Abstract
The center of mass of a protein is an artificial point useful for detecting important and simple features of proteins structure, shape and association.
CALCOM is a software which calculates the center of mass of a protein, starting from PDB protein structure files. In the case of protein complexes and of protein-small ligand complexes, the position of protein residues or of ligand atoms respect to each protein subunit can be evaluated, as well as the distance among the center of mass of the protein subunits, in order to compare different conformations and evaluate the relative motion of subunits.
Availability
The service is available at the URL: http://bioinformatica.isa.cnr.it/CALCOM/.
Keywords: center of mass, proteins, protein subunits distance, web-software
Background
The center of mass of a protein was used for defining costraints useful to predict protein tertiary models, to assess the global shape of proteins in protein-protein complexes and to measure their distance and the opening of the dimer complexes [1-3]. In fact, the inspection of the experimental structures showed that the amino acids can be assigned as interior or surface when the distance from the center of mass to the Cα atoms of residues with a relative surface accessibility of <33% and >66% was of about 7.5±5Å and 12±5Å, respectively [1]. A recent study assessed the global shape of complexed proteins by measuring the distance of the surface residues to the protein's center of mass and showed that on average the binding site residues are closer to the center of mass than the non-binding surface residues [2]. Moreover, the center of mass distances between two chains complexed in a dimer along the trajectories of molecular dynamics simulations was used to measure the opening of the two subunits of the dimer. [3] Given a protein composed by N atoms, each of the three cartesian coordinates of its center of mass are calculated as given in formula (1) (in supplementary material).
CALCOM is an user-friendly tool to calculate the cartesian coordinates of the center of mass of a single chain, as well as complexed chains and ligands, to evaluate their distance in protein-protein and protein-small ligand complexes, and to measure the distance of selected amino acids and atoms to the center of mass.
Methodology
Program input
The user can indicate the protein name, paste the file PDB in the box and choose seven possible analysing options to calculate: (1) the center of mass of a protein chain; (2) the center of mass of two protein chains and their distance; (3) for multi-chains complexes the centers of mass and the distance for each pair of chains; (4) the distance to the center of mass for specific protein amino acid(s); (5) the average distance to the center of mass for a list of protein residues; (6) the distance between the centers of mass of a ligand and a protein chain; (7) the distance to the protein center of mass for specific ligand atom(s). In the fourth and seventh options the user may write “the protein residue number” or “the ligand atom name” for which the distance to the protein center of mass may be calculated or “all” for the distance of all residues or atoms to the center of mass and also can select the residues or the atoms at distance shorter or longer than a chosen value respect to the protein center of mass and choose if the protein residues may be reported in black or coloured according to the classification reported in 1998 [1].
Program output
In the first analysis option CALCOM shows the cartesian coordinates of the center of mass of the protein chain chosen from the user. In the results pages of the second and third options CALCOM will report the cartesian coordinates of the centers of mass of two or multi protein chains chosen from the user and their distance (Figure 1a). These two options allow the user to know the opening of the several subunits composing the complex. When the user chooses the fourth option, CALCOM reports the cartesian coordinates of the center of mass of the protein chain chosen from the user and the distance of the Cα atom of the selected amino acid to calculated center of mass or a table with all the residues names and numbers and the distance of their Cα atoms to the center of mass of selected chain (Figure 1b). Then, this option allows to identify the amino acids at shortest or longest distance respect to the center of mass. In the fifth option CALCOM shows the cartesian coordinates of the center of mass of the selected protein chain and the distances of the Cα atoms of the amino acids chosen from the user to calculated center of mass and the related average distance (Figure 1c). With this analysis it is possible to study protein binding sites, protein interfaces, and structural characteristics of protein shapes and interactions. When the user chooses the sixth and seventh options, CALCOM analyses the protein-small ligand complexes and reports in the first case the cartesian coordinates of the centers of mass of ligand and protein chains and their distance, and in the second case the distance of the selected atoms of the ligand to calculated center of mass of the protein selecting those at shortest/longest distance.
Figure 1.
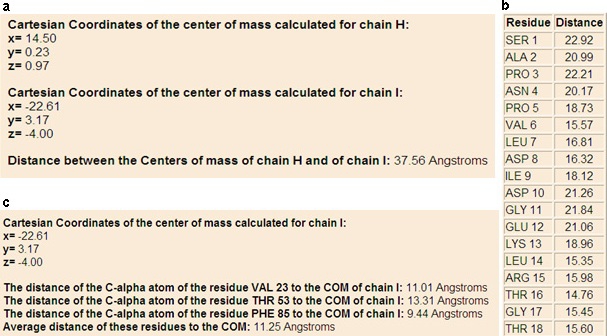
Example of output results for second (a), fourth (b) and fifth (c) options.
The results appear in few seconds and are presented in a HTML page. The user can also save directly the results in text format.
Discussion
CALCOM has been applied to evaluate the distance between two subunits of the miraculin dimer during the molecular dynamics simulations performed at different pH conditions [3]. The calculated distances have indicated that at acidic pH the dimer assumes a widely open conformation, in agreement with the hypotheses coming from other studies. Major details are reported on the web pages of Output Example (i.e. example 5).
Caveats and future development
CALCOM is a CGI script written in Perl language. We are developing other analysing options.
Supplementary material
Acknowledgments
This work was partially supported by the CNR-Bioinformatics Project and by “RNBIO - Rete Nazionale di Bioinformatica Oncologica”.
References
- 1.Chelvanayagam G, et al. Folding and Design. 1998;3:149. doi: 10.1016/S1359-0278(98)00023-6. [DOI] [PubMed] [Google Scholar]
- 2.Nicola G, Vakser IA. Bioinformatics. 2007;23:789. doi: 10.1093/bioinformatics/btm018. [DOI] [PubMed] [Google Scholar]
- 3.Paladino A, et al. BBRC. 2008;367:26. doi: 10.1016/j.bbrc.2007.12.102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


