Abstract
Background
In Arabidopsis, INDOLE-3-BUTYRIC ACID RESPONSE5 (IBR5), a putative dual-specificity protein phosphatase, is a positive regulator of auxin response. Mutations in IBR5 result in decreased plant height, defective vascular development, increased leaf serration, fewer lateral roots, and resistance to the phytohormones auxin and abscisic acid. However, the pathways through which IBR5 influences auxin responses are not fully understood.
Results
We analyzed double mutants of ibr5 with other mutants that dampen auxin responses and found that combining ibr5 with an auxin receptor mutant, tir1, enhanced auxin resistance relative to either parent. Like other auxin-response mutants, auxin-responsive reporter accumulation was reduced in ibr5. Unlike other auxin-resistant mutants, the Aux/IAA repressor reporter protein AXR3NT-GUS was not stabilized in ibr5. Similarly, the Aux/IAA repressor IAA28 was less abundant in ibr5 than in wild type. ibr5 defects were not fully rescued by overexpression of a mutant form of IBR5 lacking the catalytic cysteine residue.
Conclusion
Our genetic and molecular evidence suggests that IBR5 is a phosphatase that promotes auxin responses, including auxin-inducible transcription, differently than the TIR1 auxin receptor and without destabilizing Aux/IAA repressor proteins. Our data are consistent with the possibility that auxin-responsive transcription can be modulated downstream of TIR1-mediated repressor degradation.
Background
The phytohormone auxin is critical for plant growth and development, regulating vascular development, apical dominance, tropic responses, and organ patterning by modulating cell division and elongation [1,2]. Changes in gene expression are among the earliest molecular responses to auxin. Many auxin-responsive transcripts fall into one of three classes: GH3-related, Auxin/INDOLE-3-ACETIC ACID (Aux/IAA), and SMALL AUXIN-UP RNA (SAUR) transcripts [3-8]. Common to many of these auxin-responsive genes is a sequence in the upstream regulatory region termed the Auxin-Responsive Element (AuxRE; [9]).
AUXIN RESPONSE FACTOR (ARF) proteins are transcription factors that bind AuxREs (reviewed in [10]). Depending on the nature of the central domain, ARF family members can either activate or repress transcription [11,12]. ARF proteins can form homodimers, dimers with other ARF proteins, or dimers with transcriptionally repressive Aux/IAA proteins [13,14]. Many Aux/IAA proteins directly prevent transcriptional activation by interacting with activating ARF proteins [12,15].
Many Aux/IAA transcriptional repressors are unstable [5] and are degraded even more rapidly following auxin application [16,17]. Rapid Aux/IAA degradation following an auxin stimulus is thought to free activating ARF proteins from repression, allowing auxin-responsive gene expression. Mutant screens for decreased auxin sensitivity have identified several Aux/IAA proteins with stabilizing mutations (reviewed in [18]). Also isolated from auxin-response screens were trans-acting mutations that likewise stabilize Aux/IAA proteins, revealing the degradation mechanism for these repressors. Several auxin-resistant mutants have defects in the SCFTIR1 E3 ubiquitin ligase complex, as well as its regulatory components (reviewed in [2]).
TRANSPORT INHIBITOR RESPONSE1 (TIR1) and the other AUXIN SIGNALING F-BOX (AFB) family members are the substrate-recognition components of SCF complexes that bind auxin and promote the degradation of Aux/IAA repressor proteins [19-21]. Auxin is trapped in the TIR1 auxin-binding pocket by an interacting Aux/IAA protein [22]. Subsequent 26S proteasomal degradation of Aux/IAA proteins relieves the repression of the ARF protein, allowing auxin-responsive transcription [16,17,23]. This novel receptor-ligand interaction allows a very short signal transduction chain that may facilitate rapid transcriptional responses to auxin. In addition, RUB (RELATED TO UBIQUITIN) modification of the CULLIN subunit of SCFTIR1 is necessary for auxin response [24,25]. Mutations in AXR1 and ECR1, which encode subunits of the RUB-activating enzyme [26,27], result in decreased auxin responses accompanied by slowed Aux/IAA protein degradation [16,25,28-30], presumably because of reduced SCFTIR1 efficacy in targeting these proteins for degradation.
Normal auxin responses require active movement of auxin through the plant, which is controlled by specialized influx and efflux carriers (reviewed in [31]). AUXIN RESISTANT1 (AUX1) is an auxin influx carrier protein that allows certain auxins to enter cells [32-35]. Mutations in AUX1 result in resistance to IAA and 2,4-dichlorophenoxyacetic acid (2,4-D) [34], which are substrates of the AUX1 transporter [35].
A variety of natural and synthetic auxins and auxin precursors have activity in auxin bioassays [2]. A mutation in IBA RESPONSE5 (IBR5) was identified in a screen for resistance to the inhibitory effects of the auxin precursor indole-3-butyric acid (IBA) on root growth [36]. Subsequent analyses revealed that ibr5 mutants are less sensitive not only to IBA, but also to all tested forms of auxin and to the phytohormone abscisic acid (ABA) [37]. IBR5 encodes a putative dual-specificity protein phosphatase, and the ibr5-1 mutation causes a premature stop codon that would result in a truncated product lacking the conserved phosphatase domain [37]. Here, we examined the role of IBR5 as a phosphatase in vivo by expressing a mutant version of IBR5 predicted to be catalytically inactive in the ibr5 mutant and found that phosphatase activity is likely required for full IBR5 function. Through double mutant analyses, we found that ibr5 enhanced most tir1 defects and a subset of axr1 and aux1 defects. Further, we demonstrated that ibr5 is defective in accumulation of an auxin-responsive reporter following auxin treatment. Because this reporter accumulates after degradation of Aux/IAA transcriptional repressors, we examined the effect of the ibr5 lesion on a reporter of Aux/IAA stability and an epitope-tagged Aux/IAA protein and, interestingly, found that these reporters were not stabilized in ibr5.
Results
ibr5 enhances tir1 auxin-response defects
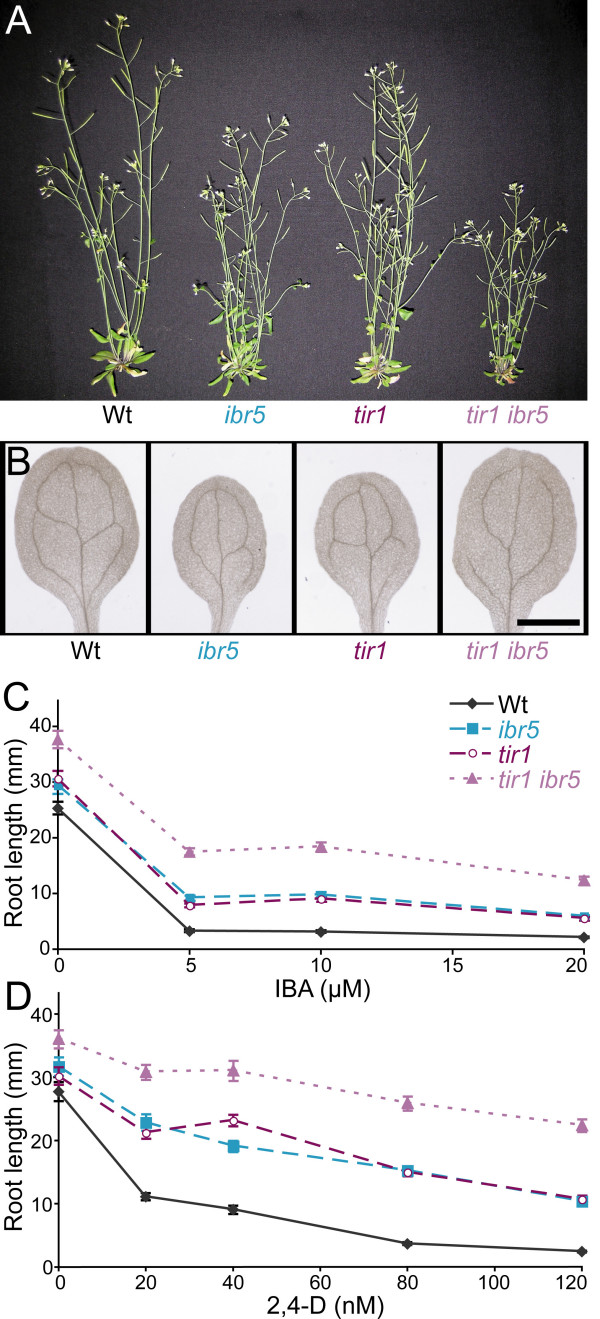
The tir1 mutant, like ibr5 [37], is less responsive to auxin in primary root elongation inhibition and lateral root formation assays [38]. To examine the genetic interaction between ibr5 and tir1, we crossed tir1-1 to ibr5-1 and examined the phenotypes of the resulting double mutant. We found that tir1 ibr5 plants were shorter than either parent (Figure 1A). ibr5 cotyledon vascularization defects were sometimes mildly enhanced by tir1 (Figure 1B). In addition, the tir1 ibr5 double mutant displayed enhanced resistance to root elongation inhibition by 2,4-D and IBA (Figures 1C and 1D, Additional File 1), fewer lateral roots in response to IBA treatment (Figure 2A), and greater resistance to IBA inhibition of hypocotyl elongation in the dark (Figure 2B, Additional File 2) than either parent.
Figure 1.
tir1 ibr5 morphological phenotypes and auxin response. (A) Adult morphologies of wild-type, ibr5, tir1, and tir1 ibr5 plants. Six-week-old Col-0 (Wt), ibr5-1, tir1-1, and tir1-1 ibr5-1 grown in continuous light are shown. (B) Vascular patterning defects. Cleared cotyledons of 8-day-old Col-0 (Wt), ibr5-1, tir1-1, and tir1-1 ibr5-1 seedlings are shown. Scale bar = 1 mm. (C, D) tir1-1 ibr5-1 auxin-response defects. Lengths of primary roots of 8-day-old seedlings grown under yellow-filtered light at 22°C on medium supplemented with various concentrations of IBA (C) or 2,4-D (D) are shown. tir1 ibr5 roots were significantly longer than tir1 and ibr5 roots on control media and on all auxins tested (P ≤ 0.001) in t-tests assuming unequal variance. Error bars represent standard errors of the means (n ≥ 18).
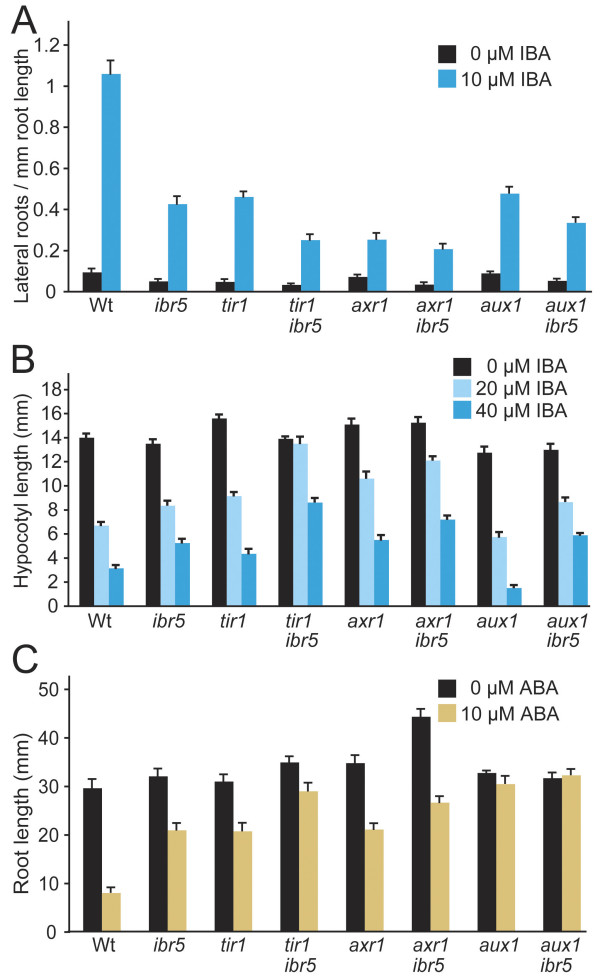
Figure 2.
Auxin-response mutant defects in lateral root induction by IBA, hypocotyl elongation inhibition by IBA, and root elongation inhibition by ABA. Hormone response of Col-0 (Wt), ibr5-1, tir1-1, tir1-1 ibr5-1, axr1-3, axr1-3 ibr5-1, aux1-7, and aux1-7 ibr5-1 were examined. (A) Lateral roots were counted 4 days after transfer of 4-day-old seedlings to medium supplemented with either 0 (ethanol control) or 10 μM IBA. Primordia emerged from the main root were counted as lateral roots. Error bars represent standard errors of the means (n ≥ 14). tir1 ibr5 had significantly fewer lateral roots in response to IBA than either tir1 or ibr5 (P ≤ 0.001 in two-tailed t-tests assuming unequal variance). (B) Lengths of hypocotyls were measured 4 days after transfer of 1-day-old seedlings to the dark. Error bars represent standard errors of the means (n = 20). tir1 ibr5 hypocotyls were significantly longer than tir1 and ibr5 hypocotyls on 20 or 40 μM IBA (P ≤ 0.0001 in two-tailed t-tests assuming unequal variance). axr1 ibr5 hypocotyls were significantly longer than axr1 and ibr5 on 20 (P ≤ 0.01) or 40 μM IBA (P ≤ 0.001) in two-tailed t-tests assuming unequal variance. (C) Length of primary roots 4 days after transfer of 4-day-old seedlings to medium supplemented with either 0 (ethanol control) or 10 μM ABA. tir1 ibr5 roots were significantly longer than tir1 and ibr5 roots on ABA (P ≤ 0.001) in two-tailed t-tests assuming unequal variance. axr1 ibr5 roots were significantly longer than axr1 and ibr5 roots following control (P ≤ 0.001) or ABA (P ≤ 0.01) treatments in two-tailed t-tests assuming unequal variance. Error bars represent standard errors of the means (n ≥ 14).
In addition to auxin resistance, ibr5 mutant roots are resistant to the phytohormone ABA [37]. We found that tir1 also exhibited ABA resistance, and that tir1 ibr5 roots were more ABA resistant than either single mutant (Figure 2C, Additional File 2). Because ibr5-1 is likely to be a null allele [37], these results support a model in which IBR5 and TIR1 act separately to affect auxin and ABA responsiveness.
ibr5 enhances certain axr1 auxin-response defects
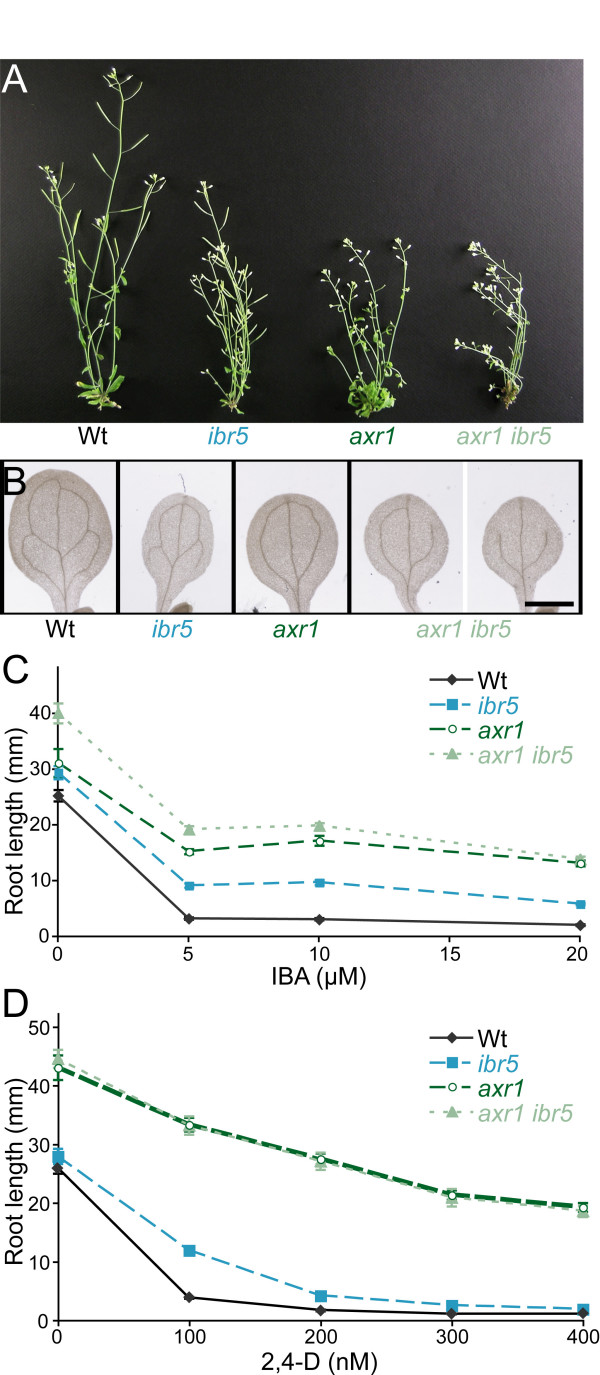
The axr1 mutant displays more extreme auxin-response defects than tir1 or ibr5, with restricted plant height, reduced apical dominance, dramatic vascularization defects, striking auxin resistance, and a longer root than wild type on unsupplemented media [29]. To examine the genetic interaction between axr1 and ibr5, we crossed axr1-3 to ibr5-1. The double mutant had similar plant height to axr1-3 (Figure 3A), but leaf epinasty (data not shown) and cotyledon vascular defects (Figure 3B) were more extreme in axr1 ibr5 compared to either parent. Further, axr1 ibr5 had a longer root on unsupplemented media than either parent (Figure 3C), consistent with the possibility that resistance to endogenous auxin was enhanced. Resistance to the auxins 2,4-D and IBA was not obviously enhanced in the double mutant when considering the longer root on unsupplemented media (Figures 3C and 3D, Additional File 1). Moreover, axr1 ibr5 did not display enhanced resistance to IBA-induced lateral root formation (Figure 2A, Additional File 2), but did exhibit slightly enhanced resistance to the inhibition by IBA of hypocotyl elongation in the dark (Figure 2B, Additional File 2). Like ibr5, axr1 is resistant to ABA inhibition of root elongation [37], and axr1 ibr5 had similar ABA resistance as both parents (Figure 2C, Additional File 2).
Figure 3.
axr1 ibr5 morphological phenotypes and auxin response. (A) Adult morphologies of wild-type, ibr5, axr1, and axr1 ibr5 plants. Six-week-old Col-0 (Wt), ibr5-1, axr1-3, and axr1-3 ibr5-1 grown in continuous light are shown. (B) Vascular patterning defects. Cleared cotyledons of 8-day-old Col-0 (Wt), ibr5-1, axr1-3, and axr1-3 ibr5-1 seedlings are shown. Scale bar = 1 mm. (C, D) axr1-3 ibr5-1 auxin-response defects. Lengths of primary roots of 8-day-old (C) or 9-day-old (D) seedlings grown under yellow-filtered light at 22°C on medium supplemented with various concentrations of IBA (C) or 2,4-D (D) are shown. axr1 ibr5 roots were significantly longer than axr1 and ibr5 on 0 (P ≤ 0.01), 5 (P ≤ 0.001), and 10 (P ≤ 0.001) μM IBA in two-tailed t-tests assuming unequal variance. axr1 ibr5 roots were not significantly different from axr1 roots on tested 2,4-D concentrations. Error bars represent standard errors of the means (n ≥ 15).
ibr5 enhances aux1 root elongation defects
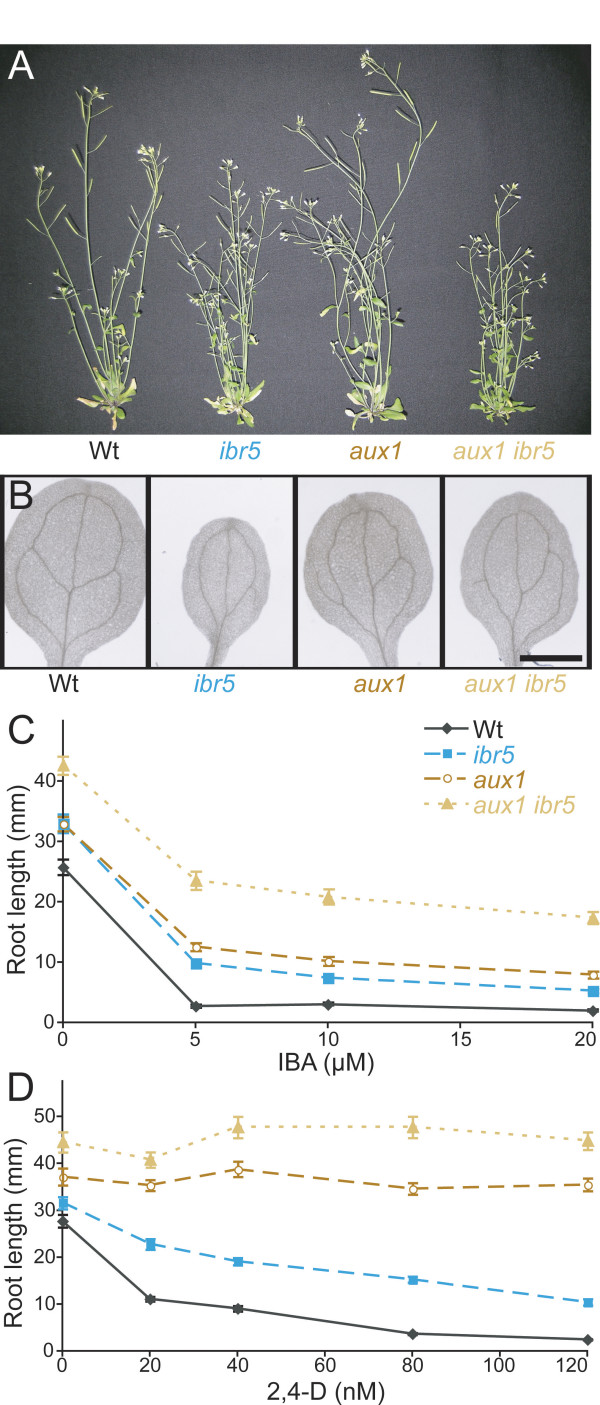
The aux1 mutant displays marked resistance to the auxins that are brought into cells by the AUX1 transporter, such as 2,4-D and IAA [33,34,39], but responds normally to 1-naphthaleneacetic acid (NAA), which is not transported by AUX1 [33-35]. Although IBA does not appear to be an AUX1 substrate [35,40], the aux1 mutant is moderately IBA resistant [36], probably because the IBA that enters cells is converted to IAA. aux1 mutant roots are agravitropic and longer than wild-type roots on unsupplemented media [39], but aux1 aerial parts resemble wild type. To examine the genetic interaction between aux1 and ibr5, we crossed ibr5-1 to aux1-7. Although aux1 plants attain normal height, adult aux1 ibr5 plants were shorter than either parent (Figure 4A). Moreover, aux1 ibr5 seedlings had longer roots on unsupplemented media than either parent (Figure 4C). Resistance to the auxins 2,4-D and IBA was not obviously enhanced in the double mutant when considering the longer root on unsupplemented media and the complete resistance of aux1 to the concentrations of 2,4-D tested (Figures 4C and 4D, Additional File 1). We examined lateral root production in aux1 ibr5 and found that aux1 did not markedly enhance ibr5 defects (Figure 2A, Additional File 2). Similarly, the ibr5 cotyledon vascular development defects did not appear to be enhanced by aux1 (Figure 4B). Hypocotyls of dark-grown aux1 responded like wild type to IBA, and aux1 ibr5 responses resembled those of ibr5 (Figure 2B, Additional File 2). aux1 was unresponsive to the ABA concentrations tested (Figure 2C, Additional File 2); thus we did not determine if ibr5 enhanced aux1 ABA resistance.
Figure 4.
aux1 ibr5 morphological phenotypes and auxin response. (A) Adult morphologies of wild-type, ibr5, aux1, and aux1 ibr5 plants. Six-week-old Col-0 (Wt), ibr5-1, aux1-7, and aux1-7 ibr5-1 grown in continuous light are shown. (B) Vascular patterning defects. Cleared cotyledons of 8-day-old Col-0 (Wt), ibr5-1, aux1-7, and aux1-7 ibr5-1 seedlings are shown. Scale bar = 1 mm. (C, D) aux1-7 ibr5-1 auxin-response defects. Lengths of primary roots of 8-day-old seedlings grown under yellow-filtered light at 22°C on medium supplemented with various concentrations of IBA (C) or 2,4-D (D) are shown. aux1 ibr5 roots were significantly longer than aux1 and ibr5 roots in the absence of hormone and on 5, 10, and 20 μM IBA (P ≤ 0.001) in two-tailed t-tests assuming unequal variance. aux1 ibr5 roots were significantly longer than aux1 and ibr5 roots on 20 (P ≤ 0.01), 40 (P ≤ 0.01), 80 (P ≤ 0.0001), and 120 (P ≤ 0.001) nM 2,4-D in two-tailed t-tests assuming unequal variance. Error bars represent standard errors of the means (n ≥ 16).
ibr5 displays reduced auxin-responsive reporter accumulation
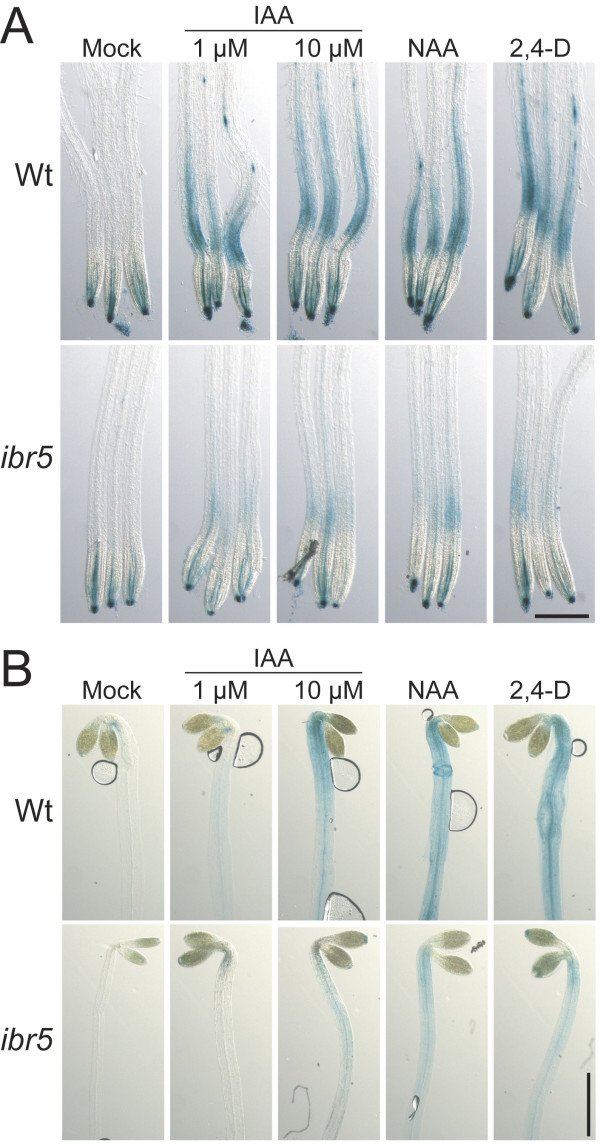
ibr5 seedlings grown on unsupplemented medium display reduced accumulation of DR5:GUS [37], a construct in which the GUS reporter is driven from a synthetic auxin-responsive promoter [13]. We compared wild-type and ibr5 DR5:GUS auxin responses in roots of light-grown seedlings and hypocotyls of dark-grown seedlings. Two-hour treatments with various auxins increased DR5:GUS activity in wild-type roots (Figure 5A) and hypocotyls (Figure 5B). In contrast, ibr5 showed reduced induction of DR5:GUS activity following auxin treatment in both roots and hypocotyls (Figures 5A and 5B).
Figure 5.
An auxin-responsive reporter is reduced in ibr5. (A) 8-day-old light-grown Col-0 (Wt) and ibr5-1 seedlings carrying the DR5:GUS construct [13, 37] were mock treated or treated with 1 μM IAA, 10 μM IAA, 10 μM NAA, or 10 μM 2,4-D for 2 hours, then stained for GUS activity. Scale bar = 0.5 mm. (B) 5-day-old dark-grown Col-0 (Wt) and ibr5-1 seedlings carrying the DR5:GUS construct [13, 37] were mock treated or treated with 1 μM IAA, 10 μM IAA, 10 μM NAA, or 10 μM 2,4-D for 2 hours, then stained for GUS activity. Scale bar = 1 mm.
This reduced induction of DR5:GUS activity suggests that ibr5 misregulates at least some auxin-regulated transcripts. Thus, we examined basal levels and auxin responsiveness of endogenous IAA1, IAA2, and GH3.3 transcripts in wild-type and ibr5 seedlings. Although we detected subtle reductions in basal levels of IAA1 and IAA2 transcripts in ibr5 in several trials (data not shown), we did not detect dramatic differences in these experiments. In any case, IAA1, IAA2, and GH3.3 transcripts eventually reached similar maximal values in wild type and ibr5 (data not shown), suggesting that IBR5 is not required for full response to high auxin levels.
ibr5 does not accumulate an AXR3/IAA17 reporter protein
Gain-of-function mutations that stabilize any of several Aux/IAA proteins can confer dominant auxin resistance (reviewed in [18]). Moreover, Aux/IAA repressor proteins or Aux/IAA-reporter fusion proteins are stabilized in numerous other auxin-resistant mutants, including tir1 [16], axr1 [16], ecr1 [28], afb1, afb2, and afb3 [21], cul1 [41], eta2/cand1 [42], eta3/sgt1b [43], and aar1 [44].
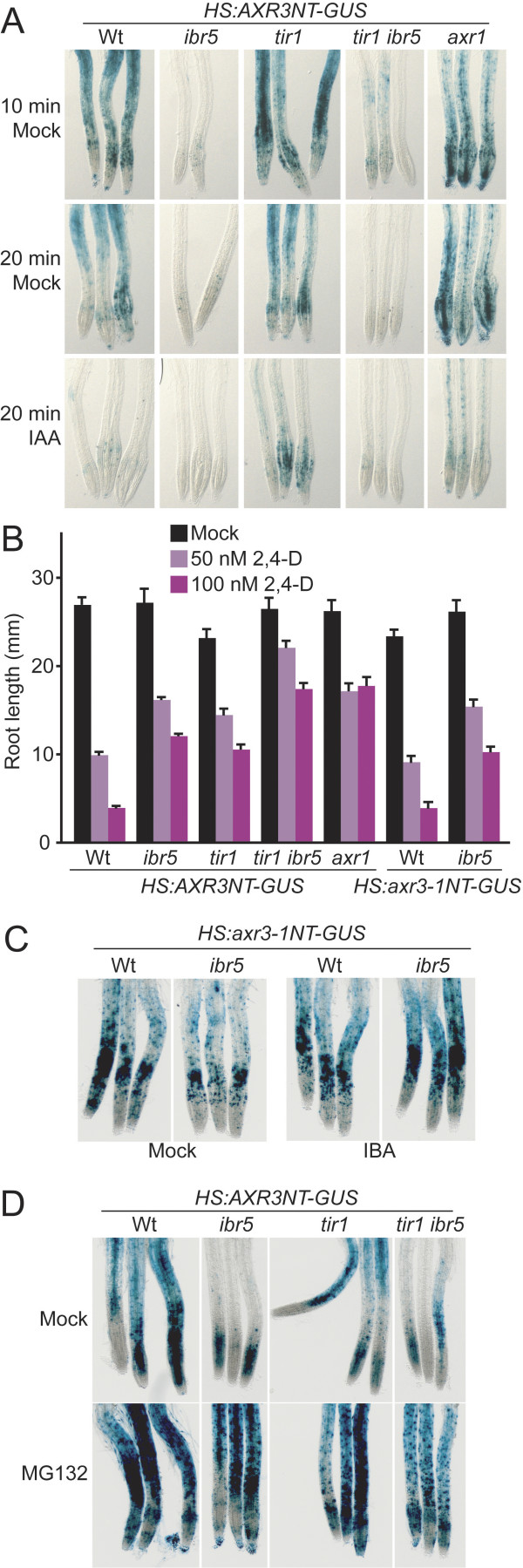
Because auxin-responsive transcripts are reduced in ibr5, we sought to analyze Aux/IAA stability in the ibr5 mutant. We crossed ibr5-1 to a line expressing the AXR3/IAA17 Aux/IAA protein N-terminal degron region fused to β-glucuronidase driven by a soybean heat-shock promoter (HS:AXR3NT-GUS; [16]). Eight-day-old seedlings were heat shocked to induce reporter transcription, and then either mock treated or auxin treated. In seedlings with intact auxin signaling, the auxin-induced disappearance of AXR3NT-GUS activity reflects the targeting of the reporter to the 26S proteasome for degradation [16]. Mutants with auxin-response defects, such as tir1 and axr1, show increased reporter activity after induction and reduced destabilization of AXR3NT-GUS upon auxin treatment [16], consistent with the axr1 defect in accumulating normal levels of auxin-responsive transcripts following auxin treatment [45,46]. Intriguingly, unlike in previously characterized auxin-response mutants, we did not detect increased AXR3NT-GUS activity in ibr5. In fact, AXR3NT-GUS appeared to be less active in ibr5 than in wild-type roots 10 or 20 minutes following heat shock (Figure 6A). This decrease in AXR3NT-GUS activity was also apparent in tir1 ibr5, suggesting that ibr5, although enhancing tir1 auxin resistance in root elongation (Figures 1C, D, and 6B), suppressed tir1 AXR3NT-GUS accumulation (Figure 6A). Response to 2,4-D for each of these lines was as expected, with tir1 ibr5 (HS:AXR3NT-GUS) showing enhanced resistance compared to the intermediate resistance of either parent (Figure 6B).
Figure 6.
ibr5 does not accumulate AXR3NT-GUS. (A) 8-day-old Col-0 (Wt), ibr5-1, tir1-1, tir1-1 ibr5-1, and axr1-3 seedlings carrying HS:AXR3NT-GUS [16] were heat shocked for 2 hours, treated with mock (ethanol) or 100 nM IAA for the indicated time, then stained for GUS activity. (B) Auxin-response defects of HS:AXR3NT-GUS lines. Lengths of primary roots of 8-day-old seedlings grown under yellow-filtered light at 22°C on medium supplemented with various concentrations of 2,4-D are shown. Error bars represent standard errors of the means (n = 20). (C) 8-day-old Col-0 (Wt) and ibr5-1 carrying HS:axr3-1NT-GUS [16] were heat-shocked for 2 hours, mock (ethanol) treated or treated with 10 μM IBA for 40 minutes, then stained for GUS activity. (D) 8-day-old Col-0 (Wt), ibr5-1, tir1-1, and tir1-1 ibr5-1 carrying HS:AXR3NT-GUS [16] were heat shocked for 2 hours. Midway through a 2-hour heat shock, DMSO (mock) or 50 μM MG132 treatment was initiated. Seedlings were stained for GUS activity 2 hours after return to room temperature. Separate experiments revealed that inclusion of DMSO during the heat shock (included as an MG132 carrier in panel D) resulted in more intense AXR3NT-GUS staining (L.C.S., unpublished), which could account for the higher apparent GUS activity in panel D when compared to panel A.
The reduced AXR3NT-GUS activity in ibr5 was apparent immediately following the 2-hour heat shock used to induce reporter expression (data not shown). Because a heat-responsive promoter drives the AXR3NT-GUS construct, we tested whether the lack of AXR3NT-GUS activity in ibr5 could be explained by a reduced transcriptional response to heat in the mutant. We introduced a construct altered to contain the stabilizing axr3-1 mutation (HS:axr3-1NT-GUS; [16]) into ibr5-1 by crossing. The proline to leucine substitution in axr3-1 [47] confers reporter stability by decreasing axr3-1NT-GUS interaction with SCFTIR1 [16]. We found similar axr3-1NT-GUS activity in wild type and ibr5 with or without auxin treatment (Figure 6C), suggesting that the transgenes were efficiently transcribed in response to the heat stimulus, and that the decreased AXR3NT-GUS activity in ibr5 was not caused by reduced transcription following heat shock. Moreover, treatment with the proteasome inhibitor MG132 restored AXR3NT-GUS activity in ibr5 and tir1 ibr5 to near wild-type levels (Figure 6D), again suggesting that the ibr5 defects in AXR3NT-GUS activity were not due to reduced transgene transcription.
ibr5 does not accumulate an IAA28 reporter protein
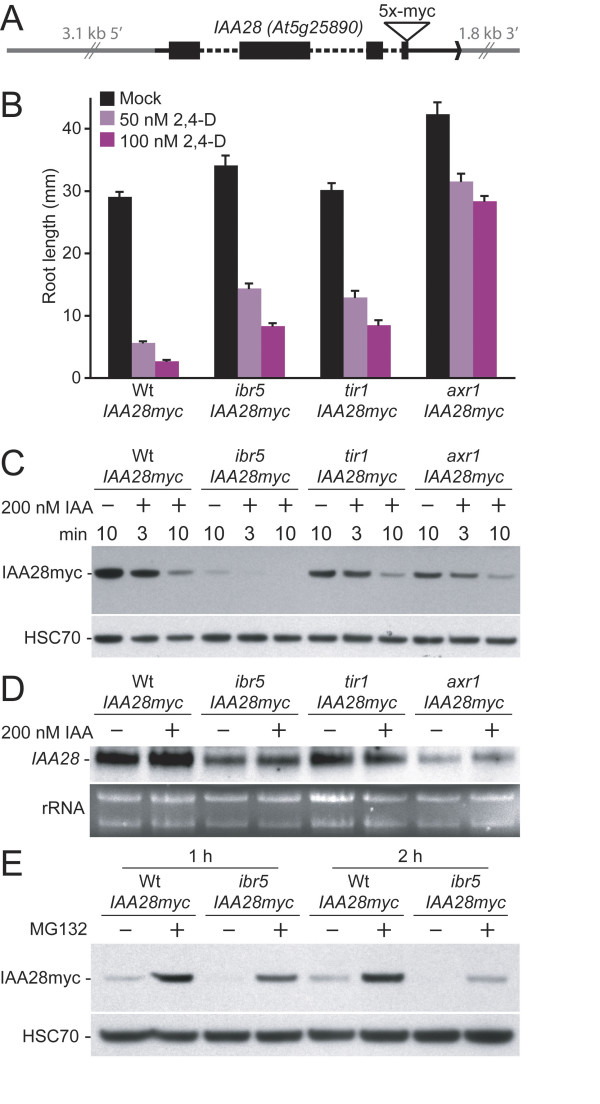
IAA28 was originally identified because the iaa28-1 gain-of-function mutation confers auxin resistance and impedes lateral root production [48]. Like typical Aux/IAA proteins, IAA28 has a transcriptional repressor domain [15] and can confer auxin-enhanced instability to a luciferase reporter [49]. To test whether the lack of AXR3NT-GUS stabilization in ibr5 was accompanied by stability effects on other Aux/IAA proteins, we crossed ibr5-1, tir1-1, and axr1-3 to a wild-type line carrying a c-Myc epitope-tagged version of IAA28 driven from IAA28 regulatory sequences (Figure 7A) and isolated homozygous mutants carrying the reporter transgene. These lines responded to 2,4-D as expected (Figure 7B). Because IAA28 is primarily expressed in roots [48], we examined accumulation of IAA28myc in wild-type and mutant roots following mock- or auxin-treatment of 10-day-old seedlings. As expected, we found that IAA28myc disappeared rapidly following auxin treatment in wild type (Figure 7C). We found lower IAA28myc levels in ibr5 than wild type in the absence of added auxin; this protein disappeared rapidly upon auxin treatment and was not detected after 10 minutes of treatment (Figure 7C). Although we had expected IAA28myc to be stabilized in tir1 and axr1, we found IAA28myc levels similar to wild-type levels that decreased in response to auxin treatment (Figure 7C) in both mutants, consistent with the observation that neither of these mutants is completely insensitive to auxin (e.g., Figure 7B). We examined IAA28 mRNA levels in these lines and found reduced IAA28 transcript levels in axr1 and, to a lesser extent, in tir1 (Figure 7D). Transcript levels were not perfectly correlated with IAA28 protein levels, suggesting differences in protein stability in these mutants. Treatment with MG132 increased IAA28myc protein levels in both wild type and ibr5 (Figure 7E), suggesting that IAA28myc is degraded via the 26S proteasome in wild type and consistent with the possibility that the proteasome contributes to the reduced IAA28myc levels in ibr5. However, the inability of MG132 to fully restore IAA28myc to wild-type levels in ibr5 (Figure 7E) suggests that the reduced IAA28 mRNA level in ibr5 (Figure 7D) contributes to the reduced IAA28myc accumulation in this mutant (Figure 7C). The striking lack of AXR3NT-GUS and IAA28myc stabilization in ibr5 is consistent with the possibility that auxin-regulated transcription is reduced in ibr5 via a mechanism that does not involve Aux/IAA protein stabilization.
Figure 7.
ibr5 does not accumulate IAA28myc. (A) An illustration of the IAA28myc construct. The region of the IAA28 transcript is shown in black, with introns designated with dashed lines and coding sequence with black boxes. (B) Auxin-response defects of indicated mutants carrying the IAA28myc construct. Lengths of primary roots of 8-day-old seedlings grown under yellow-filtered light at 22°C on medium supplemented with the indicated concentrations of 2,4-D are shown. Error bars represent standard errors of the means (n = 17). (C) IAA28myc accumulation in wild type and auxin-response mutants. Anti-myc (top panel; Santa Cruz Biotechnology) and anti-HSC70 antibodies (bottom panel; Stressgen Bioreagents) were used on immunoblots of protein prepared from roots of light-grown 10-day-old Col-0 (Wt), ibr5-1, tir1-1, and axr1-3 seedlings expressing IAA28myc that had been mock (ethanol) treated for 10 minutes or treated for 3 or 10 minutes with 200 nM IAA. (D) IAA28 mRNA accumulation in wild type and auxin response mutants. Total RNA from seedlings that had been mock (ethanol) treated or treated with 200 nM IAA for 10 minutes was separated by electrophoresis (bottom panel, ethidium bromide-stained gel), transferred to a membrane, and probed with an IAA28 probe (top panel). IAA28 and IAA28myc transcripts are not resolved from one another, and therefore are seen as a single band. (E) IAA28myc accumulation in response to MG132 treatment. Anti-myc (top panel) and anti-HSC70 (bottom panel) antibodies were used on immunoblots of protein prepared from 3-day-old light-grown Col-0 (Wt) and ibr5-1 seedlings expressing IAA28myc that had been mock (DMSO) treated or treated with 300 μM MG132 for 1 or 2 hours.
An IBR5 substitution variant (IBR5C129S) does not fully rescue ibr5 defects
Dual specificity protein phosphatase (DSP) proteins dephosphorylate both threonine and tyrosine residues of phosphorylated proteins, often thereby inactivating them (reviewed in [50]). DSP proteins contain a conserved aspartate residue and a separate, highly conserved signature motif of VxVHCx2GxSRSx5AYLM, with the cysteine and arginine residues participating with the conserved aspartate in catalysis. The cysteine of this signature begins the dephosphorylation process with a nucleophilic attack on the phosphorus atom of the phosphotyrosine or phosphothreonine substrate. Thus, disruption of this conserved cysteine results in catalytic inactivity in many DSP proteins (reviewed in [51]), including the IBR5 relative, DsPTP1 [52].
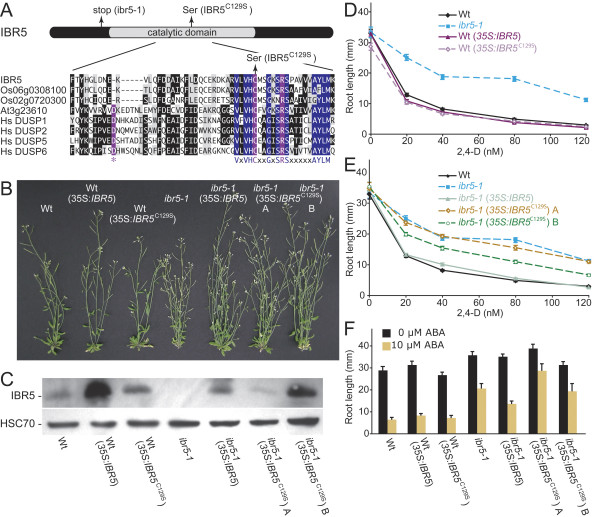
The DSP active site motif VxVHCx2GxSRSx5AYLM is present in IBR5 (Figure 8A), allowing us to identify the presumptive active site cysteine (C129) in IBR5. To test whether IBR5 phosphatase activity is required for normal auxin responses in vivo, we generated transgenic Wt and ibr5-1 lines expressing a Cys129 → Ser129 (C129S) substitution variant of IBR5 (IBR5C129S) under the control of the strong 35S viral promoter. We anticipated that 35S:IBR5C129S would not rescue ibr5-1 defects if IBR5 phosphatase activity were required to promote auxin responsiveness. We assayed ibr5-1 (35S:IBR5C129S) lines expressing low (line A) and high (line B) IBR5C129S levels (Figure 10C) for mutant phenotype rescue. Wild-type and ibr5-1 lines overexpressing unmodified IBR5 (35S:IBR5; [37]) were used for comparison.
Figure 8.
An IBR5C129S substitution variant does not fully rescue ibr5 defects. (A) A schematic showing the positions of the ibr5-1 premature stop codon relative to the conserved catalytic domain and an alignment of part of the phosphatase catalytic domains of Arabidopsis IBR5 and several putative and confirmed DSP proteins. Sequences shown are the closest IBR5 homologs from rice (Os06g0308100 and Os02g0720300), an IBR5 relative from Arabidopsis with demonstrated DSP activity (At3g23610/DsPTP1; [52]) and three human (Hs) DSP enzymes. Sequences were aligned with the MegAlign program (DNAStar, Madison, WI) using the CLUSTAL W method. Catalytic residues are shaded in purple, conserved DSP signature residues are shaded in blue, residues identical in at least four sequences are shaded in black, similar residues are shaded in gray, and dashes indicate gaps introduced to maximize alignment. (B) Six-week-old Col-0 (Wt), Wt (35S:IBR5), Wt (35S:IBR5C129S), ibr5-1, ibr5-1 (35S:IBR5), ibr5-1 (35S:IBR5C129S) line A, and ibr5-1 (35S:IBR5C129S) line B grown in continuous light are shown. (C) Immunoblot analysis with an anti-IBR5 antibody [37]; top panel) and anti-HSC70 antibody (Stressgen Bioreagents) of protein prepared from 2-day-old seedlings of the lines shown in panel B. Positions of IBR5 and HSC70 are indicated at left. (D) Wt (35S:IBR5) and Wt (35S:IBR5C129S) display similar 2,4-D response as Wt. Lengths of primary roots of 8-day-old seedlings grown under yellow-filtered light at 22°C on medium supplemented with various concentrations of 2,4-D are shown. Error bars represent standard errors of the means (n ≥ 18). (E) ibr5-1 (35S:IBR5C129S) line A, and ibr5-1 (35S:IBR5C129S) line B fail to fully rescue ibr5-1 2,4-D resistance. Seedlings were measured as in (D). Error bars represent standard errors of the means (n ≥ 17). (F) Length of primary roots 4 days after transfer of 4-day-old seedlings to either 0 (ethanol control) or 10 μM ABA medium. Error bars represent standard errors of the means (n ≥ 8).
We found that IBR5 or IBR5C129S overexpression did not noticeably alter wild-type adult morphology (Figure 8B). Moreover, IBR5 or IBR5C129S overexpression in wild type did not alter any of the hormone-response phenotypes examined (Figure 8D, F). As expected, we found that ibr5-1 (35S:IBR5) plants were restored to wild-type height (Figure 8B) and had normal root responses to auxin and ABA (Figure 8E, F). ibr5-1 (35S:IBR5C129S) line A (low expressor) exhibited restored plant height but had similar leaf epinasty to ibr5-1, whereas ibr5-1 (35S:IBR5C129S) line B (high expressor) displayed rescue of ibr5-1 plant height (Figure 8B) and partial rescue of leaf epinasty (data not shown). Resistance to the inhibitory effects of 2,4-D and ABA on root elongation was not rescued in ibr5-1 (35S:IBR5C129S) line A and only partially restored in ibr5-1 (35S:IBR5C129S) line B (Figures 8E, F). The lack of full ibr5-1 rescue by IBR5C129S is consistent with the possibility that IBR5 phosphatase activity is required for full auxin and ABA responsiveness. The partial ibr5-1 rescue observed when IBR5C129S accumulates to high levels suggests that certain IBR5 functions do not require phosphatase activity. For example, IBR5C129S may bind and sequester its substrate(s), thereby dampening normal substrate activity.
Discussion
Loss-of-function mutations in IBR5, which encodes a putative dual-specificity protein phosphatase (DSP), result in decreased auxin and abscisic acid responses [37]. DSP proteins often regulate mitogen-activated protein kinase (MAPK) proteins. Arabidopsis has 20 predicted MAPK proteins [53] but only five predicted DSP proteins, suggesting that some DSP enzymes may regulate more than one MAPK. In Arabidopsis, two of the five predicted Arabidopsis DSP proteins have been demonstrated to regulate MAPK activity. DsPTP1 (At3g23610) dephosphorylates MPK4 [52], and MAPK PHOSPHATASE2 (MKP2; At3g06110) dephosphorylates MPK3 and MPK6 [54].
Although a MAPK regulated by IBR5 has not been reported, MAPK signaling has been implicated in both auxin and ABA responses, and both of these pathways are defective in ibr5. For example, transient expression in protoplasts of constitutively active MAPK kinase kinase protein ANP1 or the tobacco homolog NPK1 results in decreased auxin-responsive transcription and activation of AtMPK3 and AtMPK6 [55,56], the targets of the MKP2 phosphatase [54]. Additionally, auxin treatment activates a ~44-kD MAPK in Arabidopsis [57]. The failure of IBR5C129S to fully restore ibr5-1 mutant phenotypes indicates that IBR5 phosphatase activity is required for full auxin and ABA responsiveness, and it will be interesting to learn if any of the MAP kinases implicated in hormone responsiveness are IBR5 substrates. The partial rescue of ibr5 defects observed when IBR5C129S was overexpressed might result from the IBR5C129S protein binding to and thus sequestering IBR5 substrate(s), as has been suggested for overexpression of the catalytically inactive MAPK phosphatase Pyp1C470S, which results in a phenotype similar to a loss-of-function allele of the substrate MAPK Spc1 in Schizosaccharomyces pombe [58,59].
In an effort to clarify the pathways through which IBR5 affects auxin responses, we examined genetic interactions between ibr5 and the auxin-response mutants tir1, axr1, and aux1. The TIR1 F-box protein acts with Aux/IAA proteins as an auxin receptor [19,20,22]. tir1 appeared to enhance all ibr5 auxin-related physiological phenotypes examined, including response defects to applied natural and synthetic auxins (Figure 1C, D) and auxin transport inhibitors (data not shown). In addition, the tir1 ibr5 mutant displayed a longer primary root (Figures 1C, D) and fewer lateral roots than either single mutant on unsupplemented medium (Figure 2A), suggesting that the double mutant has enhanced resistance to endogenous auxin as well.
In addition to influencing auxin responses, IBR5 modulates certain ABA responses [37]. We found not only that tir1 enhanced ibr5 ABA resistance, but also that tir1 itself exhibited substantial ABA resistance in the root elongation assay (Figure 2C). Although tir1 has not previously been reported to be ABA resistant, it has been characterized as glucose resistant [60], a phenotype common to many ABA-resistant mutants (reviewed in [61,62]). Because tir1 ibr5 displays enhanced resistance to both auxin and ABA, and because the ibr5-1 allele is a likely null [37], these results are consistent with the possibility that TIR1 and IBR5 promote auxin and ABA responsiveness independently of one another.
AXR1 is a subunit of the RUB-activating enzyme involved in RUB modification of CULLIN proteins. CULLIN is the backbone for the over 600 putative SCF complexes in Arabidopsis [63], including SCFTIR1 [64]; therefore, axr1 mutants have pleiotropic phenotypes, including auxin resistance. The only examined phenotypes that appeared to be additive in axr1 ibr5 were the long primary root on unsupplemented media and aberrant vascular development (Figure 3). Unlike ibr5, tir1-1 enhances the root elongation resistance of axr1-12 to 2,4-D [38]. The pleiotropic nature of the axr1 defect and the extreme auxin resistance of the axr1 single mutant complicate the interpretation of our results. Regardless, it is interesting that axr1 and ibr5 mutants, which are both less sensitive to the inhibitory effects of auxin on root elongation, seem to have opposite effects on AXR3NT-GUS stability (Figure 6A; [16]).
We also examined double mutants of ibr5 with aux1, a mutant defective in an auxin influx carrier [32,34,35]. The shoots of the aux1 ibr5 mutant resembled ibr5, whereas the roots of aux1 ibr5 were most similar to aux1. Because aux1 lacks shoot phenotypes and most aux1 root phenotypes are more dramatic than ibr5 root defects, these results are consistent with results expected from additive defects, although the extreme resistance of aux1 to 2,4-D and ABA prevented us from determining whether aux1 ibr5 had additive defects in response to these hormones.
ibr5 has decreased DR5:GUS activity when grown on unsupplemented media [37], and in this study we found defects in auxin-induced DR5:GUS activity in particular tissues (Figure 5). However, microarray analysis of mRNA accumulation in 7-day-old ibr5 and wild-type seedlings did not reveal any dramatic (> 2.5-fold) alterations in transcripts represented in the analysis [37]. These results suggested that any gene expression changes in ibr5 might be subtle or local, as in the DR5:GUS analysis, and therefore not apparent in whole seedling RNA.
Because auxin-response transcripts are regulated by Aux/IAA protein stability, we sought to analyze Aux/IAA degradation in ibr5. Auxin promotes Aux/IAA protein degradation by mediating interaction of these repressors with SCFTIR1 [19-21,65]. Several auxin-resistant mutants exhibit stabilized Aux/IAA proteins or Aux/IAA reporters, including mutants defective in the auxin receptors TIR1 [16] and the related AFB1, AFB2, and AFB3 F-box proteins [21]; other SCF components such as CUL1 [41]; and proteins that modify SCF activity, such as AXR1 [16], ECR1 [28], ETA3/SGT1b [43], and ETA2/CAND1 [42]. This stabilization probably accounts for the reduced levels of auxin-responsive transcripts reported in many of these mutants [21,25,42,45,46,66]. Like previously characterized auxin-response mutants, ibr5 exhibits reduced accumulation of the auxin-responsive DR5:GUS reporter. Unlike these other mutants, however, AXR3NT-GUS activity was not increased in ibr5. Moreover, IAA28myc protein was less abundant in ibr5, which is not expected if Aux/IAA proteins are generally stabilized, also suggesting that IBR5 modulates auxin-responsive transcription without stabilizing Aux/IAA proteins. Of course, there are 29 Arabidopsis members of the Aux/IAA family [7], and it remains possible that ibr5 specifically stabilizes certain Aux/IAA family members to reduce transcriptional activation without affecting AXR3/IAA17 or IAA28 stability.
Regardless of whether Aux/IAA proteins that remain to be assessed turn out to be stabilized in ibr5, ibr5 is the only examined auxin-response mutant [16,21,28,41-44] that does not exhibit AXR3NT-GUS stabilization. This phenotypic bifurcation may be useful in dissecting the roles of additional auxin-response mutants. Recent experiments suggest that the MYB77 transcription factor promotes auxin responses and can dimerize with ARF proteins [67]. It will be interesting to learn whether Aux/IAA proteins are stabilized in the myb77 mutant and whether IBR5 regulates this or some other factor needed to promote auxin-responsive transcription once the Aux/IAA repressors have been degraded. One of many possible scenarios is that IBR5 normally dephosphorylates and inactivates a MAPK that negatively regulates a transcription factor needed for auxin responses, providing a mechanism to fine-tune auxin responses without modulating Aux/IAA stability.
Conclusion
IBR5 resembles dual-specificity phosphatases, and in this work we provide evidence that IBR5 phosphatase activity is necessary for full auxin and ABA responsiveness. Analysis of double mutants between ibr5-1 and several other auxin-response mutants revealed that IBR5 appears to affect auxin responses independently of the TIR1 auxin receptor. Because transcriptional repression of auxin-responsive genes is relieved by Aux/IAA protein degradation, we examined the stability of two Aux/IAA reporters in ibr5 and found that these proteins were not stabilized in ibr5, suggesting that IBR5 acts downstream of auxin recognition by the SCFTIR1/AFB-Aux/IAA complexes. Future determination of IBR5 substrates may allow a more detailed understanding of how this apparent dual-specificity phosphatase is able to promote auxin responses without destabilizing Aux/IAA proteins.
Methods
Plant materials and growth conditions
Arabidopsis thaliana accession Colombia (Col-0) was the wild type used for all experiments. The ibr5-1 mutant contains a nonsense mutation at IBR5 amino acid 42, resulting in a truncated product lacking the catalytic domain [37]. The aux1-7 mutant contains a missense mutation resulting in glycine 459 being replaced by aspartic acid [32]. The tir1-1 mutant contains a missense mutation resulting in glycine 147 being replaced by an aspartic acid [38]. The axr1-3 mutant contains a missense mutation resulting in cysteine 154 being replaced by a tyrosine [26].
Surface-sterilized [68] seeds were plated on PNS (plant nutrient medium with 0.5% [w/v] sucrose) [69] solidified with 0.6% (w/v) agar. Hormones used were from 0.1-, 1.0-, or 100-mM stocks in ethanol, with ethanol-supplemented media used as controls, and all treatments normalized to the same ethanol content (less than 0.1 μL ethanol/mL medium). Seedlings were grown at 22°C under continuous light. Unless indicated otherwise, plates were incubated under yellow long-pass filters to slow the breakdown of indolic compounds [70]. Plants were grown in soil (Metromix 200; Scotts, Marysville, OH) at 22 to 25°C under continuous illumination by cool-white fluorescent bulbs (Sylvania, Danvers, MA).
IBR5C129S construct
The pKSIBR5c construct [37] was mutated using oligonucleotide-directed mutagenesis [71] to alter the presumptive catalytic cysteine at amino acid position 129 to a serine using a primer 5'-CTTTCCCAGACATCGAATGCACAAGAAC-3' (altered residues underlined) designed to contain a Taq1α restriction site. The mutant cDNA was then excised using NotI and subcloned into the plant transformation vector 35SpBARN [72] between the Cauliflower mosaic virus 35S promoter and the nos terminator. The 35S:IBR5C129S plasmid was electroporated [71] into Agrobacterium tumefaciens GV3101, which was used to transform wild-type Col-0 and ibr5-1. Transformants were identified on PNS medium supplemented with 10 μg/mL BASTA (glufosinate-ammonium) after 10 days under white light. Homozygous lines were identified in subsequent generations by examining the pattern of BASTA resistance.
IAA28myc construct
The IAA28:IAA28myc construct [73] contains the IAA28 genomic region, including 3.1 kb of DNA 5' of the IAA28 coding sequence, subcloned into the pBIN19 plant transformation vector. The last exon of IAA28 has been modified in this construct to encode five copies of the c-Myc epitope immediately upstream of the IAA28 termination codon. The IAA28:IAA28myc plasmid was electroporated [71] into Agrobacterium tumefaciens GV3101, which was used to transform wild-type Col-0. Transformed seedlings were identified on PN medium supplemented with 12 μg/ml kanamycin after growth under white light. Homozygous single-insert lines were identified by examining the pattern of kanamycin resistance in subsequent generations. ibr5-1, tir1-1, and axr1-3 were crossed to a wild-type (Col-0) single-insert line to generate ibr5, tir1, and axr1 carrying the IAA28:IAA28myc construct.
Phenotypic analyses
All assays were conducted at least three times with similar results. For auxin-response root-elongation assays, seedlings were grown for 8 or 9 days on PNS with the indicated auxin concentrations and removed from the agar, and the lengths of the primary roots were measured. For ABA response root-elongation assays, seedlings were grown for 4 days on PNS to allow efficient germination, then were transferred to PNS supplemented with either ethanol or ABA. After an additional 4 days of growth, primary root lengths were measured.
In lateral root assays, seedlings were grown for 4 days on PNS, transferred to PNS supplemented with either ethanol or 10 μM IBA, and grown for an additional 4 days, after which lateral roots were examined under a dissecting microscope. Primordia emerged from the primary root were counted as lateral roots.
For hypocotyl elongation assays, seeds were plated on media supplemented with either ethanol or 20 μM IBA. After 1 day in the light, plates were wrapped with aluminum foil and incubated for an additional 4 days in the dark, after which seedlings were removed from the agar and hypocotyls lengths were measured.
To examine cotyledon vascular patterns, seedlings were grown for 8 days on PNS. Chlorophyll was removed using an ethanol series, and seedlings were cleared by incubating for one week at room temperature in chloral hydrate solution (80 g chloral hydrate, 20 mL glycerol, and 10 mL water). Cleared seedlings were mounted and photographed through a dissecting microscope.
Double mutant isolation
The ibr5-1 mutant was crossed to aux1-7 [39], axr1-3 [30], and tir1-1 [38], all in the Col-0 accession. Double mutants were identified by PCR analysis of F2 plants. Amplification of AUX1 with AUX1-3 (5'-CATGGGTCAACAAAGCTTTGGATTTTGTCC-3') and AUX1-4 (5'-TTCGTGACTTTTACTCCCTTCACGTATACG-3') yields a 464-bp product with two DpnII restriction sites in wild type and three in aux1-7. Amplification of AXR1 with the derived cleaved amplified polymorphic sequence primer [74,75] AXR1-Acc1 (5'-AAACCAACTTAACGTTTGCATGTCG-3'; altered residue underlined) and AXR1-15 (5'-TCTCATATGTACTTTTCCTCGTCCTCTTCAC-3') yields a 185-bp product with one Acc1 restriction site in wild type and none in axr1-3. Amplification of TIR1 with TIR1-3 (5'-TTGAAGAGATAAGGCTGAAGAGGATGG-3') and TIR1-4 (5'-TACACCACCGTTAAATAAGACCCACCAGAAAG-3') yields a 488-bp product with one DpnII restriction site in wild type and two restriction sites in tir1-1. PCR-based identification of ibr5-1 was as described previously [37].
Northern analysis
Surface-sterilized [68] seeds were plated on filter paper-lined PNS and grown under continuous illumination. After 6 days, the filter paper with seedlings was lifted off the agar surface. Seedlings were floated in liquid PN supplemented with mock (ethanol) or 200 nM IAA for 10 minutes. Seedlings were collected and ground with a mortar and pestle in liquid nitrogen. RNA was extracted using TriReagent (Sigma) according to the manufacturer's instructions. Total RNA (10 μg) was electrophoresed on a 1% agarose gel containing 0.37 M formaldehyde [71] and transferred to a positively-charged nylon 66 membrane (Roche Applied Science, Indianapolis, IN) in 20× SSC using capillary action. DIG-labeled probe was hybridized in DIG Easy Hyb buffer (Roche) overnight at 50°C and washed at moderate stringency according to the manufacturer's instructions. Probes were detected using an alkaline phosphatase-conjugated anti-DIG antibody (Roche) diluted 1:20,000 in Blocking Buffer (Roche), then visualized using a 1:200 dilution of CDP-Star (Roche).
DIG-labeled IAA28 probe was synthesized by PCR amplification using a PCR DIG Probe Synthesis Kit (Roche) according to the manufacturer's instructions from a cDNA template [48] using T1N24-20 (5'-CCATCGAACTGATGATTTTGGCC-3') and T1N24-21 (5'-CCTCCTTGTCACCAATTCACTTCC-3'), yielding a 525-bp product.
Immunoblot analysis
To visualize IBR5, protein was extracted from entire 2-day-old seedlings grown in 0.1% agar under white light by grinding frozen tissue with a pestle and adding one volume NuPAGE 2X LDS buffer (Invitrogen, Carlsbad, CA). Debris was pelleted by centrifugation for 4 minutes. The supernatant was heated to 100°C for 5 minutes. Protein extracts were separated by SDS-polyacrylamide gel electrophoresis beside Cruz markers (Santa Cruz Biotechnology, Santa Cruz, CA) using a NuPAGE 10% Bis-Tris gel and MES running buffer (Invitrogen).
Protein was transferred for 35 min at 24 V to a Hybond ECL nitrocellulose membrane (Amersham Pharmacia Biotech, Piscataway, NJ) using NuPAGE transfer buffer (Invitrogen). After blocking for 1 hour in 8% powdered milk in Tween Tris-buffered saline (TTBS; [71]), the membrane was incubated overnight at 4°C with affinity purified IBR5 antibody [37] diluted 1:20 in blocking buffer. The membrane was then washed three times with TTBS and incubated with HRP-conjugated goat anti-rabbit secondary antibody (Santa Cruz Biotechnology) diluted 1:2000 for 1 hour, washed again as described before, then visualized using LumiGLO reagent (Cell Signaling, Beverly, MA). Membranes subsequently were incubated with an antibody against spinach (Spinacia oleracea) HSC70 (Stressgen Bioreagents SPA-817) diluted 1:5000, followed by HRP-conjugated goat anti-mouse secondary antibody (Santa Cruz Biotechnology) diluted 1:2000, and visualized using LumiGLO reagent (Cell Signaling).
To visualize IAA28myc, 10-day-old light-grown seedlings were removed from PNS medium and floated in liquid PN supplemented with either ethanol (mock) or 200 nM IAA. At the indicated time points, roots were excised and protein was extracted. For MG132 treatment, 3-day-old light-grown seedlings were removed from PNS medium and suspended in water containing either DMSO (mock) or 300 μM MG132. After 1 or 2 hours, protein was extracted from whole seedlings. In both experiments, protein was separated by SDS-polyacrylamide gel electrophoresis and transferred to a membrane as described above. After blocking for 1 hour in 8% powdered milk in TTBS, the membrane was incubated overnight at 4°C with monoclonal 9E10 anti-c-Myc antibody (Santa Cruz Biotechnology SC-40) diluted 1:500 and anti-HSC70 antibody (Stressgen Bioreagents SPA-817) diluted 1:250,000 in blocking buffer. The membrane was then washed three times with TTBS and incubated with HRP-conjugated goat anti-mouse secondary antibody (Santa Cruz Biotechnology) diluted 1:2000 for 4 hours, washed again as described before, then visualized using LumiGLO reagent (Cell Signaling).
HS:AXR3NT-GUS analysis
Wild-type (Col-0) lines carrying HS:AXR3NT-GUS and HS:axr3-1NT-GUS and axr1-3 carrying HS:AXR3NT-GUS were described previously [16]. To obtain additional lines carrying these reporters, ibr5-1 was crossed to wild type carrying HS:AXR3NT-GUS and to wild type carrying HS:axr3-1NT-GUS. tir1 ibr5 was crossed to wild type carrying HS:AXR3NT-GUS to obtain tir1-1 HS:AXR3NT-GUS and tir1 ibr5 HS:AXR3NT-GUS. Mutants were identified by PCR analysis of the F2 plants, as described above. The presence of HS:AXR3NT-GUS or HS:axr3-1NT-GUS was assayed by resistance to kanamycin and confirmed by GUS staining.
For histochemical assays, 8-day-old light-grown seedlings were removed from PNS plates and floated in 0.5 mL liquid PN medium diluted six-fold with water (for hormone response assays) or sterile water (for MG132 assays) contained in a 12-well plate. Two milliliters of prewarmed (37°C) one-sixth liquid PN or prewarmed water was added to each well, and the plates were incubated at 37°C for 2 hours. For the hormone response assays, seedlings were transferred to room temperature for 20 minutes prior to 10- and 20-minute mock (ethanol) or 100 nM IAA treatments, or 40-minute mock (ethanol) or 10 μM IBA treatment. For MG132 assays, seedlings were incubated in the dark, and either DMSO (mock) or MG132 (to 50 μM) was added to each well 1 hour into the heat treatment. After heat treatment, plates were transferred to room temperature for 2 hours. Seedlings were stained for GUS activity as previously described [76].
Authors' contributions
LCS isolated and characterized double mutants, analyzed transcripts, performed GUS experiments, IAA28 western analyses, and characterized the phenotypes of 35S:IBR5C129S lines. MMA made 35S:IBR5C129S constructs, transformed them into plants, and performed IBR5 western analyses. BB conceived and coordinated the study. All authors participated in drafting and editing the manuscript, and read and approved the final manuscript.
Supplementary Material
Normalized ibr5 double mutant auxin response defects. The auxin responses of ibr5 and double mutants are represented after normalization to root lengths of mock-treated seedlings.
Normalized auxin-response mutant defects in lateral root induction by IBA, hypocotyl elongation inhibition by IBA, and root elongation inhibition by ABA. The IBA and ABA responses of ibr5 and double mutants are represented after normalization to mock-treated seedlings.
Acknowledgments
Acknowledgements
We thank Luise Rogg for wild type transformed with the IAA28:IAA28myc construct, Ottoline Leyser and Stefan Kepinski for wild-type and axr1-3 lines carrying HS:AXR3NT-GUS and HS:axr3-1NT-GUS, Natasha Raikhel for the c-Myc epitope-tagging plasmid, Mark Estelle for axr1-3 and aux1-7, the ABRC for tir1-1 and cDNA clones, Mary Ellen Lane for microscope use, Kristen Rogers and Erin Beisner for technical assistance, and Matthew Lingard, Naxhiely Martinez, and Andrew Woodward for critical comments on the manuscript. This research was supported by the National Science Foundation (IBN-0315596), the Robert A. Welch Foundation (C-1309), and the National Institutes of Health (F32-GM075689 to LCS).
Contributor Information
Lucia C Strader, Email: strader@rice.edu.
Melanie Monroe-Augustus, Email: melaniem@rice.edu.
Bonnie Bartel, Email: bartel@rice.edu.
References
- Davies PJ. Introduction. In: Davies PJ, editor. Plant Hormones. 2nd. Dordrecht , Kluwer Academic Publishers; 1995. pp. 1–38. [Google Scholar]
- Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Ann Bot. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JC, Key JL. Isolation of cloned cDNAs to auxin-responsive poly(A)+RNAs of elongating soybean hypocotyl. Proc Natl Acad Sci USA. 1982;79:7185–7189. doi: 10.1073/pnas.79.23.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Kleinschmidt A, Guilfoyle T. Auxin-regulated gene expression in intact soybean hypocotyl and excised hypocotyl sections. Planta. 1984;162:147–153. doi: 10.1007/BF00410211. [DOI] [PubMed] [Google Scholar]
- Abel S, Oeller PW, Theologis A. Early auxin-induced genes encode short-lived nuclear proteins. Proc Natl Acad Sci USA. 1994;91:326–330. doi: 10.1073/pnas.91.1.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil P, Liu Y, Orbovi´c V, Verkamp E, Poff KL, Green PJ. Characterization of the auxin-inducible SAUR-AC1 gene for use as a molecular genetic tool in Arabidopsis. Plant Physiol. 1994;104:777–784. doi: 10.1104/pp.104.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, Theologis A. Early genes and auxin action. Plant Physiol. 1996;111:9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure BA, Hagen G, Brown CS, Gee MA, Guilfoyle TJ. Transcription, organization, and sequence of an auxin-regulated gene cluster in soybean. Plant Cell. 1989;1:229–239. doi: 10.1105/tpc.1.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Wong LM, Theologis A. Identification of the auxin-responsive element, AuxRE, in the primary indoleacetic acid-inducible gene, PS-IAA4/5, of pea (Pisum sativum) J Mol Biol. 1993;233:580–596. doi: 10.1006/jmbi.1993.1537. [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Ulmasov T, Hagen G. The ARF family of transcription factors and their role in plant hormone-responsive transcription. Cell Mol Life Sci. 1998;54:619–627. doi: 10.1007/s000180050190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci USA. 1999;96:5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15:533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. Plant J. 1999;19:309–319. doi: 10.1046/j.1365-313X.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell. 2004;16:533–543. doi: 10.1105/tpc.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCFTIR1-dependent degradation of Aux/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- Zenser N, Ellsmore A, Leasure C, Callis J. Auxin modulates the degradation rate of Aux/IAA proteins. Proc Natl Acad Sci USA. 2001;98:11795–11800. doi: 10.1073/pnas.211312798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW. Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 2001;6:420–425. doi: 10.1016/S1360-1385(01)02042-8. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. Aux/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell. 2001;13:2809–2822. doi: 10.1105/tpc.13.12.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Estelle M. The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc Natl Acad Sci USA. 1999;96:15342–15347. doi: 10.1073/pnas.96.26.15342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Dharmasiri S, Hellmann H, Walker L, Gray WM, Estelle M. AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis Cullin AtCUL1 is required for auxin response. Plant Cell. 2002;14:421–433. doi: 10.1105/tpc.010282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser HMO, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature. 1993;364:161–164. doi: 10.1038/364161a0. [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Timpte C, Tan S, Callis J, Estelle M. The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science. 1998;280:1760–1763. doi: 10.1126/science.280.5370.1760. [DOI] [PubMed] [Google Scholar]
- Woodward AW, Ratzel SE, Woodward EE, Shamoo Y, Bartel B. Mutation of E1-CONJUGATING ENZYME-RELATED1 decreases RELATED TO UBIQUITIN conjugation and alters auxin response and development. Plant Physiol. 2007;144:976–987. doi: 10.1104/pp.107.100404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 1990;2:1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle MA, Somerville C. Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol Gen Genet. 1987;206:200–206. doi: 10.1007/BF00333575. [DOI] [Google Scholar]
- Vieten A, Sauer M, Brewer PB, Friml J. Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci. 2007;12:160–168. doi: 10.1016/j.tplants.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schultz B, Feldmann KA. Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science. 1996;273:948–950. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Yamamoto KT. Differential effects of 1-naphthaleneacetic acid, indole-3-acetic acid and 2,4-dichlorophenoxyacetic acid on the gravitropic response of roots in an auxin-resistant mutant of Arabidopsis, aux1. Plant Cell Physiol. 1998;39:660–664. doi: 10.1093/oxfordjournals.pcp.a029419. [DOI] [PubMed] [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 1999;18:2066–2073. doi: 10.1093/emboj/18.8.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E. High-affinity auxin transport by the AUX1 influx carrier protein. Curr Biol. 2006;16:1123–1127. doi: 10.1016/j.cub.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Zolman BK, Yoder A, Bartel B. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics. 2000;156:1323–1337. doi: 10.1093/genetics/156.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe-Augustus M, Zolman BK, Bartel B. IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell. 2003;15:2979–2991. doi: 10.1105/tpc.017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 1998;12:198–207. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher EP, Martindale SJB. Mutants of Arabidopsis thaliana with altered responses to auxins and gravity. Biochem Genet. 1980;18:1041–1053. doi: 10.1007/BF00484337. [DOI] [PubMed] [Google Scholar]
- Rashotte AM, Poupart J, Waddell CS, Muday GK. Transport of the two natural auxins, indole-3-butyric acid and indole-3-acetic acid, in Arabidopsis. Plant Physiol. 2003;133:761–772. doi: 10.1104/pp.103.022582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Ito H, Zhang W, Gray WM. Characterization of a novel temperature-sensitive allele of the CUL1/AXR6 subunit of SCF ubiquitin-ligases. Plant J. 2005;43:371–383. doi: 10.1111/j.1365-313X.2005.02449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang HW, Zhang W, Gray WM. Arabidopsis ETA2, an apparent ortholog of the human cullin-interacting protein CAND1, is required for auxin responses mediated by the SCFTIR1 ubiquitin ligase. Plant Cell. 2004;16:1883–1897. doi: 10.1105/tpc.021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Muskett PR, Chuang HW, Parker JE. Arabidopsis SGT1b is required for SCFTIR1-mediated auxin response. Plant Cell. 2003;15:1310–1319. doi: 10.1105/tpc.010884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Nakasone A, Chhun T, Ooura C, Biswas KK, Uchimiya H, Tsurumi S, Baskin TI, Tanaka A, Oono Y. A small acidic protein 1 (SMAP1) mediates responses of the Arabidopsis root to the synthetic auxin 2,4-dichlorophenoxyacetic acid. Plant J. 2006;47:788–801. doi: 10.1111/j.1365-313X.2006.02832.x. [DOI] [PubMed] [Google Scholar]
- Abel S, Nguyen MD, Theologis A. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol. 1995;251:533–549. doi: 10.1006/jmbi.1995.0454. [DOI] [PubMed] [Google Scholar]
- Timpte C, Lincoln C, Pickett FB, Turner J, Estelle M. The AXR1 and AUX1 genes of Arabidopsis function in separate auxin-response pathways. Plant J. 1995;8:561–569. doi: 10.1046/j.1365-313X.1995.8040561.x. [DOI] [PubMed] [Google Scholar]
- Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O. Changes in auxin response from mutations in an AUX/IAA gene. Science. 1998;279:1371–1373. doi: 10.1126/science.279.5355.1371. [DOI] [PubMed] [Google Scholar]
- Rogg LE, Lasswell J, Bartel B. A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell. 2001;13:465–480. doi: 10.1105/tpc.13.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher KA, Brown J, Saw RE, Callis J. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell. 2006;18:699–714. doi: 10.1105/tpc.105.039172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq A, Zhou MM. Structure and regulation of MAPK phosphatases. Cell Signal. 2004;16:769–779. doi: 10.1016/j.cellsig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Neel BG, Tonks NK. Protein tyrosine phosphatases in signal transduction. Curr Opin Cell Biol. 1997;9:193–204. doi: 10.1016/S0955-0674(97)80063-4. [DOI] [PubMed] [Google Scholar]
- Gupta R, Huang Y, Kieber J, Luan S. Identification of a dual-specificity protein phosphatase that inactivates a MAP kinase from Arabidopsis. Plant J. 1998;16:581–589. doi: 10.1046/j.1365-313x.1998.00327.x. [DOI] [PubMed] [Google Scholar]
- Ichimura K, Shinozaki K, Tena G, Sheen J, Henry Y, Champion A, Kreis M, Zhang S, Hirt H, Wilson C, Heberle-Bors E, Ellis B, Morris PC, Innes RW, Ecker JR, Scheel D, Klessig D, Machida Y, Mundy J, Ohashi Y, Walker JC. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002;7:301–308. doi: 10.1016/S1360-1385(02)02302-6. [DOI] [PubMed] [Google Scholar]
- Lee JS, Ellis BE. Arabidopsis MAPK phosphatase 2 (MKP2) positively regulates oxidative stress tolerance and inactivates the MPK3 and MPK6 MAPKs. J Biol Chem. 2007;282:25020–25029. doi: 10.1074/jbc.M701888200. [DOI] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Zeng W, Sheen J. Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature. 1998;395:716–720. doi: 10.1038/27240. [DOI] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Tena G, Sheen J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA. 2000;97:2940–2945. doi: 10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis K, Howell SH. Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. Plant J. 2000;24:785–796. doi: 10.1046/j.1365-313x.2000.00921.x. [DOI] [PubMed] [Google Scholar]
- Shiozaki K, Russell P. Cell-cycle control linked to extracellular environment by MAP kinase pathway in fission yeast. Nature. 1995;378:739–743. doi: 10.1038/378739a0. [DOI] [PubMed] [Google Scholar]
- Hannig G, Ottilie S, Erikson RL. Negative regulation of mitosis in fission yeast by catalytically inactive pyp1 and pyp2 mutants. Proc Natl Acad Sci U S A. 1994;91:10084–10088. doi: 10.1073/pnas.91.21.10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science. 2003;300:332–336. doi: 10.1126/science.1080585. [DOI] [PubMed] [Google Scholar]
- Fedoroff NV. Cross-talk in abscisic acid signaling. Science's STKE. 2002;2002:RE10. doi: 10.1126/stke.2002.140.re10. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, McCourt P. Cross-talk in plant hormone signalling: What Arabidopsis mutants are telling us. Annals of Botany. 2003;91:605–612. doi: 10.1093/aob/mcg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:11519–11524. doi: 10.1073/pnas.162339999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 1999;13:1678–1691. doi: 10.1101/gad.13.13.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. Auxin-induced SCFTIR1-Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proc Natl Acad Sci USA. 2004;101:12381–12386. doi: 10.1073/pnas.0402868101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Zhao Y, Dai X, Zhang W, Gray WM, Huq E, Estelle M. A new CULLIN 1 mutant has altered responses to hormones and light in Arabidopsis. Plant Physiol. 2007;143:684–696. doi: 10.1104/pp.106.091439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell. 2007;19:2440–2453. doi: 10.1105/tpc.107.050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last RL, Fink GR. Tryptophan-requiring mutants of the plant Arabidopsis thaliana. Science. 1988;240:305–310. doi: 10.1126/science.240.4850.305. [DOI] [PubMed] [Google Scholar]
- Haughn GW, Somerville C. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet. 1986;204:430–434. doi: 10.1007/BF00331020. [DOI] [Google Scholar]
- Stasinopoulos TC, Hangarter RP. Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol. 1990;93:1365–1369. doi: 10.1104/pp.93.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York , Greene Publishing Associates and Wiley-Interscience; 1999. [Google Scholar]
- LeClere S, Bartel B. A library of Arabidopsis 35S-cDNA lines for identifying novel mutants. Plant Mol Biol. 2001;46:695–703. doi: 10.1023/A:1011699722052. [DOI] [PubMed] [Google Scholar]
- Rogg LE. Biochemistry and Cell Biology. Houston , Rice University; 2001. Cloning and characterization of IAA28, a gene involved in suppressing lateral root development and mediating auxin responses in Arabidopsis thaliana; p. 211. [Google Scholar]
- Neff MM, Neff JD, Chory J, Pepper AE. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 1998;14:387–392. doi: 10.1046/j.1365-313X.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. A robust method for detecting single-nucleotide changes as polymorphic markers by PCR. Plant J. 1998;14:381–385. doi: 10.1046/j.1365-313X.1998.00123.x. [DOI] [PubMed] [Google Scholar]
- Bartel B, Fink GR. Differential regulation of an auxin-producing nitrilase gene family in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1994;91:6649–6653. doi: 10.1073/pnas.91.14.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Normalized ibr5 double mutant auxin response defects. The auxin responses of ibr5 and double mutants are represented after normalization to root lengths of mock-treated seedlings.
Normalized auxin-response mutant defects in lateral root induction by IBA, hypocotyl elongation inhibition by IBA, and root elongation inhibition by ABA. The IBA and ABA responses of ibr5 and double mutants are represented after normalization to mock-treated seedlings.