Crystallization and X-ray data collection of the C-terminus of gp36 from bacteriophage ϕKMV (KMV36C) are reported.
Keywords: phage infection, endolysin, T4 lysozyme, ϕKMV
Abstract
The C-terminus of gp36 of bacteriophage ϕKMV (KMV36C) functions as a particle-associated muramidase, presumably as part of the injection needle of the ϕKMV genome during infection. Crystals of KMV36C were obtained by hanging-drop vapour diffusion and diffracted to a resolution of 1.6 Å. The crystals belong to the cubic space group P432, with unit-cell parameters a = b = c = 102.52 Å. KMV36C shows 30% sequence identity to T4 lysozyme (PDB code 1l56).
1. Introduction
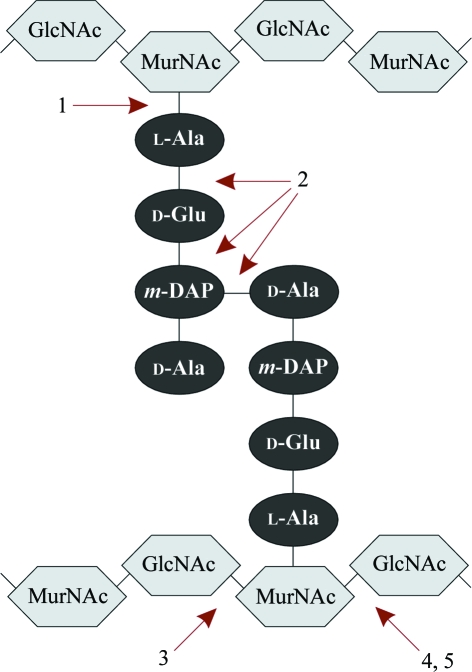
Bacteriophages are viruses with bacterial host cells. After infection, they multiply in the cytosol and this is followed by release of the newly formed progeny virions. In their lytic replication cycle, bacteriophages have to degrade the bacterial cell wall twice, i.e. during initial infection and upon lysis induction of the host cell. The major barrier to overcome is the peptidoglycan (PDG), a rigid layer within the cell wall that is responsible for cell shape and integrity. Most dsDNA bacteriophages encode specific PDG lytic enzymes (lysins) to facilitate infection (as part of the mature phage particle) and to accomplish lysis through a holin–endolysin system (Young et al., 2000 ▶). These PDG lysins are classified according to the bond that is hydrolyzed (Fig. 1 ▶).
Figure 1.
Biochemical classification of PDG lysins: 1, amidases; 2, endopeptidases; 3, glucosaminidases; 4, lysozymes (or muramidases); 5, transglycosylases. The latter hydrolyze the β-1,4-glycosidic bond to form a 1,6-anhydro bond. GlcNAc, N-acetylglucosamine; MurNAc, N-acetylmuramic acid; m-DAP, meso-diaminopimelic acid.
The ϕKMV-like phages, with ϕKMV as the hallmark representative of the genus, belong to the Podoviridae and infect Pseudomonas aeruginosa, an important opportunistic Gram-negative pathogen that is resistant to many commonly used antibiotics (Lavigne et al., 2003 ▶; Ceyssens et al., 2006 ▶).
In silico analysis of ϕKMV and mass spectrometry of phage particles revealed that the C-terminus (amino acids 737–898) of gp36 (KMV36C) represents a muramidase which is presumably part of the injection needle of ϕKMV during infection (Lavigne et al., 2003 ▶, 2006 ▶). KMV36C shows sequence identity to T4 lysozyme (gp e) and is apparently highly thermoresistant (Lavigne et al., 2004 ▶). The remarkable thermoresistance of KMV36C might be explained by oligomerization during the unfolding of the protein, which protects KMV36C from further irreversible denaturation. Furthermore, KMV36C is able to refold while conserving its (partially) intact structure (Briers et al., 2006 ▶). All this is in agreement with the carpenter’s rule model, a hypothesis proposed for the functional analogue gp16 of bacteriophage T7, i.e. gp16 migrates through the head–tail connection and the tail in an unfolded conformation and refolds within the cell wall, thereby forming a channel for DNA injection (Molineux, 2001 ▶). The in vivo existence of a functional unfolded conformation of KMV36C is in agreement with the strong thermoresistance to irreversible denaturation.
Careful analysis of the crystal structure of KMV36C should allow a search for the determinants responsible for its exceptional thermoresistance. Furthermore, a start could be made in refining a model system for the infection mechanism of the Podoviridae in general and the new ϕKMV-like genus in particular.
2. Materials and methods
2.1. Protein expression and purification
KMV36C was recombinantly expressed in Escherichia coli with a C-terminal E-tag and 6×His tag as described previously (Lavigne et al., 2004 ▶). The E-tag is an epitope that is recognized by the commercially available anti-E-tag antibody (Amersham Biosciences, Uppsala, Sweden). The cell pellet from a 1.5 l expression culture was resuspended in 60 ml lysis buffer (20 mM NaH2PO4, 0.5 M NaCl, 10 mM imidazole pH 7.4). After three freeze–thaw cycles, the lysate was clarified by sonication, centrifugation (3313g, 30 min, 277 K) and filtration (0.45 and 0.22 µm pore size). The protein was purified using a two-step chromatography protocol. The filtrate was applied onto a 1 ml Ni–NTA HisTrap column (GE Healthcare, Buckinghamshire, England). Metal-affinity purification was performed according to the manufacturer’s instructions using 50 mM imidazole in the equilibration and wash buffer. The protocol was run as three loops of 20 ml filtrate in order to prevent column saturation. The volume of the pooled fractions was reduced to 8 ml using a centrifugal filter device (Centriplus YM-30, Amicon, Millipore, Massachusetts, USA) and loaded onto a HiPrep 26/60 Sephacryl S-100 gel-filtration column (GE Healthcare, Buckinghamshire, England). The elution buffer was 0.05 M NaH2PO4, 0.15 M NaCl pH 7.2 with a flow rate of 1 ml min−1. 2 ml fractions of purified recombinant protein were pooled up to 38 ml (1.024 mg ml−1). The molecular weight of the recombinant KMV36C was 20 884 Da as determined by SELDI–TOF analysis.
MALDI–TOF mass spectrometry was used to control the purity of the protein sample prior to the crystallization process. The MALDI–TOF MS was performed on an ABI-4800 (Applied Biosystems) proteomic analyzer operating in linear mode. Sinapinic acid (5 mg ml−1) was used as the matrix.
2.2. Crystallization
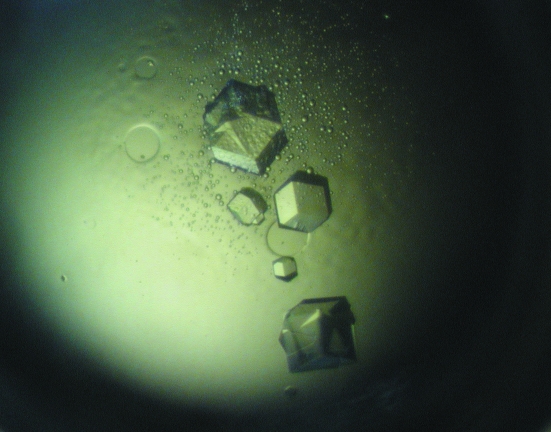
The protein was concentrated to approximately 20 mg ml−1 using a centrifugal filter device (Centriplus YM-30, Amicon, Millipore, Massachusetts, USA) and buffered in 0.05 M NaH2PO4–NaOH, 0.15 M NaCl pH 7.2. The concentration was estimated spectrophotometrically and was considered to be an optimal value for the initial crystallization screens as determined using PCT from Hampton Research (Watson & O’Callaghan, 2005 ▶). Crystallization trials were performed in Linbro plates (Hampton Research, Aliso Viejo, USA) using the hanging-drop vapour-diffusion method at 289 K. Crystal Screens I and II from Hampton Research (Aliso Viejo, USA) and Structure Screens 1 and 2 from MDL (Molecular Dimensions Ltd, Suffolk, England) were tested as precipitants. Crystals suitable for X-ray diffraction analysis appeared after approximately one week in condition No. 26 and No. 32 of Crystal Screen II [0.1 M sodium chloride, 0.1 M HEPES pH 7.5, 1.6 M ammonium sulfate and 0.2 M ammonium sulfate, 0.1 M MES pH 6.5, 30%(w/v) PEG monomethylether 5000, respectively] as well as in the same conditions from Structure Screen 2 (No. 14 and No. 26). Therefore, 1.5 µl protein solution was mixed with 1.5 µl screening solution using a 0.5 ml reservoir of the same screening solution. The quality of the crystals obtained using Crystal Screen II (Fig. 2 ▶) was tested on a Bruker Smart 6000 CCD system with cryocooling, showing the necessity of using synchrotron radiation. Notably, the diffraction pattern of crystals obtained using condition No. 32 showed ice-ring formation, which could be removed by using 30%(w/v) PEG 6000 as a cryoprotectant. Crystals obtained using condition No. 26 showed no ice-ring formation, but were of poorer diffraction quality.
Figure 2.
Typical crystals of KMV36C obtained by hanging-drop vapour diffusion using condition No. 32 from Crystal Screen II. The largest crystal at the top of the picture is approximately 0.3 mm in length.
2.3. Data collection and processing
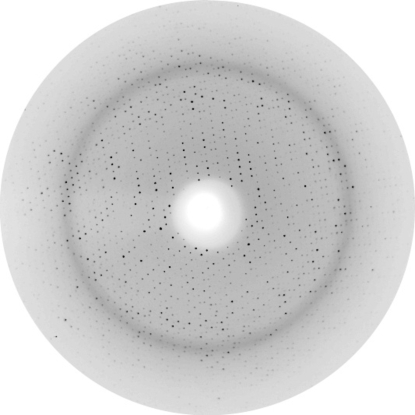
A single crystal with dimensions of 0.3 × 0.3 × 0.2 mm (condition No. 32) was used to collect a 96.7% complete data set at EMBL beamline BW7b of the DESY synchrotron in Hamburg. Data were collected on a MAR345 imaging-plate detector with λ = 0.8423 Å, a 190° ϕ range, a 1° increment and a crystal-to-detector distance of 220 mm. The crystal was cryoprotected by quickly washing the crystal in a 30%(w/v) PEG 6000 solution and was immediately flash-cooled in a liquid-nitrogen cryostream at 100 K. Overloaded reflection data were collected using the same crystal with a crystal-to-detector distance of 310 mm. A total of 24 126 unique reflections were observed in the resolution range 34.2–1.6 Å (R merge = 0.107). Although the resolution edge of the diffraction pattern was 1.43 Å (Fig. 3 ▶), careful analysis showed that the data in the outermost shells were of much lower quality; therefore, the resolution cutoff was chosen at 1.6 Å. Data were processed with MOSFLM v.7.0.0 (Leslie, 1992 ▶) and scaled using SCALA v.3.2.25 (Evans, 1997 ▶). The latter was used as part of the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶). Data-collection statistics for KMV36C are given in Table 1 ▶.
Figure 3.
1° oscillation image of a crystal of KMV36C taken on a MAR Research image plate on beamline BW7b (EMBL, Hamburg). The resolution edge of the image is 1.43 Å.
Table 1. Data-collection statistics for KMV36C.
Values in parentheses are for the outermost resolution shell.
| Resolution range (Å) | 34.20–1.60 (1.69–1.60) |
| Wavelength (Å) | 0.8423 |
| Space group | P432 |
| Unit-cell parameters (Å, °) | a = b = c = 102.52, α = β = γ = 90 |
| No. of reflections (full and summed partials) | 1321888 (135314) |
| No. of unique reflections | 24126 (3553) |
| Mosaicity (°) | 0.25 |
| Completeness (%) | 96.7 (100.0) |
| Rmerge (%) | 10.7 (63.0) |
| Mean I/σ(I) | 40.6 (7.5) |
| Multiplicity | 54.8 (38.1) |
| Solvent content (%) | 42.9 |
3. Results and discussion
The crystals most likely belong to the primitive cubic space group P432, with unit-cell parameters a = b = c = 102.52 Å, which was confirmed using POINTLESS v.1.2.0 (Collaborative Computational Project, Number 4, 1994 ▶). The Matthews coefficient (V M) is 2.15 Å3 Da−1 for one protein molecule in the asymmetric unit, resulting in a solvent fraction of 42.9%. When performing a BLASTP search (Altschul et al., 2005 ▶), 30% sequence identity (47% positives) and an E value of 1 × 10−10 were found for T4 lysozyme with PDB code 1l56 (Nicholson & Matthews, 1992 ▶).
Structure determination by molecular replacement (MR) is currently in progress using T4 lysozyme structures as MR models. However, as many attempts to date have failed to solve the structure by MR and the unit-cell and space-group assignments have already indicated a possibly significantly different structure, as an alternative MAD/SAD phasing will be performed using the anomalous signal from selenomethionine variants, which are presently in preparation. A total of four methionine residues are present in the 162-amino-acid sequence of KMV36C.
Acknowledgments
The authors thank the staff of the EMBL Hamburg Outstation for their support during the synchrotron experiments. YB holds a predoctoral fellowship from the ‘Instituut voor aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen’ (IWT, Belgium). RL holds a postdoctoral fellowship from the ‘Fonds voor Wetenschappelijk Onderzoek-Vlaanderen’ (FWO-Vlaanderen, Belgium). Support from the Flemish FWO (research grant G.0308.05) and from the European Community Research Infrastructure Action under the FP6 ‘Structuring the European Research Area Programme’, contract No. RII3-CT-2004-506008, are gratefully acknowledged. BioMacS, the K. U. Leuven Interfacultary Centre for Biomolecular Structure, is supported by the Impulse Project of the K. U. Leuven.
References
- Altschul, S. F., Wootton, J. C., Gertz, E. M, Agarwala, R., Morgulis, A., Schäffer, A. A. & Yu, Y.-K. (2005). FEBS J.272, 5101–5109. [DOI] [PMC free article] [PubMed]
- Briers, Y., Lavigne, R., Plessers, P., Hertveldt, K., Hanssens, I., Engelborghs, Y. & Volckaert, G. (2006). Cell. Mol. Life Sci.63, 1899–1905. [DOI] [PMC free article] [PubMed]
- Ceyssens, P.-J., Lavigne, R., Mattheus, W., Chibeu, A., Hertveldt, K., Mast, J., Robben, J. & Volckaert, G. (2006). J. Bacteriol.188, 6924–6931. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Evans, P. R. (1997). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.33, 22–24.
- Lavigne, R., Briers, Y., Hertveldt, K., Robben, J. & Volckaert, G. (2004). Cell. Mol. Life Sci.61, 2753–2759. [DOI] [PubMed]
- Lavigne, R., Burkal’tseva, M. V., Robben, J., Sykilinda, N. N., Kurochkina, L. P., Grymonprez, B., Jonckx, B., Krylov, V. N., Mesyanzhinov, V. V. & Volckaert, G. (2003). Virology, 312, 49–59. [DOI] [PubMed]
- Lavigne, R., Noben, J.-P., Hertveldt, K., Ceyssens, P.-J., Briers, Y., Dumont, D., Roucourt, B., Krylov, V. N., Mesyanzhinov, V. V., Robben, J. & Volckaert, G. (2006). Microbiology, 152, 529–534. [DOI] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Molineux, I. J. (2001). Mol. Microbiol.40, 1–8. [DOI] [PubMed]
- Nicholson, H. & Matthews, B. W. (1992). Biopolymers, 32, 1431–1441. [DOI] [PubMed]
- Watson, A. A. & O’Callaghan, C. A. (2005). Acta Cryst. F61, 1094–1096. [DOI] [PMC free article] [PubMed]
- Young, R. Y., Wang, I.-N. & Roof, W. D. (2000). Trends Microbiol.8, 120–128. [DOI] [PubMed]