 10 mM EDTA at pH 8.0, at 25°C. Precipitated proteins were removed by centrifugation and the supernatant was applied to Sephadex G-75 column equilibrated with 0.1 M ammonium bicarbonate at pH 8.0.
The β-galactosidase moiety of the fusion protein βgalCRor/K was removed by limited tryptic digestion using TPCK-treated trypsin (Sigma). βgalCRor/K (1 mg/mL) was incubated with trypsin (2 μg/mL) in 0.1 M ammonium bicarbonate at pH 8.0 for 30 min at 25°C. Reaction was arrested with 1mM phenylmethylsulfonyl fluoride (Serva) and the kringle domain was separated from the digested β-galactosidase peptides on a Sephadex G-50 colunm equilibrated with 0.1 M NH4CO3 at pH 8.0.
N-terminal sequencing of the truncated protein (CrorK) was performed with an Applied Biosystems 471A protein sequencer with an on-line ABI 120A phenyltiohydantoin (PTH) analyzer. Protein CrorK had a unique N-terminal sequence (LAAHPPFASHTHWCYVNS), confirming that residues 1–27 of the β-galactosidase moiety of the fusion protein have been removed. The concentration of the Cror/K protein was determined using the extinction coefficient 3 |qx 104M−1 cm−1, calculated according to a described method (Mach et al. 1992).
The kringle of rat neurotrypsin
The recombinant kringle-domain of rat neurotrypsin was produced by expression in E. coli as follows. The DNA segment coding for the kringle domain of rat neurotrypsin was amplified with the 5`CCGTCCCGGGGACGATTCCACGCCGCTGCGGG 3' sense, and 5`CCAGAAGCTTTACCCTTGACCACAATCGCAGTAGC 3` antisense primers from a rat fetal brain cDNA library (Clontech). The amplified DNA was digested with SmaI and HindIII restriction endonucleases and ligated into M13mp19 digested with the same enzymes. The sequence of the resulting recombinant plasmid (M13mp19/NT/K) was determined by dideoxy sequencing (Sanger et al. 1977). The DNA encoding the kringle domain was excised from M13mp19/NTK with SmaI–HindIII digestion and cloned into PvuII–HindIII digested pmed23 bacterial expression vector (Lukacsovich et al. 1987). The pmed23/NT/K plasmid expresses the kringle fragment fused to the N-terminal 35 residues of β-galactosidase under the control of the lac operator. The fusion protein has the sequence: MTMITDSLAVVLQRRDWENPGVTQLNRLAAHPPFARGTIP RRCGAGEPWGNATNLGVPCLHWDEVPPFLERSPPASWAE LRGQPHNFCRSPDGAGRPWCFYRNAQGKVDWGYCDCGQG*
E. coli cells carrying recombinant pmed23NT/K plasmids were grown, and expression and isolation of the recombinant fusion protein βgalNT/K was carried out essentially as described above for the βgalCRor/K protein. The β-galactosidase moiety of the βgalNT/K fusion protein was removed by incubating βgalNTK (1 mg/mL) with trypsin (10 μg/mL) in 0.1 M ammonium bicarbonate at pH 8.0 for 30 min at 25°C. Reaction was arrested with 1mM PMSF, the kringle domain was isolated by chromatography on a Sephadex G-50 fine colunm equilibrated with 0.1 M NH4CO3 at pH 8.0 and the protein was lyophilized.
The N-terminal sequence of the truncated protein (NT/K) was determined using an Applied Biosystems 471A protein sequencer with an on-line ABI 120A phenyltiohydantoin (PTH) analyzer. N-terminal sequencing has yielded a unique sequence (RCGAGEPWGN), confirming that the β-galactosidase moiety of the fusion proteins has been removed. The concentration of the NT/K protein was determined using the extinction coefficient of 31370 M−1cm−1, calculated according to a described method (Mach et al. 1992).
The kringle 4 domain of human plasminogen, the second FN2 domain of bull seminal plasma PDC-109
The kringle 4 domain of human plasminogen and the second FN2 domain of bull seminal plasma PDC-109 were generated via proteolysis of the parent proteins and belonged to batches described previously (Rejante et al. 1991 a,b; l1Constantine et al. 1991,1992).
Gel electrophoresis
The composition of protein samples was analyzed by SDS-PAGE using 11–22% linear polyacrylamide gradient slab gels under both reducing and nonreducing conditions (Laemmli et al. 1970).
NMR spectroscopy
NMR spectra were acquired on a Bruker Avance DRX spectrometer at 500 MHz. The probe temperature was maintained at 300 K. Dioxane was used as internal standard. COSY, TOCSY (τmix = 70 ms), and NOESY (τmix = 100 ms) spectra were collected in the phase-sensitive detection mode using standard pulse sequences. Solvent signal suppression was achieved by low-power irradiation at the water frequency or with pulsed field gradient "WATERGATE" technique incorporating the 3–9–19 pulse sequence (Sklenar et al. 1993). All data processing was performed on a Silicon Graphics O2 workstation using FELIX 98.0 software (MSI).
Acknowledgments
This work was supported by NIH grant HL-19409 of Miguel Llinas, by the ICGEB (International Centre for Genetic Engineering and Biotechnology, Trieste, Italy) grant CRP/HUN 98–03 of László Patthy and the OTKA grant (Hungarian Research Fund, Budapest, Hungary) T034317 of László Patthy. We thank András Patthy (Agricultural Biotechnology Center, Gödöllo, Hungary) for sequence analysis of recombinant proteins.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
CD, circular dichroism
COSY, two-dimensional NMR chemical shift correlated spectroscopy
CRor, Ror-type receptor tyrosine kinase of C. elegans
CRor/K, the kringle domain of the Ror receptor tyrosine kinase of C. elegans
FN2, fibronectin type II domain
IPTG, isopropyl-β-D-thiogalactopyranoside
K, kringle domain
NMR, nuclear magnetic resonance
NOESY, two-dimensional NMR nuclear Overhauser effect correlated spectroscopy
NT/K, the kringle domain of neurotrypsin
Pgn/K4, human plasminogen kringle 4
PDC-109/b, second fibronectin type II domain of bovine PDC-109
ppm, parts-per-million
PMSF, phenylmethyl sulfonyl fluride
SDS-PAGE, sodium dodecylsulfate polyacrylamide gel electrophoresis
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/ps.15801.
References
- Arnold, J.M., Kennett, C., and Lavin, M.F. 1997. Transient expression of a novel serine protease in the ectoderm of the ascidian Herdmania momus during development. Dev. Genes Evol. 206 455–463. [DOI] [PubMed] [Google Scholar]
- Atkinson, R.A. and Williams, R.J.P. 1990. Solution structure of the kringle 4 domain from human plasminogen by 1H nuclear magnetic resonance spectroscopy and distance geometry. J. Mol. Biol. 212 541–552. [DOI] [PubMed] [Google Scholar]
- Bányai, L. and Patthy, L. 1991. Evidence for the involvement of type II domains in collagen binding by 72 kDa type IV procollagenase. FEBS Letters 282 23–25. [DOI] [PubMed] [Google Scholar]
- Biunno, I., Bernard, L., Dear, P., Cattaneo, M., Volorio, S., Zannini, L., Bankier, A., and Zollo, M. 2000. SEL1L, the human homolog of C. elegans sel-1: Refined physical mapping, gene structure and ideication of polymorphic markers. Hum. Genet. 106 227–235. [DOI] [PubMed] [Google Scholar]
- Bokman, A.M., Jiménez-Barbero, J., and Llinás, M. 1993. 1H-NMR characterization of the urokinase kringle module: Structural, but not functional, relatedness to homologous domains. J. Biol. Chem. 26813858–13868. [PubMed] [Google Scholar]
- Briknarová, K., Grishaev, A., Bányai, L., Tordai, H., Patthy, L., and Llinás, M. 1999. The second type II module from human matrix metalloproteinase 2: Structure, function and dynamics. Structure Folding Design 7 1235–1245. [DOI] [PubMed] [Google Scholar]
- Byeon, I.J.L. and Llinás, M. 1991. Solution structure of the tissue-type plasminogen activator kringle 2 domain complexed to 6-aminohexanoic acid, an aibrinolytic drug. J. Mol. Biol. 222 1035–1051. [DOI] [PubMed] [Google Scholar]
- Byeon, I.J.L., Kelley, R.F., Mulkerrin, M.G., An, S.S.A., and Llinás, M. 1995. Ligand binding to the tissue-type plasminogen activator kringle 2 domain: structural characterization by 1H-NMR. Biochemistry 34 2739–2750. [DOI] [PubMed] [Google Scholar]
- Choi-Miura, N.H., Tobe, T., Sumiya, J., Nakano, Y., Sano, Y., Mazda, T., and Tomita, M. 1996. Purification and characterization of a novel hyaluronan-binding protein (PHBP) from human plasma: It has three EGF, a kringle and a serine protease domain, similar to hepatocyte growth factor activator. J. Biochem. (Tokyo) 119 1157–1165. [DOI] [PubMed] [Google Scholar]
- Collier, I.E., Wilhelm, S.M., Eisen, A.Z., Marmer, B.L., Grant, G.A., Seltzer, J.L., Kronberger, A., He, C.S., Bauer, E.A., and Goldberg, G.I. 1988. H-ras oncogene-transformed human bronchial epithelial cells (TBE-1) secrete a single metalloprotease capable of degrading basement membrane collagen. J. Biol. Chem. 263 6579–6587. [PubMed] [Google Scholar]
- Constantine, K.L., Ramesh, V., Bányai, L., Trexler, M., Patthy, L., and Llinás, M. 1991. Sequence specific 1H-NMR assignments and structural characterization of bovine seminal fluid protein PDC-109 domain b. Biochemistry 30 1663–1672. [DOI] [PubMed] [Google Scholar]
- Constantine, K.L., Madrid, M., Bányai, L., Trexler, M., Patthy, L., and Llinás, M. 1992. Refined solution structure and ligand-binding properties of PDC-109 domain b: A collagen binding domain. J. Mol. Biol. 223 281–298. [DOI] [PubMed] [Google Scholar]
- Cox, M., Schaller, J., Boelens, R., Kaptein, R., Rickli, E., and Llinás, M. 1994. Kringle solution structures via NMR: Two-dimensional 1H-NMR analysis of horse plasminogen kringle 4. Chem. Phys. Lipids. 67/68 43–58. [DOI] [PubMed] [Google Scholar]
- De Marco, A., Laursen, R.A., and Llinás, M. 1985. Proton Overhauser experiments on kringle 4 from human plasminogen. Implications for the structure of the kringles' hydrophobic core. Biochim. Biophys. Acta 827 369–380. [DOI] [PubMed] [Google Scholar]
- De Marco, A., Petros, A.M., Llinás, M., Kaptein,R., and Boelens, R. 1989. Ligand-binding effects on the kringle 4 domain from human plasminogen: A study by laser photo-CIDNP 1H-NMR spectroscopy. Biochim. Biophys. Acta 994 121–137. [DOI] [PubMed] [Google Scholar]
- Esch, F.S., Ling, N.C., Bohlen, P., Ying, S.Y., and Guillemin, R. 1983. Primary structure of PDC-109, a major protein constituent of bovine seminal plasma. Biochem. Biophys. Res. Commun. 113 861–867. [DOI] [PubMed] [Google Scholar]
- Forrester, W.C., Dell, M., Perens, E., and Garriga, G.A. 1999. C. elegans Ror receptor tyrosine kinase regulates cell motility and asymmetric cell division. Nature 400 881–885. [DOI] [PubMed] [Google Scholar]
- Fu, A.K., Smith, F.D., Zhou, H., Chu, A.H., Tsim, K.W., Peng, B.H., and Ip, N.Y. 1999. Xenopus muscle-specific kinase: molecular cloning and prominent expression in neural tissues during early embryonic development. Eur. J. Neurosci. 11 373–382. [DOI] [PubMed] [Google Scholar]
- Gschwend, T.P., Krueger, S.R., Kozlov, S.V., Wolfer, D.P., and Sonderegger, P. 1997. Neurotrypsin, a novel multidomain serine protease expressed in the nervous system. Mol. Cell. Neurosci. 9 207–219. [DOI] [PubMed] [Google Scholar]
- Günzler, W.A., Steffens, G.J., Ötting, F., Kim, S.M.A., Frankus, E., and Flohé, L. 1982. The primary structure of high molecular mass urokinase from human urine: The complete amino acid sequence of the A chain. Hoppe-Seyler's Z Physiol. Chem. 363 1155–1165. [DOI] [PubMed] [Google Scholar]
- Han, S., Stuart, L.A., and Friezner-Degen, S.J. 1991. Characterization of the DNF15S2 locus on human chromosome 3: Ideication of a gene coding for four kringle domains with homology to hepatocyte growth factor. Biochemistry 30 9768–9780. [DOI] [PubMed] [Google Scholar]
- Hansen, A.P., Petros, A.M., Meadows, R.P., Nettesheim, D.G., Mazar, A.P., Olejniczak, E.T., Xu, R.X., Pederson, T.M., Henkin, J., and Fesik, S.W. 1994. Solution structure of the amino-terminal fragment of urokinase-type plasminogen activator. Biochemistry 33 4847–4864. [DOI] [PubMed] [Google Scholar]
- Harada, Y., Ozaki, K., Suzuki, M., Fujiwara, T., Takahashi, E., Nakamura, Y., and Tanigami, A. 1999. Complete cDNA sequence and genomic organization of a human pancreas-specific gene homologous to Caenorhabditis elegans sel-1. J. Hum. Genet. 44 330–336. [DOI] [PubMed] [Google Scholar]
- Hochschwender, S.M. and Laursen, R.A. 1981. The lysine binding sites of human plasminogen: Evidence for a critical tryptophan in the binding site of kringle 4. J. Biol. Chem. 256 11172–11176. [PubMed] [Google Scholar]
- Iijima, N., Tanaka, M., Mitsui, S., Yamamura, Y., Yamaguchi, N., and Ibata, Y. 1999. Expression of a serine protease (motopsin PRSS12) mRNA in the mouse brain: in situ hybridization histochemical study. Brain Res. Mol. Brain Res. 66 141–149. [DOI] [PubMed] [Google Scholar]
- Ishizaki, J., Hanasaki, K., Higashino, K., Kishino, J., Kikuchi, N., Ohara, O., and Arita, H. 1994. Molecular cloning of pancreatic group I phospholipase A2 receptor. J. Biol. Chem. 269 5897–5904. [PubMed] [Google Scholar]
- Jennings, C.G., Dyer, S.M., and Burden, S.J. 1993. Muscle-specific trk-related receptor with a kringle domain defines a distinct class of receptor tyrosine kinases. Proc. Natl. Acad. Sci. 90 2895–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W. and Nussenzweig, M.C. 1995. The receptor DEC-205 expressed by dendritic cells and thymic epithelial cells is involved in antigen processing Nature 375 151–155. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Li, X., Bokman, A.M., Llinás, M., Smith, R.A.G., and Dobson, C.M. 1994. Solution structure of the kringle domain from urokinase-type plasminogen activator. J. Mol. Biol. 2351548–1559. [DOI] [PubMed] [Google Scholar]
- Llinás, M., De Marco, A., Hochschwender, S.M., and Laursen, R.A. 1983. A 1H-NMR study of isolated domains from human plasminogen: Structural homology between kringles 1 and 4. Eur. J. Biochem. 135 379–391. [DOI] [PubMed] [Google Scholar]
- Lukacsovich, T., Boros, I., and Venetianer, P. 1987. New regulatory features of the promoters of an Escherichia coli rRNA gene. J. Bacteriol. 169 272–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach, H., Middaugh, C.R., and Lewis, R.V. 1992. Statistical determination of the average values of the extinction coefficients of tryptophan and tyrosine in native proteins. Anal. Biochem. 200 74–80. [DOI] [PubMed] [Google Scholar]
- Magnusson, S., Petersen, T.E., Sottrup-Jensen, L., and Claeys, H. 1975. Complete primary structure of prothrombin: Isolation, structure and reactivity of ten carboxylated glutamic acid residues and regulation of prothrombin activation by thrombin. In: Proteases and Biological Control. ( eds. E. Reich et al.), Cold Spring Harbor Laboratory Conferences on Cell Proliferation, vol. 2. pp. 123–149. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Masiakowski, P. and Carroll, R.D. 1992. A novel family of cell surface receptors with tyrosine kinase-like domain. J. Biol. Chem. 267 26181–26190. [PubMed] [Google Scholar]
- McMullen, B.A. and Fujikawa, K. 1985. Amino acid sequence of the heavy chain of human alpha-factor XIIa (activated Hageman factor). J. Biol. Chem. 260 5328–5341. [PubMed] [Google Scholar]
- Miyazawa, K., Shimomura, T., Kitamura, A., Kondo, J., Morimoto, Y., and Kitamura, N. 1993. Molecular cloning and sequence analysis of the cDNA for a human serine protease responsible for activation of hepatocyte growth factor. J. Biol. Chem. 268 10024–10028. [PubMed] [Google Scholar]
- Morgan, D.O., Edman, J.C., Standring, D.N., Fried, V.A., Smith, M.C., Roth, R.A., and Rutter, W.J. 1987. Insulin-like growth factor II receptor as a muunctional binding protein. Nature 329 301–307. [DOI] [PubMed] [Google Scholar]
- Morgunova, E., Tuuttila, A., Bergmann, U., Isupov, M., Lindqvist, Y., Schneider, G., and Tryggvason, K. 1999. Structure of human pro-matrix metalloproteinase-2: Activation mechanism revealed. Science 284 1667–1670. [DOI] [PubMed] [Google Scholar]
- Nakamura, T., Nishizawa, T., Hagiya, M., Seki, T., Shimonishi, M., Sugimura, A., Tashiro, K., and Shimizu, S. 1989. Molecular cloning and expression of human hepatocyte growth factor. Nature 342 440–443. [DOI] [PubMed] [Google Scholar]
- Oishi, I., Sugiyama, S., Liu, Z.J., Yamamura, H., Nishida, Y. and Minami, Y. 1997. A novel Drosophila receptor tyrosine kinase expressed specifically in the nervous system. Unique structural features and implication in developmental signaling. J. Biol. Chem. 27 11916–11923. [DOI] [PubMed] [Google Scholar]
- Patthy, L. 1985. Evolution of the proteases of blood coagulation and fibrinolysis by assembly from modules. Cell 41 657–663. [DOI] [PubMed] [Google Scholar]
- Patthy, L., Trexler, M., Váli, Z., Bányai, L., and Váradi, A. 1984. Kringles: Modules specialized for protein binding. Homology of the gelatin-binding region of fibronectin with the kringle structures of proteases. FEBS Letters 171 131–136. [DOI] [PubMed] [Google Scholar]
- Pennica, D., Holmes, W.E., Kohr, W.J., Harkins, R.N., Vehar, G.A., Ward, C.A., Bennett, W.F., Yelverton, E., Seeburg, P.H., Heyneker, H.L., et al. 1983. Cloning and expression of human tissue-type plasminogen activator cDNA in E. coli. Nature 301 214–221. [DOI] [PubMed] [Google Scholar]
- Pickford, A.R., Potts, J.R., Bright, J.R., Phan, I., and Campbell, I.D. 1997. Solution structure of a type 2 module from fibronectin: Implications for the structure and function of the gelatin-binding domain. Structure 5 359–370. [DOI] [PubMed] [Google Scholar]
- Proba, K., Thomas, P., Gschwend, T.P.,and Sonderegger, P. 1998. Cloning and sequencing of the cDNA encoding human neurotrypsin. Biochim. Biophys. Acta 1396 143–147. [DOI] [PubMed] [Google Scholar]
- Ramesh, V., Petros, A.M., Llinás, M., Tulinsky, A., and Park, C.H. 1987. Proton magnetic resonance study of lysine-binding to the kringle 4 domain of human plasminogen: The structure of the binding site. J. Mol. Biol. 198 481–498. [DOI] [PubMed] [Google Scholar]
- Rejante, M.R., Byeon, I.J.L., and Llinás, M. 1991a. Ligand specificity of human plasminogen kringle 4. Biochemistry 30 11081–11091. [DOI] [PubMed] [Google Scholar]
- Rejante, M., Elliott, J.B.W., and Llinás, M. 1991b. A 1H-NMR study of plasminogen kringle 4 interactions with intact and partially digested fibrinogen. Fibrinolysis 5 87–92. [Google Scholar]
- Rejante, M.R. and Llinás, M. 1994a. 1H-NMR assignments and secondary structure of human plasminogen kringle 1. Eur. J. Biochem. 221 927–937. [DOI] [PubMed] [Google Scholar]
- ———.1994b. Solution structure of the ω-aminohexanoic acid complex of human plasminogen kringle 1. Eur. J. Biochem. 221 939–949. [DOI] [PubMed] [Google Scholar]
- Sanger, F., Nicklen, S., and Coulson, A.R. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. 74 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, J., Milpetz, F., Bork, P., and Ponting, C.P. 1998. SMART, a simple modular architecture research tool: Ideication of signalling domains. Proc. Natl. Acad. Sci. 95 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, J., Copley, R.R., Doerks, T., Ponting, C.P., and Bork, P. 2000. SMART: A Web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidah, N.G., Manjunath, P., Rochemont, J., Sairam, M.R., and Chretien, M. 1987. Complete amino acid sequence of BSP-A3 from bovine seminal plasma. Homology to PDC-109 and to the collagen-binding domain of fibronectin. Biochem J. 243195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenar, V., Piotto, M., Leppik, R., and Saudek, V. 1993. Gradient tailored water-suppression for 1H-15N HSQC experiments optimized to retain full sensitivity. J. Magn. Reson. Ser. A 102 241–245. [Google Scholar]
- Skorstengaard, K., Jensen, M.S., Sahl, P., Petersen, T.E., and Magnusson, S. 1986. Complete primary structure of bovine plasma fibronectin. Eur. J. Biochem. 161 441–453. [DOI] [PubMed] [Google Scholar]
- Sonnhammer, E.L.L., Eddy, S.R., and Durbin, R. 1997. Pfam: A comprehensive database of protein families based on seed alignments. Proteins 28 405–420. [DOI] [PubMed] [Google Scholar]
- Sottrup-Jensen, L., Claeys, H., Zajdel, M., Petersen, T.E., and Magnusson, S. 1978. The primary structure of human plasminogen: Isolation of two lysine-binding fragments and one "mini-"plasminogen (MW, 38,000) by elastase-catalyzed-specific limited proteolysis. In Progress in Chemical Fibrinolysis and Thrombolysis (eds J.F. Davidson et al.), vol. 3, pp. 191–209. Raven Press, NY.
- Sticht, H., Pickford, A.R., Potts, J.R., and Campbell, I.D. 1998. Solution structure of the glycosylated second type 2 module of fibronectin. J. Mol. Biol. 276177–187. [DOI] [PubMed] [Google Scholar]
- Taylor, M.E., Conary, J.T., Lennartz, M.R., Stahl, P.D., and Drickamer, K. 1990. Primary structure of the mannose receptor contains multiple ms resembling carbohydrate-recognition domains. J. Biol. Chem. 265 12156–12162. [PubMed] [Google Scholar]
- Thewes, T., Ramesh, V., Simplaceanu, E.L., and Llinás, M. 1988. Analysis of the aromatic 1H-NMR spectrum of the kringle 5 domain from human plasminogen: Evidence for a conserved kringle fold. Eur. J. Biochem. 175 237–249. [DOI] [PubMed] [Google Scholar]
- Tordai, H. and Patthy, L. 1999. The gelatin-binding site of the second type-II domain of gelatinase A/MMP-2. Eur. J. Biochem. 259 513–518. [DOI] [PubMed] [Google Scholar]
- Trexler, M., Bányai, L., Patthy, L., Pluck, N.D., and Williams, R.J.P. 1983. The solution structure of kringle 4. NMR studies on native and several chemically modified kringle 4 species of human plasminogen. FEBS Letters 154311–318. [Google Scholar]
- Trexler, M. and Patthy, L. 1983. Folding autonomy of the kringle 4 fragment of human plasminogen. Proc. Natl. Acad. Sci. 80 2457–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm, S.M., Collier, I.E., Marmer, B.L., Eisen, A.Z., Grant, G.A., and Goldberg, G.I. 1989. SV40-transformed human lung fibroblasts secrete a 92-kDa type IV collagenase which is identical to that secreted by normal human macrophages. J. Biol. Chem. 264 17213–17221. [PubMed] [Google Scholar]
- Wilson, C., Goberdhan, D.C., and Steller, H. 1993. Dror, a potential neurotrophic receptor gene, encodes a Drosophila homolog of the vertebrate Ror family of Trk-related receptor tyrosine kinases. Proc. Natl. Acad. Sci. 90 7109–7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, T.P., Padmanabhan, K., Tulinsky, A., and Mulichak, A.M. 1991. The refined structure of the ω-aminocaproic acid complex of human plasminogen kringle 4. Biochemistry 30 10589–10594. [DOI] [PubMed] [Google Scholar]
- Yamamura, Y., Yamashiro, K., Tsuruoka, N., Nakazato, H., Tsujimura, A., and Yamaguchi, N. 1997. Molecular cloning of a novel brain-specific serine protease with a kringle-like structure and three scavenger receptor cysteine-rich ms. Biochem. Biophys. Res. Commun. 239 386–392. [DOI] [PubMed] [Google Scholar]