Abstract
Brazzein protein comes from an edible fruit, which has a long history of being a staple in the local human diet in Africa. The attractive features of brazzein as a potential commercial sweetener include its small size (53 amino acid residues), its stability over wide ranges of temperature and pH, and the similarity of its sweetness to sucrose. Heterologous production of brazzein is complicated by the fact that the protein contains four disulfide bridges and requires a specific N-terminal sequence. Our previous protocol for producing the protein from Escherichia coli involved several steps with low overall yield: expression as a fusion protein, denaturation and renaturation, oxidation of the cysteines, and cleavage by cyanogen bromide at an engineered methionine adjacent to the desired N-terminus. The new protocol described here, which is much faster and leads to a higher yield of native protein, involves the production of brazzein in E. coli as a fusion with SUMO. The isolated protein product contains the brazzein domain folded with correct disulfide bonds formed and is then cleaved with a specific SUMO protease to liberate native brazzein. This protocol represents an important advancement that will enable more efficient research into the interaction between brazzein and the receptor as well as investigations to test the potential of brazzein as a commercially viable natural low calorie sweetener.
Keywords: brazzein, sweet protein, low calorie protein sweetener, disulfide bonds, Csαβ family, bacterial expression, SUMO fusion, soluble fusion, Escherichia coli
Introduction
The affluent countries of the world are suffering from an epidemic of obesity, insulin-resistance, and type II diabetes. In our evolutionary past, a strong drive to find rich energy sources and high carbohydrate foods was an advantage for survival. Today, with lives of a more sedentary nature, this sweet-seeking behavior has now become a liability, and low-calorie sweeteners with good taste properties are becoming more sought after, particularly naturally occurring ones, such as proteins.
Over the last 30 years, high potency sweet proteins have been identified in a variety of African and South Asian fruits. The first one discovered was thaumatin (22,206 Da) [1]. It was closely followed by monellin (11,086 Da) [2], mabinlin (12,441 Da) [3], and more recently brazzein (6,473 Da) [4]. Although these three sweet proteins exhibit no sequence or structural homology, they appear to require the presence of charged residues on the protein surface over a non-contiguous area [5]; [6–10]. It is thus likely that protein sweeteners share similar receptor binding interaction sites. Among the natural, low-calorie sweeteners, brazzein is the most promising because of its superior taste quality and its physical properties. Brazzein contains no carbohydrate and bears no structural resemblance to sucrose. Recombinant brazzein is 2000 times sweeter than sucrose solution on a weight-weight comparison (17,000 times higher on a per-molecule basis) [11].
Although taste perception and taste preferences have a rich history of study, it is only in the last few years that the receptors thought to underlie this behavior have been identified. Brazzein is perceived as sweet by humans, old world monkeys, and apes, but not new world monkeys or other tested species. This species difference was exploited in the discovery of the human sweet receptor when it was found that substitution of human T1R2 in mice generated animals with humanized sweet taste preferences [12]. Two receptor gene products, T1R2 and T1R3, are required for an animal’s preference for sweet tasting molecules [13].
To understand structural and chemical properties responsible for brazzein sweetness, we engineered a synthetic gene to express the brazzein molecule and developed the first production system for brazzein in bacteria using nuclease fusion system developed in our laboratory [11]. We used this approach to discover mutants with sweet-taste properties that appear to be different from the wild-type protein [5]. The major drawbacks of nuclease fusion system are lack of protein solubility, the requirement for refolding brazzein, and inefficiency in removal of the fusion tag. We report here a new method based on the SUMO fusion system that supports more efficient production of brazzein. The major advantage of the SUMO fusion system is that it yields brazzein in folded and soluble form in high yield. This method of production will allow us to more rapidly produce brazzein with lower cost for both research studies and future large-scale production. By learning more about its mechanism of action and by developing more potent brazzein variants, we will be in a better position to evaluate it as a potential sugar substitute as a means for fighting problems related to obesity and diabetes.
Methods
Construction of Sumo-brazzein expression vector
The linearized pSUMO vector with BsaI and BamHI restriction sites and T7 promotor and kanomycin resistance [14] was purchased from LifeSensors (LifeSensors, Malvern, PA). The codon optimized synthetic gene designed for bacterial expression of brazzein [11] was cloned between the BsaI (5' -GGTCTC) unique cloning site, downstream and in-frame with the SUMO gene, and the BamHI (3' -GGATCC) site in the multiple cloning site of the pSUMO vector. The pSUMO vector is derived from pET24d and contains T7 promotor induced with IPTG. The fusion gene coded for a His6-tag at the N-terminus of the SUMO protein followed by brazzein at the C-terminus. The new pSUMO-brazzein expression vector was transformed into E. coli DH5α strain (Invitrogen). The correct gene sequence was confirmed by sequencing at the University of Wisconsin-Madison Biotechnology Center.
Protein expression of SUMO-brazzein
A variety of expression strains were tested: Rosetta (DE3) from Novagen and BL21 (pLysS), BL21-CodonPlus (DE3)RI, BL21-CodonPlus (DE3)RIP, and BL21-CodonPlus (DE3)RIPL from Stratagene. Cells were grown on Luria broth (LB) growth medium [15] supplemented with an antibiotic (34 µg/mL kanamycin). Of the host cells studied, only BL21-CodonPlus (DE3) RIPL produced high amounts of the SUMO-brazzein fusion protein as detected on Tris-Tricine SDS-PAGE 16% (Invitrogen), and this strain was used for high level protein production.
The SUMO-brazzein fusion protein was expressed by induction with 0.5 mM IPTG at about mid-log phase (OD600nm ~0.6). The optimum induction time was 24 h, and the temperature was shifted to 25 °C after IPTG addition. Cells from the rich media were harvested at a final OD600nm of 5–6, and yielded 5 to 6 g of wet cell paste per liter culture. Cells were frozen and kept at −80°C for later purification procedures.
To verify protein expression, we used SDS-PAGE (Novagen) gel electrophoresis: 1.0-ml aliquots were removed before IPTG induction (at OD600nm ~0.6) and at 24 h post induction (the latter aliquot was diluted 1:6 or 1:8 to adjust the cell concentration to that of the earlier sample). Cells were centrifuged at 6,000 rpm for 2 min, and the isolated pellet was resuspended by adding 50 µl distilled water and 50 µl 2X Tricine sample buffer (contains 200 mM Tris-HCl, 2% SDS, 40% glycerol, and 0.04%Coomassie Brilliant Blue G-250 (pH 6.8) and β-mercaptoethanol). Cell disruption, denaturation and disulfide reduction were allowed to proceed for 5 min before the sample was loaded onto the gel. The running buffer was Tris-tricine; gel staining was with R250 Coomassie blue.
Purification of the His6-SUMO-brazzein fusion
We used Ni-NTA superflow resin (Qiagen, Valencia, CA) under native conditions to quickly purify the soluble His6-SUMO-brazzein fusion from the cell lysate. Cells were first suspended by adding either BugBuster (Novagen) according to the manual or lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 6.8) complemented with 300 µg/mL lysozyme and 90 µg/mL PMSF. The suspensions were kept on ice to facilitate cell lysis. For complete lysis, cells were sonicated on ice by 2 periods each of 4 min (cycles of 10 s on followed by 30 s off). Cells were centrifuged at 10,000 g for 20 min at 4°C. The supernatant was applied to the pre-equilibrated Ni-NTA column (QIAexpressionist, Qiagen) with lysis buffer (as described above), followed by 5X wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 6.8). The fusion protein was eluted with buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 7.8). Most of the fusion protein eluted in the second fraction with more than 98% purity, according to the RP-HPLC chromatography (data not shown). Pooled fractions were dialyzed against 10 mM Tris-HCl containing 150 mM NaCl, pH 8.0 and concentrated to a final 5–10 ml volume. The product was reacted with 1 unit SUMO protease per 50 µg fusion protein for a period of 1 h at 30 °C. Interestingly, the fusion protein fractions were reddish-brown in color indicating that some of Cys residues in brazzein were incompletely oxidized (see Discussion). However, after cleavage, both the brazzein and SUMO fractions were white in dried powder form.
RP-HPLC column chromatography was used for the final brazzein purification step. Tris-Tricine SDS-PAGE 16% was used to assay the extent of proteolysis protease activity and the recovery of brazzein product.
Characterization of the brazzein product
The protein product was further characterized by electrospray ionization mass spectroscopy (ESIMS) at the Biotechnology Center, University of Wisconsin-Madison, and by NMR spectroscopy at the National Magnetic Resonance Facility at Madison. A human panel determined the taste properties of the protein (HSC protocol #2002-244).
Results
Expression of the fusion protein
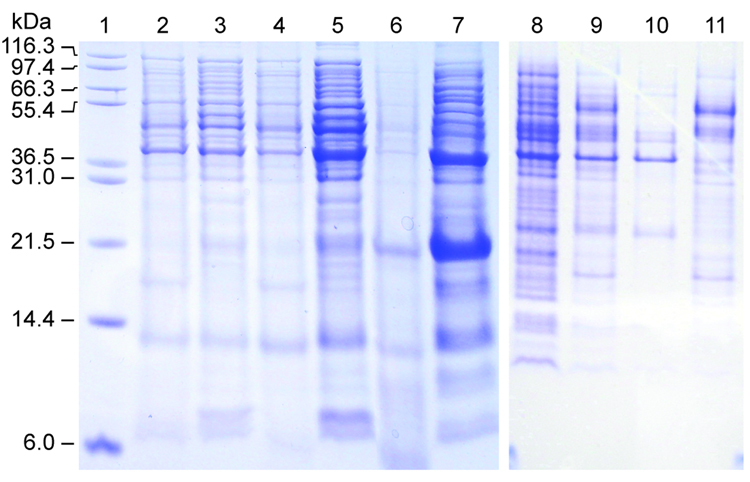
We found that expression levels of His6-SUMO-brazzein fusion protein were very low in conventional E. coli strains, e.g. BL21(DE3)pLysS, possibly because of complications of codon bias, because the SUMO gene was derived from a eukaryotic system. In an attempt to improve expression levels, we tested several other bacterial strains recommended for T7 driven expression systems. Although we observed no expression of the fusion construct from Rosetta (DE3)pLysS cells, we found increasing levels of expression from the cell series, BL21-CodonPlus (DE3)RP, BL21-CodonPlus (DE3)RIL, BL21-CodonPlus (DE3)RIPL (RP<RIL<RIPL) (Fig. 1).
Figure 1.
SDS-PAGE showing the expression of the His6-SUMO-brazzein fusion in various host strains. Lane 1: Molecular weight markers (Invitrogen). Lanes 2/3: −IPTG / +IPTG (BL21 (DE3)/RI Codon Plus). Lanes 4/5: −IPTG / +IPTG (BL21 (DE3)/RIL Codon Plus). Lanes 6/7: −IPTG / +IPTG (BL21 (DE3)/RILP Codon Plus). Lanes 8/9: −IPTG / +IPTG (BL21(DE3)/pLysS). Lanes 10/11: −IPTG / +IPTG (Rosetta (DE3)). The His6-SUMO-brazzein fusion is observed at 23.5 kD and brazzein at 6.4 KD.
Fusion protein production and purification and recovery of brazzein
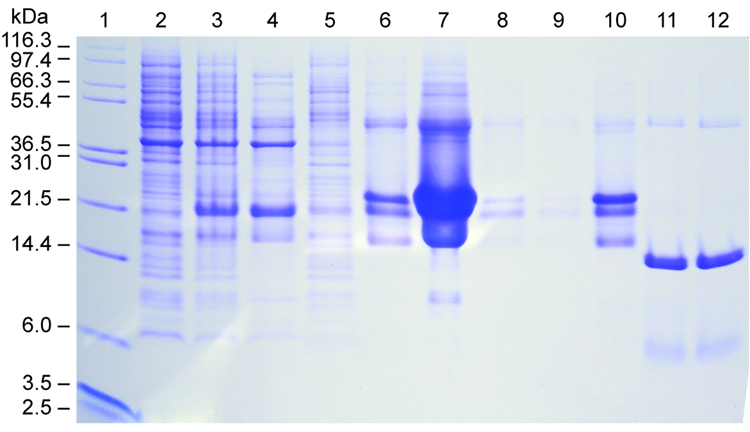
To achieve high yields, we expressed the His6-SUMO-brazzein fusion from BL21-CodonPlus (DE3)RIPL cells under the control of a T7 promoter with induction by 0.5 mM IPTG followed by 24 h growth at 25 °C. The fusion protein was efficiently produced in soluble form and purified to greater than 90% purity by Ni-NTA column chromatography in a single step (Fig. 2). The proper ration of SUMO protease 1 to fusion protein was determined by small-scale trials. Starting with the protocol recommended by LifeSensors (1 unit protease/10–100 µg fusion protein in the presence of 5 mM DTT), we found by monitoring the expected 6.4 kDa band on SDS PAGE that the optimal conditions for cleavage are 1 unit protease to 50 µg fusion protein at either 5 mM DTT or <0.5 mM DTT. However, the protein products produced by cleavage in 5 mM DTT and <0.5 mM DTT behaved differently on RP-HPLC. In either case, the digested His6-SUMO protein eluted at 34–35 min (at 58% gradient mixture), however, brazzein produced by cleavage in the presence of 5 mM DTT eluted at 29.6 min (at 42% gradient mixture) (Fig. 3a), whereas brazzein produced by cleavage in the presence of <0.5 mM DTT eluted at 23.2 min (at 38% gradient mixture) (Fig. 3b). Brazzein produced by cleavage at the higher DTT concentration was not sweet. With the concentration of DTT in the protease cleavage mixture kept below <0.5 mM (the SUMO protease 1 preparation contains 5 mM DTT), elution position corresponded to that of native brazzein [5], and the protein was sweet. At this low level of DTT and with 1 unit SUMO protease 1/50 µg fusion protein, the cleavage was >90% after 60 min at 30 °C. The peak at 23 min was characterized by ESI-MS with mass of 6,366 consistent with calculated value of 6,365.8 for brazzein. The cleavage product contained small amounts of protein eluting at 28.4, and 29.7 min (Fig. 3b); these fractions were analyzed by SDS PAGE and ESI-MS and judged to be partially folded or incompletely oxidized brazzein and were found not to be sweet.
Figure 2.
Medium-scale expression and purification of His6-SUMO-brazzein produced from BL21 (DE3)/ PRIL Codon Plus cells. Lane 1: Molecular weight markers (Invitrogen). Lane 2: −IPTG. Lane 3: total cell lysates. Lane 4: supernatant. Lane 5: pellet. Lanes 6–9: Ni-NTA column fractions (2 to 5). Lane 10: purified fusion protein. Lane 11: SUMO 1 protease cleavage product after 30 min. Lane 12: cleavage product after 60 min.
Figure 3.
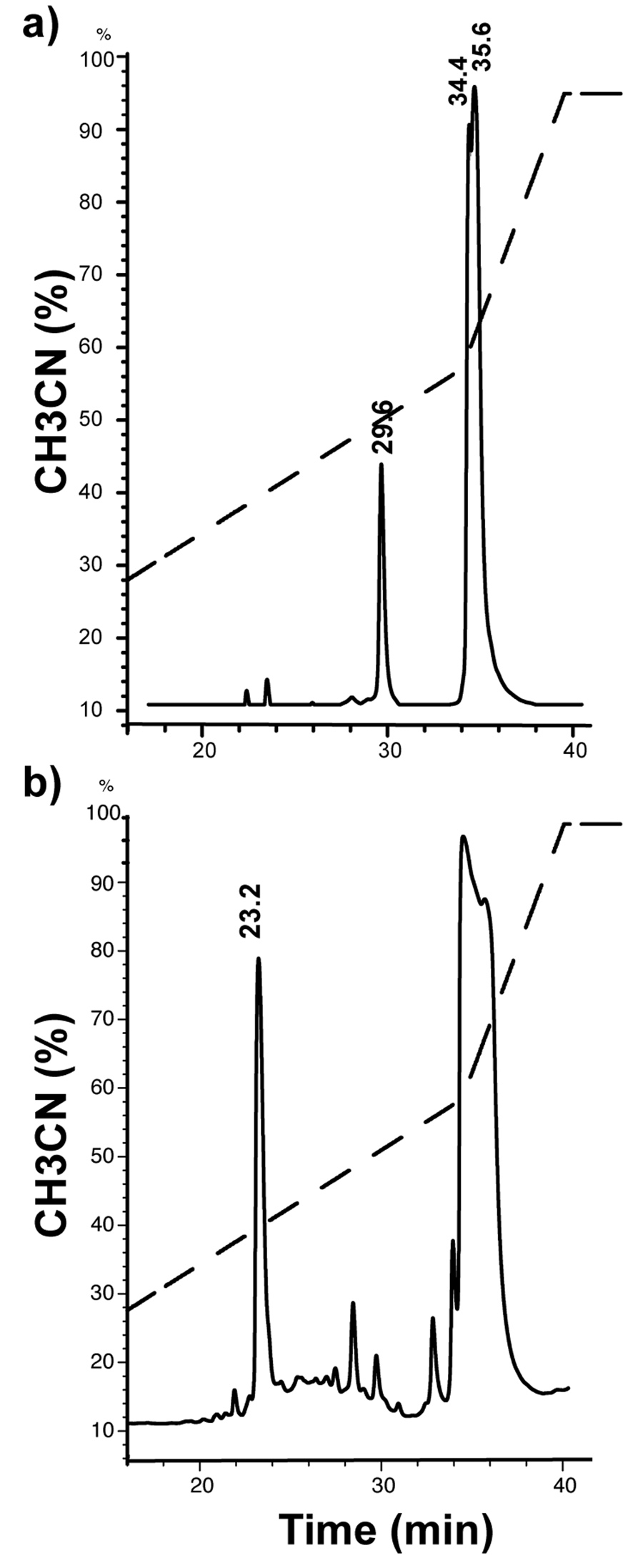
RP-HPLC traces of mixture resulting from SUMO 1 cleavage of the His6-SUMO-brazzein fusion protein. (a) His6-brazzein-SUMO fusion + 5 mM DTT. (b) His6-brazzein-SUMO fusion + 0.1 mM DTT. Brazzein elution times are marked in folded/or unfolded/non-fully oxidized protein.
We used a Bradford protein assay (Bio-Rad) to quantify the amounts of protein recovered in various steps in the protocol. The fusion protein was found to represent 26% of the total cell lysate protein, and the amount of brazzein produced represented 9% of the total soluble protein from the cells (Table 2). Then amount of fusion protein recovered through Ni-NTA column was high, and resulted native brazzein protein following cleavage as judged by RP-HPLC.
Table 2.
Codon abundance in SUMO and brazzein proteins and codons frequently found in E. coli.
| Amino acid | Codons and their normal abundance in E. coli (more abundant codons are shaded) | Number of codons in the SUMO gene | Number of codons in the synthetic brazzein gene | |
|---|---|---|---|---|
| Arginine | CGT | 0.643 | 0 | 2 |
| CGC | 0.330 | 1 | 0 | |
| CGA | 0.011 | 0 | 0 | |
| CGG | 0.008 | 0 | 0 | |
| AGA | 0.006 | 4 | 0 | |
| AGG | 0.003 | 1 | 0 | |
| Isoleucine | ATT | 0.335 | 5 | 0 |
| ATC | 0.659 | 3 | 1 | |
| ATA | 0.006 | 0 | 0 | |
| Proline | CCT | 0.112 | 3 | 0 |
| CCC | 0.016 | 0 | 0 | |
| CCA | 0.153 | 2 | 0 | |
| CCG | 0.719 | 0 | 1 | |
| Leucine | TTA | 0.055 | 3 | 0 |
| TTG | 0.034 | 2 | 0 | |
| CTT | 0.056 | 0 | 0 | |
| CTC | 0.080 | 0 | 0 | |
| CTA | 0.008 | 0 | 0 | |
| CTG | 0.767 | 1 | 1 | |
Folding and activity
The protein folding status product was characterized further by 1D 1H NMR spectroscopy and 2D 1H-1H NOESY (Fig. 4). These spectra matched those of authentic native brazzein. Furthermore, the protein had the sweetness of native brazzein.
Figure 4.
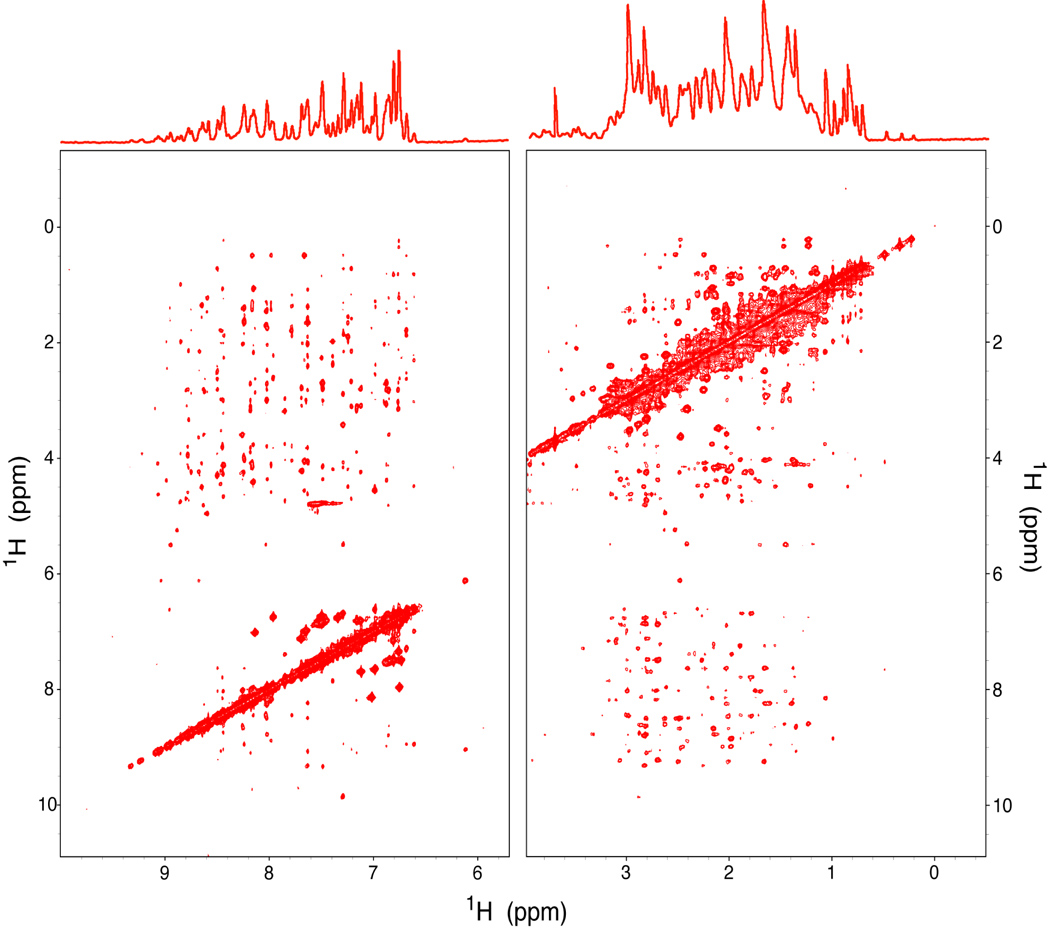
Spectra of the purified brazzein product obtained on a Varian 800 MHz NMR spectrometer at pH 5.2 and 25 °C. (Top) One-dimensional 1H NMR spectrum. (Bottom) Two-dimensional 1H-1H NOESY map.
Discussion
To date, no gene for brazzein has been isolated from its plant of origin. We used the sequence of the minor form of brazzein isolated from Pentadiplandra brazzeana fruit as the basis for designing a synthetic gene with codon frequencies optimized for protein production in E. coli [5]. We chose E. coli for protein production, because of the ease of rapid genetic manipulation. Brazzein is difficult to express on its own, and our earlier approach for producing brazzein utilized a fusion to staphylococcal nuclease (nuclease) with an engineered cyanogen bromide cleavage site (wild-type brazzein contains no Met, and the Met residues of nuclease were removed by mutagenesis) [5]. Because the nuclease-brazzein fusion protein was expressed in insoluble form, the fusion protein had to solubilized and refolded prior to cyanogen bromide cleavage. This procedure was time consuming, and the refolding and oxidation steps required manual effort. In addition, the yield from the CNBr cleavage step was only about 50% as the result of partial oxidation of the methionine residue during the oxidation processes.
The SUMO fusion system (LifeSensors, Inc.), which has been found to solubilize and stabilize difficult proteins including those with disulfide bridges [16], appeared to offer an attractive alternative. We cloned the synthetic brazzein gene designed in our laboratory at the 3′ end of the His6-SUMO gene. However, when we followed the standard recommended protocol, which specified Rosetta (DE3)pLysS host cells and Ni-NTA purification, the yield of the His6-SUMO-brazzein fusion protein was undetectable. Thus, we introduced a series of changes that led to high-level production and purification of the fusion protein. Expression of recombinant proteins in E. coli is generally difficult when the codon use in the recombinant gene differs from the codon use in the host cells. Our studies showed that the best host for expressing the His6-SUMO-brazzein fusion was BL21-CodonPlus (DE3)RIPL. This strain provides an increased supply of rare E. coli tRNAs (argU, ileY, leuW, and proL) that correspond to codons used more frequently by eukaryotes. These four amino acids constitute ~25% of SUMO, but much less of brazzein (Table 1). Also, whereas brazzein has been optimized to use more abundant E. coli codons for these amino acids, the gene coding for SUMO utilizes primarily rare codons (Table 1). This explains why the BL21-CodonPlus (DE3)RIPL was more successful than Rosetta (DE3)pLysS, which is not optimized for these particular amino acids.
Table 1.
Protein determined by Bradford protein assay from 10g wet cells. Following RP-HPLC and cleavage.
| Fraction | Mass (mg) |
|---|---|
| Total soluble protein | 636 |
| Flow-through | 376 |
| Wash | 53 |
| Insoluble1 | 44 |
| His6-SUMO-brazzein | 163 |
| Brazzein | 54 |
Precipitate formed following salt removal and concentration.
To cleave the His6-SUMO-brazzein fusion protein to yield brazzein with the correct amino terminus, we used the recommended SUMO protease 1, a small ubiquitin-like modifier protease that recognizes only the tertiary structure of SUMO and efficiently cleaves at the junction between SUMO and the protein of interest [14]. We found it necessary to reduce the amount of DTT present from the recommended level of 5 mM to less than 0.5 mM to prevent reduction of the disulfide bridges of brazzein leading to misfolded, inactive product. The lowered DTT concentration reduced the cleavage efficiency from 100% to >90%. The typical yield of pure brazzein following RP-HPLC was about 50 mg/10 g wet cells. The recombinant brazzein after purification was characterized by SDS-PAGE for its purity, and its molecular weight was confirmed by mass spectrometry. Although the isolated His6-SUMO-brazzein fusion protein had a brownish cast, this color disappeared completely following cleavage and final purification. The brown color may have resulted from the stripping of Ni2+ ions from the Ni-NTA column by reduced cysteines of brazzein. The purified brazzein product was in native form as judged from RP-HPLC, 1D 1H NMR and 2D 1H-1H NOESY analysis and taste tests.
In summary, SUMO fusion system and customized expression and purification protocol described here have greatly improved the efficiency and lowered the costs of producing brazzein. This method has three major advantages over the nuclease fusion system developed earlier [5]: 1) the SUMO fusion does not require refolding; 2) the protein is purified quickly (2 days vs. two weeks); 3) the protease cleavage efficiency is >90% vs. ~50% for CNBr cleavage. Thus the new protocol provides a higher yield of brazzein with less time and effort. It is enabling us to rapidly produce brazzein variants for ongoing investigations of the interaction between the protein and the sweet receptor. In addition, the new approach may become a springboard for industrial level production of brazzein.
Acknowledgements
Work supported in part by NIH grant P41 RR02301. The authors thank Dr. Marco Tonelli for help with NMR data collection, Dr. James Radek for cloning brazzein into the SUMO plasmid, and Dr. Ronnie Fredrick for helpful discussions.
Abbreviations
- IPTG
isopropyl-β-D-thiogalactopyranoside
- Ni-NTA
nickel-nitrilotriacetic acid
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- RP-HPLC
reversed phase high pressure liquid chromatography
- TFA
trifluoroacetic acid
- SUMO
small ubiquitin-like modifier
- DTT
dithiothreitol
- 2D-NOESY
two-dimensional nuclear Overhauser spectroscopy
- NMR
nuclear magnetic resonance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.van der Wel H, Loeve K. Isolation and Characterization of Thaumatin I and II, the Sweet- Tasting Proteins from Thaumatococcus danielli Benth. Eur. J. Biochem. 1972;31:221–225. doi: 10.1111/j.1432-1033.1972.tb02522.x. [DOI] [PubMed] [Google Scholar]
- 2.Morris JA, Cagan RH. Purification of Monellin, the Sweet Principle of Dioscoreophyllum cumminsii. Biochim. Biophys. Acta. 1972;261:114–122. doi: 10.1016/0304-4165(72)90320-0. [DOI] [PubMed] [Google Scholar]
- 3.Hu Z, He M. Studies on mabinlin, a Sweet Protein from the Seeds of Capparis masaikai Levle. I. Extraction, Purification and Certein Characteristics. Acta Botanica Yunnanica. 1983;5:207–212. [Google Scholar]
- 4.Ming D. Purification and Characterization of the Sweet Protein Brazzein. University of Wisconsin Madison; 1994. [Google Scholar]
- 5.Assadi-Porter FM, Aceti DJ, Markley JL. Sweetness Determinant Sites of Brazzein, a Small, Heat-stable, Sweet-Tasting Protein. Arch. Biochem. Biophys. 2000;376:259–265. doi: 10.1006/abbi.2000.1726. [DOI] [PubMed] [Google Scholar]
- 6.Tancredi T, Iijima H, Saviano G, Amodeo P, Temussi PA. Structural Determination of the Active Site of a Sweet Protein A 1H NMR Investigation of pMNEI. FEBS Lett. 1992;310:27–30. doi: 10.1016/0014-5793(92)81138-c. [DOI] [PubMed] [Google Scholar]
- 7.Morris RW, Cagan RH, Martenson RE, Deibler G. Methylation of the Lysine Residues of Monellin. Proc. Soc. Exp. Biol. Med. 1978;127:194–199. doi: 10.3181/00379727-157-40019. [DOI] [PubMed] [Google Scholar]
- 8.Mizukoshi T, Kohmura M, Suzuki E, Ariyoshi Y. Structure and Dynamic Studies by NMR of the Potent Sweet Protein Monellin and a Non-sweet analog. Evidence on the Importance of Residue AspB7 for Sweet Taste. FEBS Lett. 1997;413:409–416. doi: 10.1016/s0014-5793(97)00945-9. [DOI] [PubMed] [Google Scholar]
- 9.Slootstra JW, De Geus P, Haas H, Verrips CT, Meloen RH. Possible Active Site of the Sweet-tasting Protein Thaumatin. Chem. Senses. 1995;20:535–543. doi: 10.1093/chemse/20.5.535. [DOI] [PubMed] [Google Scholar]
- 10.Kaneko R, Kitabatake N. Structure-Sweetness Relationship in Thaumatin: Importance of Lysine Residues. Chem. Senses. 2001;26:167–177. doi: 10.1093/chemse/26.2.167. [DOI] [PubMed] [Google Scholar]
- 11.Assadi-Porter FM, Aceti DJ, Cheng H, Markley JL. Efficient Production of Recombinant Brazzein, a Small, Heat-Stable, Sweet-Tasting Protein of Plant Origin. Arch. Biochem. Biophys. 2000;376:252–258. doi: 10.1006/abbi.2000.1725. [DOI] [PubMed] [Google Scholar]
- 12.Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJ, Zuker CS. The Receptors for Mammalian Sweet and Umami Taste. Cell. 2003;115:255–266. doi: 10.1016/s0092-8674(03)00844-4. [DOI] [PubMed] [Google Scholar]
- 13.Damak S, Rong M, Yasumatsu K, Kokrashvili Z, Varadarajan V, Zou S, Jiang P, Ninomiya Y, Margolskee RF. Detection of Sweet and Umami Taste in the Absence of Taste Receptor T1r3. Science. 2003;301:850–853. doi: 10.1126/science.1087155. [DOI] [PubMed] [Google Scholar]
- 14.Malakhov MP, Mattern MR, Malakhova OA, Drinker M, Weeks SD, Butt TR. Sumo Fusions and SUMO-specific Protease for Efficient Expression and Purification of Proteins. J. Struct. Func. Genom. 2004;5:75–86. doi: 10.1023/B:JSFG.0000029237.70316.52. [DOI] [PubMed] [Google Scholar]
- 15.Luria SE, Burrous JW. Hybridization between Escherichia coli and Shigella. J. Bacteriol. 1957;74:461–476. doi: 10.1128/jb.74.4.461-476.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuo X, Mattern MR, Tan R, Li S, Hall J, Sterner DE, Shoo J, Tran H, Lim P, Sarafianos SG, Kazi L, Navas-Martin S, Weiss SR, Butt TR. Expression and Purification of SARS Coronavirus Proteins using SUMO-fusions. Prot. Expr. & Purif. 2005;42:100–110. doi: 10.1016/j.pep.2005.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]