The X-ray crystal structure of the cold-shock domain protein from N. meningitidis reveals a strand-exchanged dimer.
Keywords: cold-shock domain proteins, Neisseria meningitidis, domain-exchanged dimers
Abstract
The structure of the cold-shock domain protein from Neisseria meningitidis has been solved to 2.6 Å resolution and shown to comprise a dimer formed by the exchange of two β-strands between protein monomers. The overall fold of the monomer closely resembles those of other bacterial cold-shock proteins. The neisserial protein behaved as a monomer in solution and was shown to bind to a hexathymidine oligonucleotide with a stoichiometry of 1:1 and a K d of 1.25 µM.
1. Introduction
The human pathogen Neisseria meningitidis is a Gram-negative bacterium that is responsible for bacterial meningitis and septicaemia. Sequencing and annotation of the N. meningitidis serotype B genome (strain MC58) revealed a putative cold-shock gene (NMB0838) predicted to encode a 7.2 kDa acidic protein with high sequence similarity to other bacterial cold-shock proteins (Csps; Tettelin et al., 2000 ▶). Expression of Csps in bacteria is induced under a range of growth conditions, most notably in response to cold stress; they are essential for growth at low temperatures (Thieringer et al., 1998 ▶). Amongst the most well studied Csps is the major cold-induced protein in Escherichia coli, CspA, which functions as an RNA chaperone binding nonspecifically to RNA and ssDNA (Jiang et al., 1997 ▶). At low temperature, RNA forms stable secondary structures and it has been proposed that CspA (and other Csps) enhances translation by eliminating stabilized RNA secondary structures (Jiang et al., 1997 ▶). Csps also appear to function as transcription antiterminators, providing a mechanism for controlling the expression of other cold-shock-induced genes (Bae et al., 2000 ▶).
The structures of a number of Csps have been determined by either X-ray crystallography or NMR, namely E. coli CspA (Ec-CspA; Schindelin et al., 1994 ▶; Newkirk et al., 1994 ▶), Bacillus subtilis CspB (Bs-CspB; Schnuchel et al., 1993 ▶), B. caldolyticus Csp (Bc-Csp; Mueller et al., 2000 ▶) and Thermotoga maritima Csp (Kremer et al., 2001 ▶). All show an OB (oligonucleotide/oligosaccharide-binding) fold consisting of five antiparallel β-strands forming a β-barrel structure. The oligonucleotide-binding region of Csps has been mapped to two conserved sequence motifs, RNP-1 and RNP-2, located on adjacent β-strands (Schroder et al., 1995 ▶). These sequences are characteristic of many RNA-binding proteins (Landsman, 1992 ▶) and comprise a high proportion of basic and aromatic residues. The structures of CspB from Bacillus subtilis and Csp from B. caldolyticus in complex with hexathymidine (dT6) have been reported (Max et al., 2006 ▶, 2007 ▶). Both structures show that oligonucleotide binding involves stacking interactions between phenylalanine residues and the thymidine base, together with hydrogen bonds between the side chains of polar amino acids and pyrimidine bases. The residues involved in the interaction surface of Csps from Bacillus are conserved within the Neisseria NMB0838 gene product, implying that this protein may also bind ssDNA in a similar way to CspB. In this report, we describe the production of the neisserial Csp (referred to as Nm-Csp), analysis of ssDNA binding and structure determination by X-ray crystallography.
2. Materials and methods
2.1. Protein production and crystallization
The NMB0838 gene was amplified by PCR using N. meningitidis genomic DNA (strain MC58) as a template and primers designed for ligation-independent cloning via In-Fusion technology (Clontech). PCR with KOD HiFi polymerase (Merck) used the forward and reverse primers 5′-AAGTTCTGTTTCAGGGCCCGATGGCAACCGGTATCGTAAAATGG-3′ and 5′-ATGGTCTAGAAAGCTTTATTAGGCCTGAATGTTGGCGG-3′, respectively. The PCR product was purified using QIAquick 96 plates (Qiagen) and inserted into the vector pOPINB using the protocol of Berrow et al. (2007 ▶). The expression construct contained an N-terminal His tag and a 3C protease cleavage site (bold): MGSSHHHHHHSSGLEVLFQGP. Recombinant clones were identified by PCR with the gene-specific forward primer and a T7 reverse primer and were verified by DNA sequencing.
Both native and selenomethionine-labelled Nm-Csp were produced in E. coli strain B834 (DE3) using the auto-induction method (Studier, 2005 ▶). Native protein was expressed in Overnight Express Instant TB media (Merck) and selenomethionine-labelled protein was produced in a glucose-free version of the SelenoMet media (Molecular Dimensions) in conjunction with Overnight Express System 1 (Merck). Cells were grown in 2 l cultures at 310 K for 4 h and the temperature was then lowered to 298 K. Following incubation for a further 20 h, the cells were harvested by centrifugation for 15 min at 6000g and lysed using a Basic Z Cell Disruptor (Constant Systems) at 207 MPa in the presence of 50 mM Tris pH 7.5, 500 mM NaCl, 0.2% Tween-20, 10 µg ml−1 DNAse and an EDTA-free protease inhibitor cocktail tablet (Roche). The lysate was centrifuged at 16 000g for 30 min to remove cell debris before loading the soluble fraction onto a 1 ml HisTrap FF column (GE Healthcare). The column was washed with 50 mM Tris pH 7.5, 500 mM NaCl, 20 mM imidazole before elution of the protein with 50 mM Tris pH 7.5, 500 mM NaCl, 500 mM imidazole. The protein was then injected onto a 16/60 HiLoad Superdex 75 column (GE Healthcare) and eluted with 20 mM Tris pH 7.5, 200 mM NaCl. Protein-containing fractions were analysed by SDS–PAGE (NuPage, Invitrogen). The N-terminal tag was removed by incubation overnight at 277 K with His-tagged 3C protease (prepared from a pET24a/His3C expression vector kindly donated by A. Geerlof, EMBL, Hamburg). The 3C protease and any uncleaved protein were removed by nickel-affinity chromatography and the protein was concentrated to 40 mg ml−1 using a 2 kDa molecular-weight cutoff Vivaspin 15R concentrator (Sartorius). The quality of the protein sample and the selenomethionine incorporation were analysed by electrospray mass spectrometry (Geerlof et al., 2006 ▶).
Native protein crystals were obtained in a 200 nl sitting-drop vapour-diffusion crystallization experiment in 0.8 M sodium dihydrogen phosphate, 1.2 M dipotassium hydrogen phosphate, 100 mM acetate pH 4.5 with a final pH of 6.7 (condition No. 35 of Emerald Wizard II Screen; Emerald Biosystems; Walter et al., 2005 ▶). Selenomethionine-labelled protein crystals were obtained by cross-seeding with native crystals using the same crystallization condition according to the protocol of Walter et al. (2008 ▶).
2.2. Data collection, structure determination and refinement
Multiple-wavelength anomalous dispersion data were collected to 2.6 Å resolution from a selenomethionine-labelled Nm-Csp crystal on beamline BM14 at the ESRF (Grenoble, France). Following a fluorescence scan, 360° of X-ray data at the peak wavelength (0.979 Å) and 180° at a remote wavelength (0.907 Å) were collected with an oscillation of 1.0° per frame. Crystals taken directly from the crystallization drop without the addition of a cryoprotectant were flash-frozen and maintained at 100 K under a stream of nitrogen gas during data collection. Indexing and integration of data images were carried out using HKL-2000. The statistics of the X-ray data are given in Table 1 ▶.
Table 1. X-ray data-collection and refinement statistics.
(a).
Data-collection details. Values in parentheses are for the highest resolution shell.
| Data set | Peak | Remote |
|---|---|---|
| X-ray source | ESRF BM14 | |
| Wavelength (Å) | 0.979 | 0.907 |
| Space group | P41212 | |
| Unit-cell parameters (Å) | a = b = 59.37, c = 89.42 | |
| Resolution range (Å) | 30.0–2.60 (2.69–2.60) | 30.0–2.80 (2.90–2.80) |
| Unique reflections | 5271 (459) | 4299 (412) |
| Completeness (%) | 98.5 (88.8) | 99.7 (98.8) |
| Redundancy | 23.9 (13.1) | 12.7 (10.0) |
| Average I/σ(I) | 29.2 (1.7) | 17.9 (1.3) |
| Rmerge | 0.127 (0.834) | 0.137 (0.957) |
(b).
Refinement statistics.
| Resolution range (Å) | 30.0–2.60 |
| No. of reflections (working/test) | 4951/270 |
| R factor (Rwork/Rfree) | 0.233/0.296 |
| No. of atoms (protein/water) | 1014/16 |
| R.m.s. bond-length deviation (Å) | 0.008 |
| R.m.s. bond-angle deviation (°) | 1.0 |
| Mean B factor (protein/water) (Å2) | 72/63 |
The Nm-Csp crystal belongs to space group P41212, with unit-cell parameters a = b = 59.37, c = 89.42 Å. There are two monomers in the crystal asymmetric unit, with a solvent content of 53%. The SHELX program suite was used to evaluate the anomalous signal during the course of the MAD data collection and to solve the positions of the selenium sites. SOLVE/RESOLVE (Terwilliger, 2000 ▶; Terwilliger & Berendzen, 1999 ▶) were then used for refinement of the selenium positions and phase improvement combined with automated model building. The model partially built by RESOLVE was rebuilt manually using the program O (Jones et al., 1991 ▶) and then refined with CNS (Brünger et al., 1998 ▶) using simulated annealing and positional refinement with main-chain NCS restraints followed by individual isotropic B-factor refinement. There are apparent structural differences in the N-terminal part (residues 1–27) between the two monomers which were not included in the NCS restraints. The structure was finally refined with REFMAC and the statistics for the structure are given in Table 1 ▶.
2.3. Analytical size-exclusion chromatography
200 µl of 1 mg ml−1 selenomethionine-labelled protein was loaded onto a Superdex 75 10/300 GL column (GE Healthcare) equilibrated in 20 mM Tris pH 7.5, 200 mM NaCl. Elution of the protein in 20 mM Tris pH 7.5, 200 mM NaCl was detected by UV absorbance (A 280 and A 254). A further sample of protein was incubated with the single-stranded DNA fragment dT6 in a 1:1 molar ratio at room temperature for 30 min before analysis by analytical size-exclusion chromatography as described above. The column was calibrated using a Gel Filtration LMW Calibration Kit (GE Healthcare).
2.4. Fluorescence titration DNA-binding assay
The fluorescence of Trp8 in Nm-Csp was measured using a Shimadzu RF-1501 spectrofluorophotometer. Experiments were carried out at room temperature in 20 mM Tris pH 7.5, 200 mM NaCl. 0.25 µM protein solution was titrated with increasing amounts of dT6 from 0 to 20 µM. The samples were gently stirred and after 2 min incubation the fluorescence of Trp8 was excited at 280 nm and measured at 343 nm. The initial volume was 600 µl and the increase in volume after the addition of concentrated DNA solution was 2% or less. The fluorescence was corrected for buffer fluorescence, filter effects and DNA autofluorescence. The effect of random collisions to the fluorescence reading was eliminated by repeating the titration with 40 µM N-acetyltryptophanamide (data not shown). The fluorescence-quenching data revealed a binding isotherm from which K d, the dissociation constant of the complex, and n, the stoichiometry of the protein–DNA complex (the number of protein molecules bound to one ssDNA molecule), were calculated using the nonlinear regression software package Prism 5 (GraphPad Software Inc.).
3. Results and discussion
3.1. Overall structure
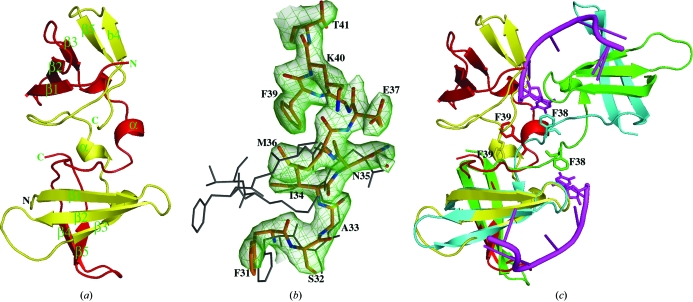
The X-ray crystal structure of the cold-shock domain protein from N. meningitidis (Nm-Csp) was determined by multiple-wavelength anomalous dispersion using selenomethionine-substituted protein. Despite high sequence homology to Ec-CspA and Bs-CspB, solution by molecular replacement did not prove to be possible owing to the novel arrangement of the protein described below. The structure reveals that two polypeptide chains from Nm-Csp form a dimer in the crystallographic asymmetric unit consisting of two five-stranded β-barrels (Fig. 1 ▶ a). Superimpositions of the monomers of Nm-Csp with those of Ec-CspA (PDB code 1mjc) and Bs-CspB (PDB code 2es2) gave r.m.s.d.s of 1.1 and 1.2 Å for 66 and 63 equivalent Cα atoms out of 67, respectively, confirming the high degree of structural conservation of the Csp fold. However, in both Ec-CspA and Bs-CspB each chain is folded into an independent three-dimensional biological unit, whereas in Nm-Csp each monomer is composed of residues from two chains: the N-terminal residues 1–36 are from one chain and the C-terminal residues 37–67 are from the other (Figs. 1 ▶ a and 1 ▶ b). At the point of transition, residues Met36, Glu37 and Gly38 of each subunit form a single α-helical turn. This results in the two phenylalanine residues at position 39 of the subunits being buried in the core of the dimer, forming a potential ring-stacking interaction (Fig. 1 ▶ b). This contrasts with the monomeric structures of Ec-CspA and Bs-CspB, in which the side chains of the equivalent residues are exposed at the surface of the proteins. Domain swapping has recently been observed in the structure of Csp from B. caldolyticus (Bc-Csp) in complex with a hexathymidine ligand (Max et al., 2007 ▶). The point of exchange was the same as in Nm-Csp, but the relative orientation and separation of the two monomers in the Bc-Csp structure is very different. As shown in Fig. 1 ▶(c), only one of the two domain-swapped monomers can be overlaid. This appears to be entirely a consequence of the methionine at position 36 in Nm-Csp, which favours the formation of the short helical turn. In contrast, the glycine at this position in Bc-Csp results in a more extended hinge-loop region between the monomers and the exposure of the equivalent of Phe39 at the surface of the protein as observed in Ec-CspA and Bs-CspB. Interestingly, as in the Bs-CspB–ssDNA complex structure, this phenylalanine residue is involved in the interaction between the protein and the oligonucleotide ligand d(T)6 (Max et al., 2007 ▶, 2006 ▶). The residues that are involved in the binding of oligonucleotides in Bs-CspB and Bc-Csp (Max et al., 2006 ▶, 2007 ▶) are highly conserved in Nm-Csp (Fig. 2 ▶), implying that the protein would bind to ssDNA oligonucleotides in the same way as the Bacillus Csps. However, the fact that one of the hydrophobic residues (Phe39) that form the binding surface is buried in the Nm-Csp structure suggests that binding to the domain-swapped dimer may be compromised. Domain swapping has been noted in a variety of different proteins, with the swapped domains showing diverse secondary structures (Liu & Eisenberg, 2002 ▶; Bennett et al., 1995 ▶). The functional significance of these phenomena depends upon the protein and in some cases arises as an artifact in vitro. However, it has been noted that the formation of secondary structure in the hinge region involved in domain exchange indicates that the dimer is relatively more stable than the monomeric form of the protein (Liu & Eisenberg, 2002 ▶).
Figure 1.
The structure of Nm-Csp and comparison with Bc-Csp. (a) Ribbon diagram showing the Nm-Csp dimer in the crystallographic asymmetric unit; the two polypeptide chains are coloured yellow and red and secondary-structure features are numbered. (b) |2F o| − |F c| map contoured at 1.2σ showing electron density for residues 31–41 where chain switching occurs between monomers of Nm-Csp. The Nm-Csp residues, which are labelled and shown in stick format, are superimposed onto the equivalent region of Bs-CspB (PDB code 2es2) shown as black lines. (c) Overlay of the Nm-Csp dimer, coloured red and yellow, with that of Bc-Csp (PDB code 2hax), coloured green and turquoise, bound to oligo-dT (coloured purple). The positions of the Phe38 residues involved in nucleotide base stacking in Bc-Csp and the corresponding Phe39 residues in Nm-Csp are shown.
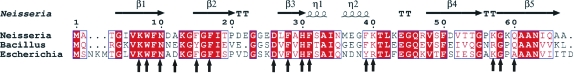
Figure 2.
Structure-based alignment of cold-shock proteins (Csps). The sequences of N. meningitis Csp (Nm-Csp), E. coli CspA (Ec-CspA) and B. caldolyticus Csp (Bc-Csp) were aligned using ClustalW and displayed with secondary structures using ESPript2.2. The key residues involved in the DNA binding of Bacillus Csps are indicated by arrows.
3.2. DNA binding
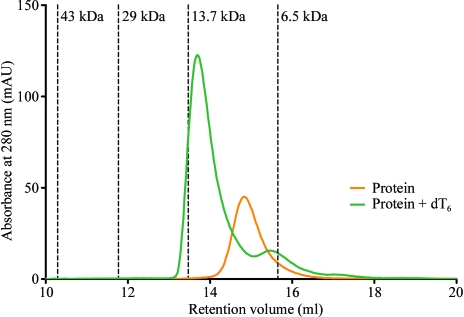
Analytical size-exclusion chromatography (SEC) and fluorescence titration experiments were used to investigate the DNA-binding properties of Nm-Csp. The oligonucleotide dT6 was chosen as Max and coworkers have reported the structures of Bs-CspB and Bc-Csp with this single-stranded DNA incorporated into the DNA-binding site (Max et al., 2006 ▶, 2007 ▶). During analytical size-exclusion chromatography, purified Nm-Csp eluted as an 8 kDa monomeric protein (Fig. 3 ▶), in contrast to the crystallized protein, which is dimeric. Following incubation of the protein with hexathymidine (dT6) in a 1:1 molar ratio, the elution peak shifted to around 13 kDa (Fig. 3 ▶), indicating the formation of a complex between Nm-Csp and the single-stranded DNA oligomer. The ratio A 254/A 280 in the eluted peak increased from 2.94 to 9.91, which is consistent with the presence of DNA in the sample after complex formation. The 5 kDa shift of the elution peak is larger than would be expected for a globular complex comprising one Nm-Csp monomer (8 kDa) and one oligomer of dT6 (corresponding to a molecular weight of 1.4 kDa). However, oligonucleotides behave on gel-filtration columns as though four to five times larger than globular proteins of corresponding molecular weight (Kim et al., 2007 ▶). Therefore, we attribute the discrepancy in the size of the complex to the presence of the oligonucleotide and conclude that Nm-Csp forms a monomeric complex with ssDNA. Further experiments, for example using analytical ultracentrifugation, would be required to confirm this result.
Figure 3.
Analysis of Nm-Csp by size-exclusion chromatography with and without single-stranded DNA (107 µg). Overlaid size-exclusion chromatograms (each using 200 µg of protein) showing the monomeric Nm-Csp (orange trace) eluting at 14.8 ml and the Nm-Csp–dT6 heterodimer eluting at 13.6 ml (green trace). The elution volumes of the calibrants along with their molecular weights are represented by vertical dotted lines. The A 254/A 280 ratios for the eluted peaks were 2.94 for the protein alone and 9.91 for the protein plus oligonucleotide.
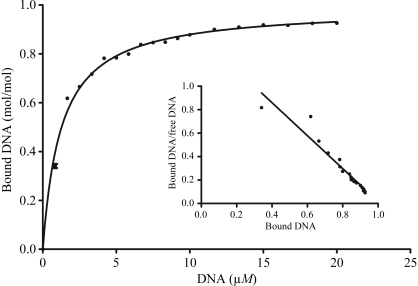
The stoichiometry n and the dissociation constant K d of the protein–oligonucleotide complex were obtained using a fluorescence titration experiment which measures the fluorescence quenching of Trp8 upon DNA binding (Max et al., 2006 ▶; Zeeb & Balbach, 2003 ▶). Trp8 forms part of the RNP1 nucleic acid binding motif (Landsman, 1992 ▶; Schroder et al., 1995 ▶). The number of protein molecules bound to each dT6 molecule, n, was calculated to be 0.99 ± 0.01, indicating 1:1 binding of the DNA to the protein (Fig. 4 ▶). The K d for the protein–oligonucleotide complex was calculated to be 1.25 ± 0.04 µM (Fig. 4 ▶). This compares with a K d of 0.32 ± 0.02 µM for the wild-type Bs-CspB–dT6 complex (Zeeb et al., 2006 ▶), indicating that Nm-Csp binds dT6 less strongly than Bs-CspB. All the hydrophobic residues that form the DNA-binding surface in the Bs-CspB complex are conserved in the Nm-Csp sequence; the only differences among the DNA-contact residues are alanine for glutamate at position 12 and lysine for arginine at position 56 (Fig. 2 ▶). It is not clear that these differences would account for the reduced binding.
Figure 4.
Binding isotherm for the interaction of Nm-Csp with single-stranded DNA along with the resulting Scatchard plot (insert). The nonlinear regression fit of the experimental data revealed a K d of 1.25 ± 0.04 µM and revealed the stoichiometry of the protein–DNA complex to be 0.99 ± 0.01.
4. Conclusions
We have described the crystallographic structure of Nm-Csp at 2.6 Å resolution. The structure comprises a dimer with each subunit built of residues from two separate chains; this is in contrast to the Ec-CspA and Bs-CspB structures reported previously (Schindelin et al., 1994 ▶; Schnuchel et al., 1993 ▶), but similar to the structure of Bc-Csp (Max et al., 2007 ▶). This suggests that Csp proteins may have a propensity to undergo domain swapping to form metastable dimeric structures. The overall structure and ssDNA binding confirm that Nm-Csp belongs to the family of bacterial cold-shock proteins. Although the function of the protein remains to be determined, it seems unlikely that oligonucleotide binding occurs in the context of cold adaptation, since N. meningitidis, in contrast to E. coli and Bacillus spp., are not free-living bacteria but are commensal organisms that are resident in the human upper respiratory tract.
Supplementary Material
PDB reference: cold-shock domain protein, 3cam, r3camsf
Acknowledgments
The authors would like to thank Dr Charles Redwood (Department of Cardiovascular Medicine, University of Oxford) for help with the fluorescence titration experiments. We also thank the staff at BM14 and the ESRF for help with data collection. The OPPF is funded by the UK Medical Research Council.
References
- Bae, W., Xia, B., Inouye, M. & Severinov, K. (2000). Proc. Natl Acad. Sci. USA, 97, 7784–7789. [DOI] [PMC free article] [PubMed]
- Bennett, M. J., Schlunegger, M. P. & Eisenberg, D. (1995). Protein Sci.4, 2455–2468. [DOI] [PMC free article] [PubMed]
- Berrow, N. S., Alderton, D., Sainsbury, S., Nettleship, J., Assenberg, R., Rahman, N., Stuart, D. I. & Owens, R. J. (2007). Nucleic Acids Res.35, e45. [DOI] [PMC free article] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Geerlof, A. et al. (2006). Acta Cryst. D62, 1125–1136. [DOI] [PMC free article] [PubMed]
- Jiang, W., Hou, Y. & Inouye, M. (1997). J. Biol. Chem.272, 196–202. [DOI] [PubMed]
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed]
- Kim, I., McKenna, S. A., Viani Puglisi, E. & Puglisi, J. D. (2007). RNA, 13, 289–294. [DOI] [PMC free article] [PubMed]
- Kremer, W., Schuler, B., Harrieder, S., Geyer, M., Gronwald, W., Welker, C., Jaenicke, R. & Kalbitzer, H. R. (2001). Eur. J. Biochem.268, 2527–2539. [DOI] [PubMed]
- Landsman, D. (1992). Nucleic Acids Res.20, 2861–2864. [DOI] [PMC free article] [PubMed]
- Liu, Y. & Eisenberg, D. (2002). Protein Sci.11, 1285–1299. [DOI] [PMC free article] [PubMed]
- Max, K. E., Zeeb, M., Bienert, R., Balbach, J. & Heinemann, U. (2006). J. Mol. Biol.360, 702–714. [DOI] [PubMed]
- Max, K. E., Zeeb, M., Bienert, R., Balbach, J. & Heinemann, U. (2007). FEBS J.274, 1265–1279. [DOI] [PubMed]
- Mueller, U., Perl, D., Schmid, F. X. & Heinemann, U. (2000). J. Mol. Biol.297, 975–988. [DOI] [PubMed]
- Newkirk, K., Feng, W., Jiang, W., Tejero, R., Emerson, S. D., Inouye, M. & Montelione, G. T. (1994). Proc. Natl Acad. Sci. USA, 91, 5114–5118. [DOI] [PMC free article] [PubMed]
- Schindelin, H., Jiang, W., Inouye, M. & Heinemann, U. (1994). Proc. Natl Acad. Sci. USA, 91, 5119–5123. [DOI] [PMC free article] [PubMed]
- Schnuchel, A., Wiltscheck, R., Czisch, M., Herrler, M., Willimsky, G., Graumann, P., Marahiel, M. A. & Holak, T. A. (1993). Nature (London), 364, 169–171. [DOI] [PubMed]
- Schroder, K., Graumann, P., Schnuchel, A., Holak, T. A. & Marahiel, M. A. (1995). Mol. Microbiol.16, 699–708. [DOI] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif.41, 207–234. [DOI] [PubMed]
- Terwilliger, T. C. (2000). Acta Cryst. D56, 965–972. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed]
- Tettelin, H. et al. (2000). Science, 287, 1809–1815. [DOI] [PubMed]
- Thieringer, H. A., Jones, P. G. & Inouye, M. (1998). Bioessays, 20, 49–57. [DOI] [PubMed]
- Walter, T. S. et al. (2005). Acta Cryst. D61, 651–657. [DOI] [PMC free article] [PubMed]
- Walter, T. S., Mancini, E. J., Kadlec, J., Graham, S. C., Assenberg, R., Ren, J., Sainsbury, S., Owens, R. J., Stuart, D. I., Grimes, J. M. & Harlos, K. (2008). Acta Cryst. F64, 14–18. [DOI] [PMC free article] [PubMed]
- Zeeb, M. & Balbach, J. (2003). Protein Sci.12, 112–123. [DOI] [PMC free article] [PubMed]
- Zeeb, M., Max, K. E., Weininger, U., Low, C., Sticht, H. & Balbach, J. (2006). Nucleic Acids Res.34, 4561–4571. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: cold-shock domain protein, 3cam, r3camsf