An intramolecular complex comprising the LIM domains of Lhx3 and the interacting domain of Isl1 tethered by a flexible linker was engineered, overexpressed in E. coli, purified and crystallized.
Keywords: Lhx3, Isl1, LIM-homeodomain proteins, protein complex, motor-neuron development
Abstract
A stable intramolecular complex comprising the LIM domains of the LIM-homeodomain protein Lhx3 tethered to a peptide region of Isl1 has been engineered, purified and crystallized. The monoclinic crystals belong to space group C2, with unit-cell parameters a = 119, b = 62.2, c = 51.9 Å, β = 91.6°, and diffract to 2.05 Å resolution.
1. Introduction
Different combinations of LIM-homeodomain (LIM-HD) transcription factors form a ‘LIM code’ that contributes to the specification of many different cell types (Lumsden, 1995 ▶). LIM-HD proteins contain two highly conserved domains: a pair of closely spaced LIM domains at the N-terminus and a centrally located homeodomain. They also contain a nonconserved and poorly characterized C-terminus. LIM domains are a class of zinc finger that ligate two zinc ions and are involved in making protein–protein interactions; the acronym derives from the first three genes in which the motif was identified, Lin-11, Isl1 and Mec-3 (reviewed in Bach, 2000 ▶). Homeodomains from LIM-HD proteins bind AT-rich DNA sequences.
The LIM code is particularly prevalent in the developing central nervous system of vertebrates, where ∼12 different LIM-HDs (as well as splice variants of these proteins) are required for the correct specification of diverse neuronal cell types. The molecular mechanisms that underlie the LIM code are likely to involve, at least in part, competition for essential protein partners, such as LIM domain-binding protein 1 (Ldb1/NLI/CLIM; Milan et al., 1998 ▶; Shoresh et al., 1998 ▶; Zeng et al., 1998 ▶). However, direct interactions of different LIM-HD proteins also appear to be involved in the molecular basis of the LIM code (Jurata et al., 1998 ▶). For example, a direct interaction between the LIM-HD proteins Lhx3 and Isl1 is required for the correct development of motor neurons in the ventral spinal cord. This interaction is mediated by the LIM domains of Lhx3 and the C-terminus of Isl1 (Thaler et al., 2002 ▶). The LIM domains of Lhx3 tend to be insoluble (Lee et al., 2005 ▶). Thus, in this study we generated a stable soluble complex of the LIM domains of Lhx3 with a peptide region from the C-terminal domain of Isl1, fusing these proteins via a flexible 11-residue linker. The purification, crystallization and preliminary X-ray crystallographic analysis of this complex are reported.
2. Methods
2.1. Cloning, expression and purification
A construct encoding residues 28–152 of Lhx3, a linker (GGSGGHMGSGG) and residues 262–291 of Isl1 was generated by overlap extension PCR and cloned into a pGEX-2T vector (GE Healthcare). This results in an Lhx3–Isl1 tethered complex with an N-terminal glutathione S-transferase (GST) tag. The plasmid was transformed into Escherichia coli BL21 (DE3) cells and protein expression was induced by the addition of 0.4 mM IPTG at 293 K in Luria Broth. GST-Lhx3–Isl1 was purified by standard glutathione-affinity chromatography on Sepharose-4B (GE Healthcare) resin. The GST tag was removed by treatment with thrombin and the eluted Lhx3–Isl1 tethered complex, which contains two additional residues (GS) at the N-terminus derived from the vector, was further purified using size-exclusion chromatography on a HiLoad 16/60 Superdex-75 column (GE Healthcare) in buffer containing 50 mM Tris–HCl, 150 mM NaCl, 1 mM dithiothreitol at pH 8.0. The purified protein was concentrated in Centricon YM-3 centrifugal filtration devices (Millipore) to a concentration of 10 mg ml−1 in the same buffer prior to crystallization.
2.2. Crystallization
Preliminary crystallization conditions were discovered using sparse-matrix and systematic salt screens and the sitting-drop vapour-diffusion method. Each of the solutions from Hampton Crystal Screens 1 and 2 and Hampton SaltRx Screen 1 (Hampton Research, CA, USA; 2 µl) was mixed with purified protein solution (2 µl) at room temperature (∼293 K).
2.3. Data collection and processing
Crystals were cryoprotected by soaking them in reservoir solution containing 15%(v/v) glycerol prior to flash-cooling in a nitrogen-gas stream at 100 K. Diffraction data were recorded on a MAR345 imaging-plate detector (MAR Reserach) using X-rays produced by a Rigaku RU200H rotating-anode generator (Cu Kα) focused with Osmic mirrors (MSC Rigaku). The diffraction data were processed and scaled with the HKL suite of programs, DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
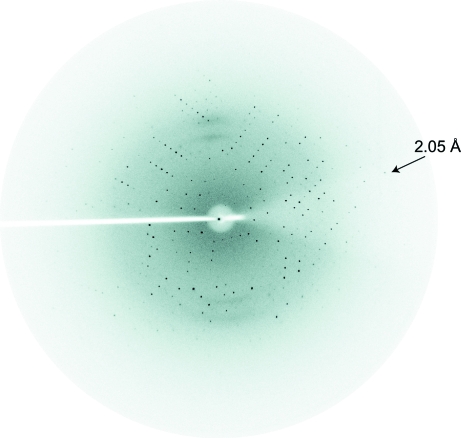
The tethered Lhx3–Isl1 complex (MW = 18 700 Da) was purified to >95% purity as determined by SDS–PAGE with Coomassie staining, with typical yields of ∼4–5 mg per litre of culture medium. Crystals of the complex were observed after two weeks under four different conditions (Hampton SaltRx Screen 1 condition Nos. 19, 20, 31 and 32) and after four weeks under two conditions (Hampton SaltRx Screen 1 condition Nos. 16 and 29). Optimization of Hampton SaltRx Screen 1 condition No. 20 (0.7 M sodium citrate dihydrate, 0.1 M Tris pH 8.5) yielded conditions that produced good-quality crystals: 1.0 M sodium citrate trihydrate, 0.1 M Tris–HCl pH 7.25. Crystals were grown using hanging drops in which 2 µl protein was mixed with 2 µl reservoir solution and equilibrated against 0.5 ml well solution at room temperature. Crystals appeared after 3 d (Fig. 1 ▶). Native diffraction data were recorded to 2.05 Å resolution (Fig. 2 ▶). Data-collection and processing statistics are summarized in Table 1 ▶.
Figure 1.
Monoclinic crystals of an Lhx3–Isl1 complex. Crystal dimensions are 300 × 100 × 100 µm.
Figure 2.
Diffraction image of an Lhx3–Isl1 complex. The arrow shows data at the limit of resolution (2.05 Å).
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 1.542 |
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 119, b = 62.2, c = 51.9, β = 91.6 |
| Resolution limit (Å) | 2.05 (2.11–2.05) |
| Mosaicity (°) | 1.05 |
| Completeness (%) | 96.4 (78.8) |
| Unique reflections | 23120 |
| Redundancy | 3.5 (2.1) |
| Rmerge† | 0.051 (0.388) |
| 〈I/σ(I)〉 | 13.4 (2.4) |
R
merge = 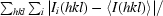
 .
.
The symmetry of and lack of systematic absences in the diffraction data reveal that the crystals are monoclinic, space group C2, with unit-cell parameters a = 119, b = 62.2, c = 51.9 Å, β = 91.6°. The asymmetric unit is estimated to contain two complexes, with a corresponding crystal volume per protein weight of 2.5 Å3 Da−1 and a solvent content of 51.3% (Matthews, 1968 ▶). Phasing using the anomalous signal of the four Zn atoms in the LIM domains of Lhx3 is currently being attempted in order to solve the structure.
Acknowledgments
MB and ML were supported by Australian Postgraduate Awards. JMM is a Senior Research Fellow of the Viertel Foundation. This work was supported by grant DP663289 from the Australian Research Council.
References
- Bach, I. (2000). Mech. Dev.91, 5–17. [DOI] [PubMed]
- Jurata, L. W., Pfaff, S. L. & Gill, G. N. (1998). J. Biol. Chem.273, 3152–3157. [DOI] [PubMed]
- Lee, C., Nancarrow, A. L., Bach, I., Mackay, J. P. & Matthews, J. M. (2005). J. Biomol. NMR, 33, 198. [DOI] [PubMed]
- Lumsden, A. (1995). Curr. Biol.5, 491–495. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Milan, M., Diaz-Benjumea, F. J. & Cohen, S. M. (1998). Genes Dev.12, 2912–2920. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Shoresh, M., Orgad, S., Shmueli, O., Werczberger, R., Gelbaum, D., Abiri, S. & Segal, D. (1998). Genetics, 150, 283–299. [DOI] [PMC free article] [PubMed]
- Thaler, J. P., Lee, S. K., Jurata, L. W., Gill, G. N. & Pfaff, S. L. (2002). Cell, 110, 237–249. [DOI] [PubMed]
- Zeng, C., Justice, N. J., Abdelilah, S., Chan, Y. M., Jan, L. Y. & Jan, Y. N. (1998). Proc. Natl Acad. Sci. USA, 95, 10637–10642. [DOI] [PMC free article] [PubMed]