The cloning, expression, crystallization and preliminary X-ray data analysis of norcoclaurine synthase from T. flavum, a protein which catalyzes the first committed step in the biosynthesis of benzylisoquinoline alkaloids, are reported.
Keywords: norcoclaurine synthase, benzylisoquinoline alkaloid biosynthesis, MAD phasing
Abstract
Norcoclaurine synthase (NCS) catalyzes the condensation of 3,4-dihydroxyphenylethylamine (dopamine) and 4-hydroxyphenylacetaldehyde (4-HPAA) as the first committed step in the biosynthesis of benzylisoquinoline alkaloids in plants. The protein was cloned, expressed and purified. Crystals were obtained at 294 K by the hanging-drop vapour-diffusion method using ammonium sulfate and sodium chloride as precipitant agents and diffract to better than 3.0 Å resolution using a synchrotron-radiation source. The crystals belong to the trigonal space group P3121, with unit-cell parameters a = b = 86.31, c = 118.36 Å. A selenomethionine derivative was overexpressed, purified and crystallized in the same space group. A complete MAD data set was collected at 2.7 Å resolution. The model is under construction.
1. Introduction
Norcoclaurine synthase (NCS) is a protein that catalyzes the first committed step in benzylisoquinoline alkaloid biosynthesis, which consists of the condensation of dopamine and 4-hydroxyphenylacetaldehyde (4-HPAA) to norcoclaurine (Samanani & Facchini, 2001 ▶). NCS plays a regulatory or rate-limiting role in controlling the rate of pathway flux in benzylisoquinoline alkaloid biosynthesis (Samanani et al., 2004 ▶).
Benzylisoquinoline alkaloids are a large and diverse group of secondary metabolites that are mainly found in five related plant families, including the Papaveraceae and Ranunculaceae. Many benzylisoquinoline alkaloids are of great medical interest because of their pharmacological activity, such as, for example, the antibiotic sanguinarine, the muscle relaxants papaverine and tubocurarine and the analgesics codeine and morphine (Weid et al., 2004 ▶).
The NCS protein contains 210 residues. A search of the NCBI protein database performed with BLAST (http://www.ncbi.nlm.nih.gov/blast) showed that the central part of the protein (residues 40–170) displays a high degree of sequence identity (28–44%) to members of class 10 of the pathogenesis-related (PR 10) proteins. The PR proteins are plant proteins that are expressed under conditions of stress (Ebner et al., 2001 ▶) and have been divided into 14 classes. The PR 10 proteins include the family of Bet v 1 homologous allergens that are of medical interest because of the effect that they have on the human immune system (Bohle, 2007 ▶). These proteins cause type I allergies involving weaker symptoms such as allergic rhinitis as well as asthma.
In this framework, determination of the X-ray structure of NCS may shed light on the evolutionary connections between the PR 10 proteins and proteins of the Bet v 1 homologous allergen family, the enzymatic activity of which has not been established.
For this purpose, His-tagged norcoclaurine synthase (NCS) from Thalictrum flavum and its SeMet-substituted variant were cloned, expressed, purified and crystallized.
2. Results and discussion
2.1. Protein expression and purification
The expression of the NCS protein was designed, synthesized and optimized for Escherichia coli codon usage using GeneOptimizer Assisted Sequence Analysis (GENART). The NCS protein truncated at the first 19 amino acids (Samanani et al., 2004 ▶) with an N-terminal MTGS sequence and a His tag at the C-terminus was subcloned into the NdeI and XhoI restriction sites of the vector pET22b. Transformation of chemically competent E. coli strain BL21DE (Invitrogen) was performed and transformed cells were plated onto LB-ampicillin plates. A single colony was picked and grown overnight at 310 K in LB medium containing 50 µg ml−1 ampicillin. The overnight culture was diluted 1:50 into fresh LB medium containing 50 µg ml−1 ampicillin and grown at 310 K to an A 600nm of 0.6; IPTG was then added to a final concentration of 0.5 mM to induce expression. The culture was incubated for 6 h at 298 K and the cell paste was harvested, frozen in liquid nitrogen and stored at 253 K.
The above transformed cells were also used to express the SeMet protein, which contains four SeMet residues. A single colony was picked and grown overnight at 310 K in LB medium containing ampicillin at a final concentration of 50 µg ml−1. The overnight culture was diluted 1:100 into M9 minimal medium containing 2 mM MgSO4, 0.1 mM FeSO4, 0.4% glucose, 1 µg ml−1 riboflavin, 1 µg ml−1 thiamine, 40 µg ml−1 amino-acid mix I (alanine, arginine, asparagine, aspartic acid, cysteine, glutamine, glutamic acid, glycine, histidine, isoleucine, leucine, lysine, proline, serine, threonine and valine), 40 µg ml−1 amino-acid mix II (phenylalanine, tryptophan and tyrosine), 40 µg ml−1 seleno-l-methionine and 50 µg ml−1 ampicillin and grown to an A 600nm of 0.6. The culture was induced for 6 h with 0.5 mM IPTG at 298 K. The cell paste was harvested, frozen in liquid nitrogen and stored at 253 K.
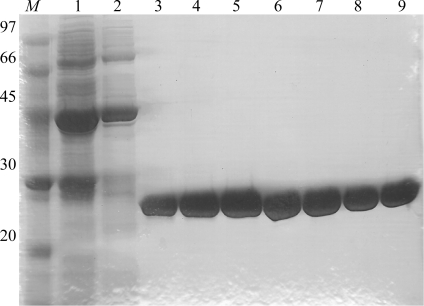
The NCS protein and the SeMet NCS variant were both purified as follows: the cell paste was defrosted and suspended in ice-cold buffer A (20 mM potassium phosphate pH 8.0) containing protease inhibitors without EDTA (Complete, GE Healthcare). Cells were disrupted by sonication and debris was removed by centrifugation at 40 000g for 20 min at 277 K. The supernatant fraction was applied onto a 5 ml HisTrap Fast Flow column (GE Healthcare) previously equilibrated with buffer A and proteins were eluted with a linear gradient of imidazole concentration from 0 to 0.5 M in buffer A on an ÄKTA FPLC system (GE Healthcare). Protein purity was determined by SDS–PAGE (Fig. 1 ▶) and protein concentration was determined by the Bradford method using BSA as standard.
Figure 1.
SDS–PAGE of fractions eluted from the HisTrap Fast Flow column. Lane M, molecular-weight markers (kDa); lanes 1–9, HisTrap elution fractions at 0.144, 0.153, 0.250, 0.259, 0.269, 0.279, 0.288, 0.298 and 0.307 M imidazole, respectively.
2.2. Crystallization
Crystallization trials using the His-tagged NCS were initially performed using the vapour-diffusion method at 293 K with various commercially available crystallization kits. The crystallization trials were performed using a protein sample concentrated to about 12 mg ml−1 and dialyzed against 20 mM Tris–HCl pH 7.5.
Crystals were grown using the hanging-drop vapour-diffusion method. The measured crystal was obtained using ammonium sulfate as a precipitant agent at a final concentration of 1.2 M, sodium chloride as an additive at a final concentration of 0.2 M and acetate buffer pH 4.5 at a final concentration of 0.1 M. 1 µl of the protein sample was mixed with an equal amount of reservoir solution and equilibrated against 500 µl reservoir solution.
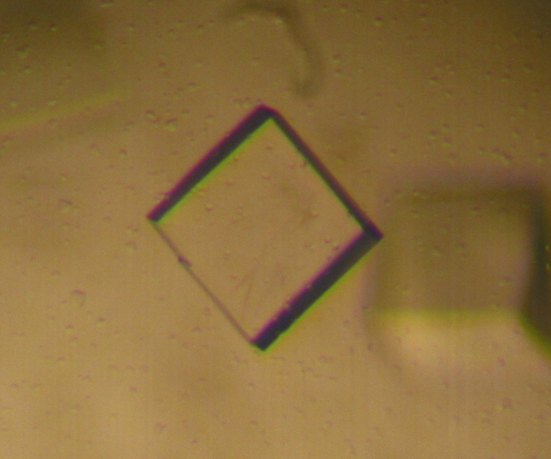
The crystals grew at a controlled temperature of 294 K in 10–15 d, reaching dimensions of 0.3 × 0.3 × 0.2 mm (Fig. 2 ▶).
Figure 2.
A typical SeMet NCS crystal (crystal dimensions 0.3 × 0.3 × 0.2 mm).
The measured SeMet NCS crystal was grown from a protein sample concentrated to about 11 mg ml−1 and dialyzed against 20 mM Tris–HCl pH 7.5 under conditions similar to those identified for the His-tagged wild-type protein: 1.4 M ammonium sulfate, 0.2 M sodium chloride and 0.1 M acetate buffer pH 4.5. The native and SeMet-derivative NCS crystals were cryoprotected in a solution containing 75%(v/v) reservoir solution and 25%(v/v) glycerol and mounted in nylon loops. The crystals were then flash-frozen by quick submersion in liquid nitrogen.
2.3. Data collection and data analysis
A single-wavelength data set (λ = 0.933 Å) was collected from NCS on the ID14-2 beamline at the ESRF synchrotron-radiation source, Grenoble, France using a CCD detector at a temperature of 100 K. The data set was processed with DENZO (Otwinowski & Minor, 1997 ▶) and scaled with SCALEPACK (Otwinowski & Minor, 1997 ▶).
The autoindexing procedure indicated that the crystals belonged to the trigonal space group P3121, with unit-cell parameters a = b = 86.91, c = 117.32 Å, α = β = 90, γ = 120°. The data set was more than 99% complete, with an R merge value of 8.3%. The statistics of the scaling procedure are reported in Table 1 ▶.
Table 1. Crystal parameters and data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| SeMet | ||||
|---|---|---|---|---|
| Native | Peak | Inflection | Remote | |
| Space group | P3121 | P3121 | ||
| Unit-cell parameters | ||||
| a (Å) | 86.91 | 86.31 | ||
| b (Å) | 86.91 | 86.31 | ||
| c (Å) | 117.32 | 118.36 | ||
| No. of molecules in ASU | 2 | 2 | ||
| Resolution (Å) | 3.0 | 2.70 | 2.85 | 2.70 |
| Total observations | 117453 | 77375 | 69000 | 60831 |
| Unique reflections | 10518 (1037) | 14034 (1344) | 12015 (1169) | 13618 (1301) |
| Completeness (%) | 99.9 (99.2) | 98.9 (97.7) | 99 (98.4) | 96 (94.1) |
| Redundancy | 11.2 (10) | 5.5 (4.8) | 5.8 (5.3) | 4.6 (4.0) |
| Rmerge† | 0.083 (0.36) | 0.114 (0.517) | 0.088 (0.55) | 0.084 (0.51) |
| χ2 | 1.5 (2.0) | 1.3 (0.9) | 1.0 (0.8) | 1.0 (0.8) |
| 〈I/σ(I)〉 | 31.0 (11.0) | 16.4 (2.5) | 17.0 (3.0) | 14. 0 (2.0) |
R
merge = 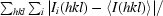
 , where I
i(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the mean intensity of the hkl reflection.
, where I
i(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the mean intensity of the hkl reflection.
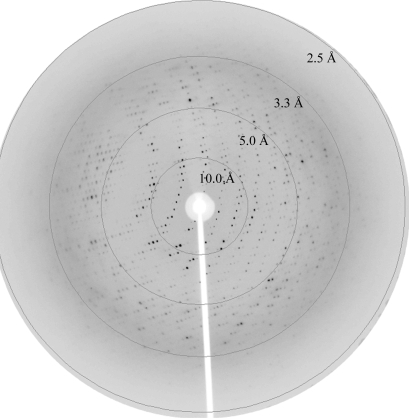
A three-wavelength MAD data set was collected from SeMet NCS on the ID14-2 beamline at the BESSY synchrotron-radiation source, Berlin, Germany using a CCD detector. Complete data sets (120° ϕ rotation each) were collected at the peak (λ = 0.97966 Å; Fig. 3 ▶), inflection (λ = 0.97984 Å) and remote (λ = 0.97800 Å) wavelengths at a temperature of 100 K. Each frame was collected with an exposure time of 2 s and a 1.0° oscillation range.
Figure 3.
X-ray diffraction of an SeMet NCS crystal to 2.7 Å at the peak wavelength (λ = 0.97966 Å). Data were collected as 1.0° oscillation frames at 100 K.
The three data sets were processed with DENZO (Otwinowski & Minor, 1997 ▶) and scaled with SCALEPACK (Otwinowski & Minor, 1997 ▶).
The three-wavelength MAD data set of SeMet NCS was further scaled and analyzed with SCALEIT from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶).
The autoindexing procedure indicated that the crystals belonged to the trigonal space group P3121, with unit-cell parameters a = b = 86.31, c = 118.36 Å, α = β = 90, γ = 120°. All three data sets were more than 96% complete, with R merge values below 10%. The statistics of the scaling procedure are reported in Table 1 ▶. A value of V M = 2.89 Å3 Da−1 was calculated according to Matthews (1968 ▶), with the assumptions that the unit cell contains six asymmetric units, that the molecular weight of the protein is about 24 kDa and that each asymmetric unit contains two monomers.
2.4. Initial phasing
Attempts to solve the three-dimensional structure by molecular replacement using the cytokine-specific binding protein from Vigna radiata (Pasternak et al., 2006 ▶; PDB code 2flh) as a search probe were unsuccessful. The failure of the molecular replacement is a consequence of the low degree of sequence identity between the search probe and NCS. A BLAST search showed that on aligning 135 of the 210 NCS residues with the corresponding residues of the cytokine-specific binding protein from V. radiata, the identity between the two proteins was only 28%.
SeMet NCS was therefore overexpressed, purified and crystallized in order to solve the protein structure using the multiple-wavelength anomalous diffraction method. A heavy-atom site search and phase determination were performed using the program SOLVE (Terwilliger & Berendzen, 1999 ▶) using data collected at three wavelengths (indicated in §2.3) in the 20–2.7 Å resolution range. A single solution was found with six of the possible eight heavy-atom sites with a Z score of 14.3 for a signal-to-noise ratio of 0.3, leading to a mean figure of merit of 0.46. The phases obtained were improved using solvent flattening and twofold-averaging DM (Cowtan, 1994 ▶; Cowtan & Main, 1996 ▶) and the electron-density maps were used to construct the main chains of the molecules using Coot (Emsley & Cowtan, 2004 ▶).
Acknowledgments
This work was supported by the European Community Research Infrastructure Action under the FP6 ‘Structuring the European Research Area’ Programme (through the Integrated Infrastructure Initiative Integrating Activity on Synchroton and Free Electron Laser Science, Contract R II 3-CT-2004-506008). We thank the European Synchrotron Radiation Facility (ESRF), Grenoble and the Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung (BESSY), Berlin, where the data were collected.
References
- Bohle, B. (2007). Allergy, 62, 3–10. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Cowtan, K. (1994). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.31, 34–38.
- Cowtan, K. D. & Main, P. (1996). Acta Cryst. D52, 43–48. [DOI] [PubMed]
- Ebner, C., Hoffmann-Sommergruber, K. & Breiteneder, H. (2001). Allergy, 56, 43–44. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pasternak, O., Bujacz, G. D., Fujimoto, Y., Hashimoto, Y., Jelen, F., Otlewski, J., Sikorski, M. M. & Jaskolski, M. (2006). Plant Cell, 18, 2622–2634. [DOI] [PMC free article] [PubMed]
- Samanani, N. & Facchini, P. J. (2001). Planta, 213, 898–906. [DOI] [PubMed]
- Samanani, N., Liscombe, D. K. & Facchini, P. J. (2004). Plant J.40, 302–313. [DOI] [PubMed]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed]
- Weid, M., Ziegler, J. & Kutchan, T. M. (2004). Proc. Natl Acad. Sci. USA, 101, 13957–13962. [DOI] [PMC free article] [PubMed]