In order to investigate the role of the protonation states of protein residues in O2 binding, large crystals of deoxy HbA (∼20 mm3) were grown in D2O under anaerobic conditions for neutron diffraction studies.
Keywords: deoxyhemoglobin, neutron diffraction
Abstract
Human hemoglobin (HbA) is an intricate system that has evolved to efficiently transport oxygen molecules (O2) from lung to tissue. Its quaternary structure can fluctuate between two conformations, T (tense or deoxy) and R (relaxed or oxy), which have low and high affinity for O2, respectively. The binding of O2 to the heme sites of HbA is regulated by protons and by inorganic anions. In order to investigate the role of the protonation states of protein residues in O2 binding, large crystals of deoxy HbA (∼20 mm3) were grown in D2O under anaerobic conditions for neutron diffraction studies. A time-of-flight neutron data set was collected to 1.8 Å resolution on the Protein Crystallography Station (PCS) at the spallation source run by Los Alamos Neutron Science Center (LANSCE). The HbA tetramer (64.6 kDa; 574 residues excluding the four heme groups) occupies the largest asymmetric unit (space group P21) from which a high-resolution neutron data set has been collected to date.
1. Introduction
Determining the various protonation states of a protein can be important for understanding the details of its biological function, since the protonation (or deprotonation) of a particular functional group may act as an initiator and/or a regulator of a chemical reaction. Although X-ray crystallography is a powerful method for determining the three-dimensional structure of proteins, it is less well suited to revealing protonation states because the X-ray atomic scattering factor of hydrogen (H) is much smaller than those of other atoms commonly found in macromolecules. In contrast, the neutron scattering lengths for H and its isotope deuterium (D) are comparable to those of non-H atoms. Neutron diffraction studies have already provided direct evidence for the protonation states of several proteins.
One of the main goals of neutron studies of the enzymes lysozyme (Mason et al., 1984 ▶), trypsin inhibitor (Kossiakoff & Spencer, 1980 ▶), endothiapepsin (Coates et al., 2001 ▶; Tuan et al., 2007 ▶), aldose reductase (Blakeley et al., 2005 ▶) and dihydrofolate reductase (Bennett et al., 2006 ▶) was to reveal the protonation states of catalytically important aspartic or glutamic acid side chains in their active sites. In the case of myoglobin (Cheng & Schoenborn, 1990 ▶), ribonuclease A (Borah et al., 1985 ▶), amicyanin (Sukumar et al., 2005 ▶), xylose isomerase (Katz et al., 2006 ▶), insulin (Ishikawa et al., 2008 ▶) and trypsin inhibitor, the protonation states of histidine side chains, either at or distal to the active site, were of interest. In addition to determining the protonation states of amino-acid side chains, neutron studies have also been used to determine whether a functionally important solvent molecule is a water molecule or hydroxide ion, as in the case of both xylose isomerase and di-isopropyl fluorophosphatase (Blum et al., 2007 ▶). We are using neutrons to study the protonation states of the human oxygen-carrier molecule hemoglobin A (HbA).
HbA is active as an α2β2 tetrameric hemoprotein containing Fe metal in the heme sites and arranged as a dimer of identical αβ dimers connected noncovalently. The ability of HbA to form two quaternary structures T (tense) and R (relaxed) of the α2β2 tetramer allows the efficient transport of O2 from lung to tissue. T and R states have low and high affinity for O2, respectively, and can interchange by conformational movement. Much of our knowledge of the control mechanisms of HbA is based on the X-ray crystal structures of deoxyhemoglobin (T state) and oxyhemoglobin (R state; Monod et al., 1965 ▶; Gelin et al., 1983 ▶; Park et al., 2006 ▶). The binding of O2 to HbA is further modulated by protons, chloride ions and inorganic phosphate, which preferentially stabilize the T state and facilitate the release of O2 from HbA at the tissues (Imai, 1982 ▶; Perutz et al., 1998 ▶). The α and β subunits of HbA include ten and nine His residues, respectively. Thus, the α2β2 tetramer contains a total of 38 His residues and we expect the protonation states of these His residues to have a direct bearing on the functional mechanism of this protein.
Some of the His residues in HbA are of particular interest. His58 and His87 sandwich the heme groups in a crevice of the α subunits. His63 and His92 act likewise in the β subunit. The proximal histidines (His87α and His92β) bind directly to the Fe2+ cation at the fifth coordination position in the heme group. The distal histidines (His58α and His63β) are positioned near the sixth coordination position of iron on the opposite side of the heme plane, but are not bonded to the Fe2+ cation. The sixth coordination position is used by the heme group as the oxygen-binding site.
In previous studies, we carried out a neutron crystallographic analysis of human deoxyhemoglobin using the ‘monochromatic’ neutron diffractometer BIX-3 in a neutron reactor at Japan Atomic Energy Agency (Chatake et al., 2007 ▶). The protonation of histidines buried in the protein could clearly be observed in 2.1 Å neutron Fourier maps. His58α and His63β were doubly protonated at both Nδ1 and N∊2, although these residues are known to exist as the neutral N∊2—H tautomer in CO-bound hemoglobin. Our results suggested that these residues were plausible candidates to contribute to the tertiary Bohr effect of HbA in the oxygen-transport system. However, because of the low completeness of the diffraction data (<50% in the highest resolution shell, 2.18–2.10 Å), we were unable to determine the protonation states of His residues at the surface of the protein.
The low completeness of the data set can be explained by a combination of factors. There is a large blind region around the rotation axis of BIX-3, which is a single-axis short cylindrical diffractometer equipped with a neutron imaging plate (40 cm internal diameter × 45 cm height; Tanaka et al., 2002 ▶). Usually, data in this blind region can be collected simply by either reorienting the capillary on the goniometer or the crystal within the capillary. However, in our particular case this was not possible because the crystal was mounted in a large airtight sealed capillary (to maintain anaerobic conditions). In addition, the crystal form used for data collection belonged to an unfavorable low-symmetry monoclinic space group (P21) that requires a large number of different crystal settings. Although an effort was made to increase the completeness by 120 d data collection at various crystal rotation settings, it was still impossible to collect a more complete data set.
We report here further neutron studies in which the Protein Crystallography Station (PCS) built at the spallation neutron source run by Los Alamos Neutron Scattering Center (LANSCE) at Los Alamos National Laboratory (LANL) (Langan et al., 2004 ▶) was used to obtain a more complete and higher resolution data set from HbA. For this particular sample, the much larger cylindrical detector of the PCS in combination with a κ-circle goniometer provided more freedom to reorient the sample. In addition, the combination of a pulsed neutron source and time-of-flight wavelength-resolved Laue diffraction methods provided us with more extensive and rapid coverage of reciprocal space. The data presented here, which were collected on the PCS in a relatively short period of 18 d, are from the largest protein asymmetric unit ever studied using neutrons at a resolution beyond 2 Å.
2. Crystallization in D2O
HbA was prepared as described previously (Shibayama et al., 1991 ▶). Purified protein was diluted about tenfold with D2O and concentrated by ultrafiltration at 278 K using an Amicon ultrafiltration unit equipped with a YM-10 membrane (Millipore). This process of dilution and concentration was repeated until the amount of free H2O was less than 1%. Crystallization of deoxyhemoglobin was carried out according to the batch method of Perutz (1968 ▶), with slight modifications, to give a D2O solution of 1.0%(w/v) deoxyhemoglobin containing 4.30 M ammonium ion, 1.94 M sulfate ion, 0.24 M phosphate ion and 5 mM sodium dithionite (pH 6.3; this was read straight from the pH meter without correction for the deuterium shift). Crystals grew to their maximum dimensions in three to four weeks.
3. Neutron data collection and reduction
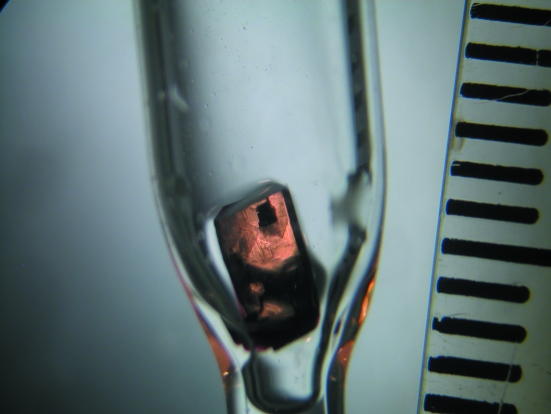
Several candidate crystals were successfully mounted in quartz capillaries (Fig. 1 ▶) and mother liquor was placed in the narrow part of the capillary. The capillaries also contained a small amount (∼100 mm3) of Oxygen Absorbing System A-500HS (ISO, Yokohama, Japan) powder placed ∼20 mm above the crystal to maintain the anaerobic conditions during the long neutron experiment. The crystals were transported personally to the PCS, with the sealed capillaries placed in a nylon plastic bag containing oxygen absorbers and oxygen-content indicators and sealed airtight. A 12 h test exposure was taken from the largest HbA crystal (∼20mm3), depicted in Fig. 1 ▶, yielding diffraction to 1.8 Å resolution. The diffraction quality and the signal-to-noise ration were deemed to be suitable for full data-set collection.
Figure 1.
Crystal of human deoxyhemoglobin used for neutron diffraction data collection. The crystal had dimensions of about 4 × 2.5 × 2 mm (∼20 mm3) and was mounted in a bottle-shaped quartz capillary. The intervals on the ruler on the right-hand side of the photo measure 1 mm.
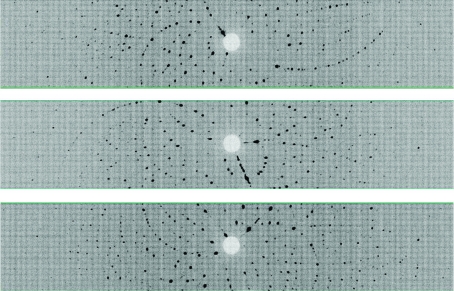
Time-of-flight wavelength-resolved Laue images were collected at room temperature on a Huber κ-circle goniometer at 36 usable settings, with approximately 12 h exposure times per diffraction image (Fig. 2 ▶). The crystal-to-detector distance was 730 mm, corresponding to the cylindrical radius of the detector. The detector was kept at 2θ = 0° during the entire experiment, effectively collecting data in the ±60° 2θ range. Because of the ±8° span of the detector in the vertical direction, the crystal was reoriented six times using the κ and ω goniometer circles and ϕ scans were performed at each crystal orientation. Each image was processed using a version of d*TREK (Pflugrath, 1999 ▶) modified for wavelength-resolved Laue neutron protein crystallography (Langan & Greene, 2004 ▶). The integrated reflections were wavelength-normalized using LAUENORM (Helliwell et al., 1989 ▶) and then merged using SCALA (Weiss, 2001 ▶) incorporated into CCP4i (Evans, 2006 ▶; Diederichs & Karplus, 1997 ▶; Weiss & Hilgenfeld, 1997 ▶; Collaborative Computational Project, Number 4, 1994 ▶). The ‘tails’ of the wavelength range were cut off slightly, with the range restricted to 0.7–6.5 Å from the original 0.6–6.5 Å wavelength distribution of the thermal neutrons, in order to remove the least accurately measured reflections. The overall completeness was 76% to 1.8 Å, with an R sym of ∼22% and a redundancy of 2.5. The multiplicity value was as expected for the low-symmetry space group P21 (Table 1 ▶).
Figure 2.
Neutron Laue diffraction pattern for the HbA crystal in three different crystal settings. In each crystal setting the three-dimensional diffraction data were projected in time-of-flight to produce a conventional two-dimensional Laue pattern.
Table 1. Neutron diffraction data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Source | PCS, LANSCE, Los Alamos National Laboratory |
| Settings | 36 |
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 63.8, b = 84.5, c = 54.4, β = 99.3 |
| Unit-cell volume (Å3) | V = 289420 |
| Resolution | 44.6–1.80 (1.90–1.80) |
| No. of reflections (measured/unique) | 100470/39926 |
| Redundancy | 2.5 (1.6) |
| Completeness (%) | 76.1 (59.0) |
| Rmerge | 0.219 (0.324) |
| Wavelength range (Å) | 0.7–6.5 |
| Mean (I)/sd | 3.9 (1.5) |
Refinement of the structure is in progress utilizing both SHELX (Sheldrick, 2008 ▶) and a version of CNS (Brünger et al., 1998 ▶) modified for neutron refinement (nCNS; Mustyakimov & Langan, 2007 ▶). A room-temperature X-ray diffraction data set is scheduled to be collected in order to enable us to perform joint X-ray/neutron refinement of the HbA structure using nCNS. Initial neutron density maps calculated after only rigid-body refinement show the clear features usually observed for neutron maps. The lysine ND3 and protonated (deuterated) His Nδ1 are visible as very strong nuclear density. Negative nuclear density for H atoms is observed around aliphatic groups of the protein residues owing to the strong incoherent scattering of H atoms. Further structural features will be elucidated during the course of the refinement and reported elsewhere.
4. Discussion
Although the d-xylose isomerase tetramer (Katz et al., 2006 ▶), with a molecular weight of 172 kDa, still represents the largest protein to be studied to high resolution (1.8 Å) using neutron crystallography, the high symmetry of its I222 space group leads to an asymmetric unit volume of only ∼120 000 Å3 (Blum et al., 2007 ▶). HbA, with a molecular weight of 64.6 kDa but with a lower symmetry P21 space group, has an asymmetric unit volume of ∼145 000 Å3, the largest yet for a neutron study of a protein to high resolution (Table 2 ▶). This achievement is to a large extent the consequence of careful crystallization and crystal mounting, which provided crystals of exceptional size and diffraction quality that were stable in the deoxy form over several weeks of data collection.
Table 2. Macromolecular structures determined by neutron diffraction and submitted to the PDB.
| Unit-cell parameters | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PDB code | α (°) | β (°) | γ (°) | a (Å) | b (Å) | c (Å) | Vasym† (Å3) | MWasym† (kDa) | Space group | Biomolecule |
| Proteins | ||||||||||
| 1lzn | 88.8 | 108.8 | 111.6 | 27.28 | 32.04 | 34.27 | 26200 | 14.3 | P1 | Lysozyme |
| 5rsa | 90 | 105.85 | 90 | 30.18 | 38.4 | 53.32 | 29700 | 13.7 | P21 | Ribonuclease A |
| 6rsa | 90 | 106.4 | 90 | 30.3 | 38.35 | 53.7 | 29900 | 13.7 | P21 | Ribonuclease A–uridine vanadate complex |
| 1cq2 | 90 | 105.82 | 90 | 64.53 | 30.91 | 34.78 | 33400 | 17.8 | P21 | Myoglobin (perdeuterated) |
| 1l2k | 90 | 105.7 | 90 | 64.53 | 30.87 | 34.87 | 33400 | 17.8 | P21 | Met-myoglobin |
| 2mb5 | 90 | 105.78 | 90 | 64.61 | 30.95 | 34.86 | 33500 | 17.8 | P21 | Carbonmonoxymyoglobin |
| 1gkt | 90 | 97 | 90 | 43.1 | 75.7 | 42.9 | 69500 | 34 | P21 | Endothiapepsin |
| This study | 90 | 99.3 | 90 | 63.8 | 84.5 | 54.4 | 144700 | 64.6 | P21 | Human deoxyhemoglobin |
| 1iu6 | 90 | 90 | 90 | 34.48 | 35.7 | 43.16 | 13300 | 5.9 | P212121 | Rubredoxin mutant (W3Y,I23V,L32I) |
| 1vcx | 90 | 90 | 90 | 34.32 | 35.31 | 44.23 | 13400 | 5.9 | P212121 | Rubredoxin wild type |
| 1wq2 | 90 | 90 | 90 | 60.5 | 65.1 | 46.5 | 45800 | 8.8 | P212121 | DsrD |
| 1ntp | 90 | 90 | 90 | 54.84 | 58.61 | 67.47 | 54200 | 23.3 | P212121 | Trypsin |
| 1io5 | 90 | 90 | 90 | 80.8 | 80.8 | 37.1 | 60500 | 14.3 | P212121 | Lysozyme |
| 1xqn | 90 | 90 | 90 | 89.16 | 86.13 | 61.59 | 59100 | 25.6 | I222 | Concanavalin A (15 K, saccharide-free) |
| 1c57 | 90 | 90 | 90 | 89.11 | 87.58 | 63.26 | 61700 | 25.6 | I222 | Concanavalin A |
| 2gve | 90 | 90 | 90 | 93.88 | 99.68 | 102.9 | 120400 | 43.3 | I222 | D-Xylose isomerase |
| 2inq | 90 | 90 | 120 | 93.12 | 93.12 | 73.87 | 92500 | 18 | P61 | DHFR |
| 3ins | 90 | 90 | 120 | 82.5 | 82.5 | 34 | 22300 | 2.4 | H3 | Insulin |
| Nucleic acids | ||||||||||
| 1v9g | 90 | 90 | 90 | 18.46 | 30.76 | 43.18 | 6130 | 1.8 | P212121 | Z-DNA CGCGCG |
| 1wqz | 90 | 90 | 120 | 32.9 | 32.9 | 96.1 | 15000 | 3.0 | P3221 | B-DNA d(CCATTAATGG)2 |
V asym and MWasym correspond to the volume and macromolecule size in the asymmetric unit. The biological unit for each macromolecule may be larger owing to the presence of multimeric forms.
The current neutron structure is of paramount importance for our understanding of how protons influence oxygen uptake and release by HbA. Furthermore, the success in obtaining and maintaining the deoxy form of HbA in the crystals at room temperature is a great achievement.
Acknowledgments
This work was supported by the Inter-University Research Program on Pulsed-neutron Scattering at Oversea Facilities and, in part, by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No. 17053011 to YM and No. 18790030 to TC), the REIMEI Research Resources of Japan Atomic Energy Research Institute (to YM) and Hyogo Science and Technology (to YM). The PCS is funded by the Office of Biological and Environmental Research of the Department of Energy. MM and PL were partly supported by an NIH–NIGMS-funded consortium (1R01GM071939-01) between LANL and LNBL to develop computational tools for neutron protein crystallography. AYK and PL were partly supported by a LANL LDRD grant (20070131ER).
References
- Bennett, B., Langan, P., Coates, L., Mustyakimov, M., Schoenborn, B., Howell, E. E. & Dealwis, C. (2006). Proc. Natl Acad. Sci. USA, 103, 18493–18498. [DOI] [PMC free article] [PubMed]
- Blakeley, M., Hazemann, I., Myles, D. A. A. & Podjarny, A. D. (2005). Hydrogen and Hydration-Sensitive Structural Biology, edited by N. Niimura, H. Mizuno, J. R. Helliwell & E. Westhof, pp. 87–102. Tokyo: KubaPro.
- Blum, M.-M., Koglin, A., Rüterjans, H., Schoenborn, B., Langan, P. & Chen, J. C.-H. (2007). Acta Cryst. F63, 42–45. [DOI] [PMC free article] [PubMed]
- Borah, B., Chen, C. W., Egan, W., Miller, M., Wlodawer, A. & Cohen, J. S. (1985). Biochemistry, 24, 2058–2067. [DOI] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Chatake, T., Shibayama, N., Park, S.-H. Y., Kurihara, K., Tamada, T., Tanaka, I., Niimura, N., Kuroki, R. & Morimoto, Y. (2007). J. Am. Chem. Soc.129, 14840–14841. [DOI] [PubMed]
- Cheng, X. & Schoenborn, B. P. (1990). Acta Cryst. B46, 195–208.
- Coates, L., Erskine, P. T., Wood, S. P., Myles, D. A. A. & Cooper, J. B. (2001). Biochemistry, 40, 13149–13157. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol.4, 269–275. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Gelin, B. R., Lee, A. W. & Karplus, M. (1983). J. Mol. Biol.25, 489–559. [DOI] [PubMed]
- Helliwell, J. R., Habash, J., Cruickshank, D. W. J., Harding, M. M., Greenhough, T. J., Campbell, J. W., Clifton, I. J., Elder, M., Machin, P. A., Papiz, M. Z. & Zurek, S. (1989). J. Appl. Cryst.22, 483-497.
- Imai, K. (1982). Allosteric Effects in Haemoglobin. Cambridge University Press.
- Ishikawa, T., Chatake, T., Ohnishi, Y., Tanaka, I., Kurihara, K., Kuroki, R. & Niimura, N. (2008). In the press.
- Katz, A. K., Li, X., Carrell, H. L., Hanson, B. L., Langan, P., Coates, L., Schoenborn, B. P., Glusker, J. P. & Bunick, G. J. (2006). Proc. Natl Acad. Sci. USA, 103, 8342–8347. [DOI] [PMC free article] [PubMed]
- Kossiakoff, A. A. & Spencer, S. A. (1980). Nature (London), 288, 414–416. [DOI] [PubMed]
- Langan, P. & Greene, G. (2004). J. Appl. Cryst.37, 253–257.
- Langan, P., Greene, G. & Schoenborn, B. P. (2004). J. Appl. Cryst.37, 24–31.
- Mason, S. A., Bentley, G. A. & McIntyre, G. J. (1984). Basic Life Sci.27, 323–334. [DOI] [PubMed]
- Monod, J., Wyman, J. & Changeux, J. P. (1965). J. Mol. Biol.12, 88–118. [DOI] [PubMed]
- Mustyakimov, M. & Langan, P. (2007). nCNS: An Open Source Distribution Patch for CNS for Macromolecular Structure Refinement. http://mnc.lanl.gov.
- Pflugrath, J. W. (1999). Acta Cryst. D55, 1718–1725. [DOI] [PubMed]
- Park, S.-H. Y., Yokoyama, T., Shibayama, N., Shiro, Y. & Tame, J. R. H. (2006). J. Mol. Biol.360, 690–701. [DOI] [PubMed]
- Perutz, M. F. (1968). J. Cryst. Growth, 2, 54–56.
- Perutz, M. F., Wilkinson, A. J., Paoli, M. & Dodson, G. G. (1998). Annu. Rev. Biophys. Biomol. Struct.27, 1–34. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shibayama, N., Imai, K., Hirata, H., Hiraiwa, H., Morimoto, H. & Saigo, S. (1991). Biochemistry, 30, 8158–8165. [DOI] [PubMed]
- Sukumar, N., Langan, P., Mathews, F. S., Jones, L. H., Thiyagarajan, P., Schoenborn, B. P. & Davidson, V. L. (2005). Acta Cryst. D61, 640–642. [DOI] [PubMed]
- Tanaka, I., Kurihara, K., Chatake, T. & Niimura, N. (2002). J. Appl. Cryst.35, 34–40.
- Tuan, H.-F., Erskine, P., Langan, P., Cooper, J. & Coates, L. (2007). Acta Cryst. F63, 1080–1083. [DOI] [PMC free article] [PubMed]
- Weiss, M. S. (2001). J. Appl. Cryst.34, 130–135.
- Weiss, M. S. & Hilgenfeld, R. (1997). J. Appl. Cryst.30, 203–205.