Abstract
Background
Three mutations in Arabidopsis thaliana strain Columbia – cpr1, snc1, and bal – map to the RPP5 locus, which contains a cluster of disease Resistance genes. The similar phenotypes, gene expression patterns, and genetic interactions observed in these mutants are related to constitutive activation of pathogen defense signaling. However, these mutant alleles respond differently to various conditions. Exposure to mutagens, such as ethyl methanesulfonate (EMS) and γ-irradiation, induce high frequency phenotypic instability of the bal allele. In addition, a fraction of the bal and cpr1 alleles segregated from bal × cpr1 F1 hybrids also show signs of phenotypic instability. To gain more insight into the mechanism of phenotypic instability of the bal and cpr1 mutations, we systematically compared the behavior of these unusual alleles with that of the missense gain-of-function snc1 allele in response to DNA damage or passage through F1 hybrids.
Results
We found that the cpr1 allele is similar to the bal allele in its unstable behavior after EMS mutagenesis. For both the bal and cpr1 mutants, destabilization of phenotypes was observed in more than 10% of EMS-treated plants in the M1 generation. In addition, exceptions to simple Mendelian inheritance were identified in the M2 generation. Like cpr1 × bal F1 hybrids, cpr1 × snc1 F1 hybrids and bal × snc1 F1 hybrids exhibited dwarf morphology. While only dwarf F2 plants were produced from bal × snc1 F1 hybrids, about 10% wild-type F2 progeny were produced from cpr1 × snc1 F1 hybrids, as well as from cpr1 × bal hybrids. Segregation analysis suggested that the cpr1 allele in cpr1 × snc1 crosses was destabilized during the late F1 generation to early F2 generation.
Conclusion
With exposure to EMS or different F1 hybrid contexts, phenotypic instability is induced for the bal and cpr1 alleles, but not for the snc1 allele. Our results suggest that the RPP5 locus can adopt different metastable genetic or epigenetic states, the stability of which is highly susceptible to mutagenesis and pairing of different alleles.
Background
The Arabidopsis RPP5 (for recognition of Peronospora parasitica 5) locus in the Columbia strain is composed of seven Resistance (R) genes that are implicated in plant innate immunity (Figure 1A) [1]. R genes in this locus encode proteins containing an N-terminal Drosophila Toll/mammalian interleukin-1 receptor (TIR) domain, in addition to nucleotide binding site (NBS) and leucine rich repeat (LRR) domains similar to those encoded by most R genes in the Arabidopsis genome [2]. Two of the R genes in the locus, RPP4 (At4g16860) and SNC1 (At4g16890) have been shown to mediate resistance to pathogens. For example, RPP4 specifies resistance to two races of the fungal pathogen Hyaloperonospora parasitica (formerly Peronospora parasitica) [3]. The activation of SNC1 (for suppressor of npr1-1, constitutive 1) causes resistance to H. parasitica and the bacterial pathogen Pseudomonas syringae [4]. Another R gene in the locus, At4g16950, shows the highest sequence similarity to RPP5, the founding member of RPP5 locus R genes in the Landsberg strain [2]. However, the function of At4g16950 might be different from that of RPP5, which is necessary for the recognition of a race of H. parasitica [5]. RPP5 locus R genes are coordinately regulated both positively and negatively [6]. RPP4, SNC1, and At4g16950 together can be transcriptionally activated by a positive feedback amplification mediated through salicylic acid accumulation [6,7]. In addition, low abundance small RNA species that can target multiple RPP5 locus R genes exist in wild-type plants, and transgenic over-expression of SNC1 can induce the cosuppression of these paralogous R genes [6].
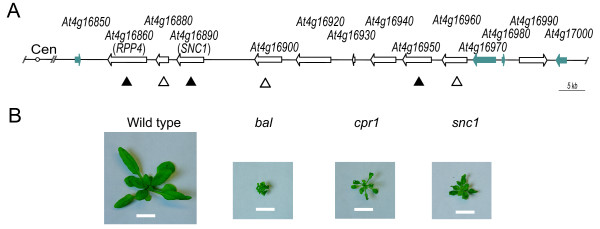
Figure 1.
Activation of RPP5 locus R genes causes similar phenotypes in bal, cpr1, and snc1 mutants. (A) Organization of the RPP5 locus in the Columbia haplotype. The R genes and a related TIR-NBS gene, At4g16990, are indicated by open arrows while non-R-genes are indicated by filled arrows. R genes that are up-regulated in all three mutants are indicated by filled upward arrowheads. Additional R genes up-regulated in the bal variant are indicated by open upward arrowheads (Additional File 1 and Data Not Shown); the expression of these genes has not yet been determined for cpr1 and snc1. Transposon-related sequences are not indicated. Cen: centromere. (B) Phenotypes of 3-week old bal, cpr1, and snc1 homozygous mutants along with a wild-type plant. Scale bar: 1 cm.
Three mutant alleles, cpr1, snc1, and bal, which cause similar dwarf phenotypes and coordinated activation of RPP5 locus R genes, map to the RPP5 locus (Figure 1 and Additional File 1) [8-10]. Constitutive activation of defense signaling in all three mutants requires both salicylic acid accumulation and EDS1 (for enhanced disease susceptibility 1) [9-12]. The snc1 gain-of-function allele is caused by a missense mutation in the region between the NBS and LRR domains that leads to elevated SNC1 activity [4]. Another mutant allele, cpr1, is a recessive allele isolated after ethyl methanesulfonate (EMS) mutagenesis [11]. In contrast, the semidominant bal allele was spontaneously generated in an inbred ddm1 (decrease in DNA methylation 1) mutant background in which genetic and epigenetic alterations accumulate over generations [13]. No mutations or epigenetic modifications responsible for the changes in phenotypes and gene expression have been reported for the cpr1 or bal allele. All three mutants exhibit elevated steady-state transcript levels of multiple RPP5 locus R genes, including RPP4, SNC1, and At4g16950, possibly through a positive feedback amplification that is initiated by SNC1 activation [6]. Consistent with the more severe phenotypes in the bal variant, a higher steady-state expression level of SNC1 was reproducibly detected in the bal variant compared to the cpr1 and snc1 mutants [6].
Many extragenic suppressor mutations have been identified for the snc1 allele after fast neutron treatment [14-18]. From ~150,000 M2 plants, Zhang and Li reported the isolation of 50 recessive mutations in 15 complementation groups that suppress snc1-dependent defense signaling. In contrast, a high degree of phenotypic instability was observed for the bal allele after EMS treatment or γ-irradiation [9]. In the M2 generation, more than 10% of the bal plants displayed signs of phenotypic suppression that were associated with a decrease in the steady-state expression level of RPP5 locus R genes. All five independent M2 lines investigated in the study carried alterations that mapped back to the RPP5 locus, suggesting that the primary mechanism was due to either a revertant or intragenic suppressor allele. In addition, hybridization-induced instability was reported for the bal and cpr1 alleles in F2 populations after the two alleles were brought together by genetic crosses in F1 hybrids [8]. By following the segregation of molecular markers linked to the bal or cpr1 allele, we determined that the cpr1 allele was destabilized to a much higher degree than the bal allele in an F1 hybrid context. Although these results revealed conditional phenotypic instability of the bal and cpr1 alleles, at least three questions remained unanswered. First, when is phenotypic instability induced in the bal variant? Second, can phenotypic instability of cpr1 be induced by EMS mutagenesis, as well as in F1 hybrids? Third, is the unusual behavior of the bal and cpr1 alleles in the F2 generation limited to the specific interaction between these two possible epigenetic alleles in F1 hybrids? In other words, can phenotypic instability of the cpr1 or bal allele be induced in cpr1 × snc1 or bal × snc1 F1 hybrids?
Here, we report that the cpr1 allele displays a high degree of phenotypic instability similar to that seen for the bal allele, while the snc1 allele does not. We observed a destabilization of phenotypes as early as the M1 generation among EMS-treated bal and cpr1 mutants. We also found that phenotypic instability of the cpr1 allele was induced in cpr1 × snc1 F1 hybrids, in which the cpr1 allele interacted with a well-defined genetic allele. Possible mechanisms to account for the unusual behavior of alleles affecting the RPP5 locus are considered.
Results
Phenotypic instability in bal and cpr1 mutants is first observed in the M1 generation after EMS treatment
Our previous work suggested that the bal allele is highly unstable in response to EMS treatment as evidenced by a high frequency of phenotypic suppression seen in the M2 generation, which is generated by self-pollination of M1 plants derived from EMS-treated seeds [9]. However, the genetic characteristics of phenotypic suppression events could not be studied in detail because each M2 pool examined in our original study was produced from a pool of ~20 M1 individuals. To gain more information regarding the instability of the bal phenotypes in response to EMS treatment, we investigated when phenotypic suppression is first established. The bal variant is characterized by small and severely curled leaves during vegetative development and short stature in later developmental stages (Figure 2A and 2B) [13]. We found that 17 of 141 EMS-treated bal M1 plants develop chimeric sectors that resemble the morphology of heterozygous bal plants, consistent with reversion of a single bal allele. In the same batch of mock- or 30 mM EMS-treated bal plants, we found that more than one third (38%: 53 out of 141 including the 17 plants that displayed chimeric sectors) of EMS-treated bal M1 plants developed sectors with a stem taller than 5 cm, another sign of phenotypic suppression in later development, while no tall stems were observed in any of the mock-treated M1 control plants (0%: 0 out of 115) (Figure 2B). Similar results were observed from more than five independent EMS treatments. In two cases, entire M1 plants displayed the phenotypes characteristic of a heterozygous bal plant with no obvious sectoring after EMS treatment. The homozygous status of a transgenic marker that we had introgressed into the bal background confirmed that these rare M1 plants were not heterozygous bal plants produced by pollen contamination (Data Not Shown) [19].
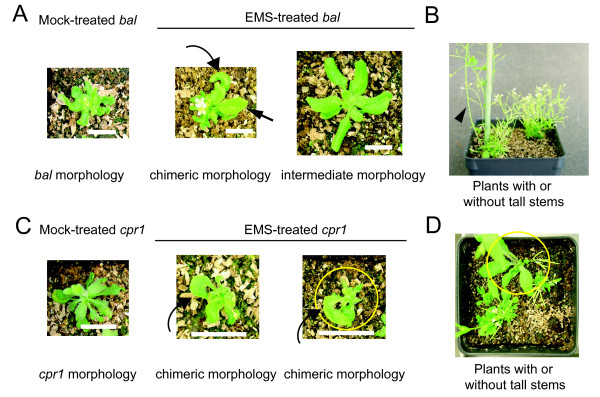
Figure 2.
Phenotypic suppression is established during the M1 generation in EMS-treated bal and cpr1 mutants. (A) Leaf phenotypes of mock- and EMS-treated bal variants. Straight and curved arrows indicate intermediately curled and crescent-shape leaves, respectively. (B) Phenotypes in two EMS-treated bal variants in the flower developmental stage. The arrowhead indicates one example of a thick and tall stem in EMS-treated bal variant. Note that the EMS-treated sibling plant on the right did not develop tall stems. (C) Leaf phenotypes of mock- and EMS-treated cpr1 mutants. Curved arrows indicate crescent-shape leaves. The plant in the yellow circle later developed a wild-type sector as shown in panel D. (D) Phenotypes of EMS-treated cpr1 mutants with or without wild-type sectors in the flower developmental stage. The plant in the yellow circle is the same plant in the yellow circle shown in panel C. Scale bar: 1 cm. Plants in panels A and C: 3.5-week old. Plants in panels B and D: 5-week old.
We found that EMS treatment also destabilized at a high frequency the narrow and slightly curled leaf phenotype caused by the cpr1 allele (Figure 2C). In about 10% of EMS-treated cpr1 plants (13/98, 5/56, and 6/46 in three independent experiments), we observed crescent-shape leaves, which likely represent phenotypic suppression on only one side of the cpr1 leaf (Figure 2C and 2D). We also noted that the majority of wild-type stems and leaves developed from regions with crescent-shape rosette leaves. Segregation of phenotypes in the following generation confirmed that the suppressed leaf phenotypes observed only in EMS-treated cpr1 mutants in the M1 generation were indeed caused by heritable changes (Figure 3).
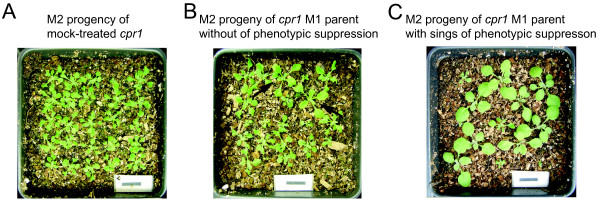
Figure 3.
Non-parental phenotypes are observed in M2 progeny of EMS-treated cpr1 mutants. Pots of 2-week old M2 plants produced from mock- (A) or EMS-treated (B and C) cpr1 parents. Two plants with intermediate morphology are marked with arrows in panel B. Note that the segregation ratios of M2 siblings in these families are included in Table 2. Black scale bar: 1 cm.
Suppression of phenotypes is found more frequently than expected in the M2 generation
We determined whether M2 plants with suppressed phenotypes were only generated from M1 plants that had previously shown signs of phenotypic changes. In all 13 independent bal M2 families examined, we discovered multiple phenotypically-suppressed plants. Six of these M2 families were derived from M1 plants with bal phenotypes, while the remaining 7 families were from M1 plants displaying a suppressed phenotype (Table 1). We mapped the changes responsible for phenotypic suppression in a total of 8 EMS-treated bal lines – 3 in this study (Data Not Shown) and five in our previous report [9], and found that the change in each case maps to the RPP5 locus [9]. Our results suggest that most EMS-treated bal individual plants carry alleles that suppress bal phenotypes and that these alleles are revertant alleles, intragenic suppressor mutations, or dominant extragenic suppressor mutations tightly linked to the bal allele.
Table 1.
Phenotypic revertants were produced from every EMS-treated bal individual tested.
| Source | Number of M2 plants phenotype | ||||
| bal | Intermediate | Wild-type | |||
| Treatment | Phenotype in M1 | Family | |||
| Experiment #1 | |||||
| Mock | bal | b1 | 77 | 0 | 0 |
| Mock | bal | b2 | 79 | 0 | 0 |
| 30 mM EMS | bal | b3 | 39 | 6 | 1 |
| 30 mM EMS | bal | b4 | 36 | 60 | 0 |
| 30 mM EMS | bal | b5 | 54 | 11 | 0 |
| 30 mM EMS | bal | b6 | 37 | 12 | 0 |
| A thick stem in 30 mM EMS-treated bal | Chimeric | b7* | 43 | 24 | 39 |
| A thick stem in 30 mM EMS-treated bal | Chimeric | b8* | 10 | 11 | 46 |
| A thick stem in 30 mM EMS-treated bal | Chimeric | b9† | 24 | 50 | 19 |
| A thick stem in 30 mM EMS-treated bal | Chimeric | b10* | 0 | 24 | 14 |
| A thick stem in 30 mM EMS-treated bal | Chimeric | b11† | 26 | 43 | 17 |
| Experiment #2 | |||||
| Mock | bal | b12 | 74 | 0 | 0 |
| Mock | bal | b13 | 54 | 0 | 0 |
| 30 mM EMS | bal | b14 | 40 | 6 | 0 |
| 30 mM EMS | bal | b15 | 48 | 4 | 0 |
| 30 mM EMS | Chimeric | b16* | 19 | 21 | 2 |
| 30 mM EMS | Chimeric | b17* | 31 | 11 | 0 |
Plants with a rosette diameter greater than the largest bal plant in the mock-treated control were counted as intermediate revertants, while ones with flat leaves were considered as wild-type revertants. M1 plants that displayed stems taller than 5 cm in addition to short and bushy stems resembling those observed in the bal variant were designated as chimeric.
†: P > 0.1 by chi square test with expected ratio of 1:2:1 and degree of freedom = 2.
*: P < 0.05 by chi square test with expected ratio of 1:2:1 or 5:2:1 and degree of freedom = 2.
We also tested whether the alleles responsible for phenotypic suppression are mitotically and meiotically stable from the late M1 to M2 generation. We determined the segregation ratio of phenotypes using seeds that were collected from siliques on stems taller than 5 cm on EMS-treated bal plants. Arabidopsis has two genetically effective cells, which are the meristematic cells that contribute to the reproductive lineages. Consequently, large M1 sectors with a presumptive heterozygous genotype are expected to give rise to M2 progeny showing Mendelian segregation ratios of 1:2:1 or 5:2:1 (dwarf:intermediate:normal) [9,13,20,21]. However, among the seven M2 families studied that were derived from large M1 sectors, only two families conformed to either expected segregation ratio (Table 1; Families b9 & b11). Of the five remaining M2 families, two (b7 & b8) had more wild-type M2 plants than expected (Table 1). In addition, suppression of bal phenotypes was evident even in the six lines derived from parents that showed no signs of phenotypic change in the M1 generation (Table 1; Families b3–b6, b14, & b15). The inheritance pattern, as well as the frequency of phenotypic suppression, demonstrates that bal phenotypes are extremely unstable.
In the case of cpr1 M2 plants, wild-type M2 progeny were identified along with cpr1-like siblings for all lines tested in which a chimeric morphology was observed in the M1 generation. We observed a deviation from the 1:3 or 5:3 segregation ratio of cpr1 and wild-type morphology in the M2 generation that was expected regardless of the potential linkage of suppressor mutations to the RPP5 locus (Table 2; Family c5). Plants with non-cpr1 morphology were derived from M1 parents that displayed cpr1 morphology (Figure 3B and Table 2; Families c3, c4 & c9). We conclude that phenotypic suppression in EMS-treated cpr1 mutants and bal variants is not limited to M1 plants with a chimeric morphology.
Table 2.
Phenotypic revertants were identified in the M2 generation among the progeny of EMS-treated cpr1 plants.
| Source | Number of M2 plants phenotype | ||||
| cpr1 | Intermediate | Wild-type | |||
| Treatment | Phenotype in M1 | Family | |||
| Experiment #1 | |||||
| Mock3A | cpr1 | c1 | 99 | 0 | 0 |
| Mock | cpr1 | c2 | 98 | 0 | 0 |
| 30 mM EMS | cpr1 | c3 | 66 | 9 | 0 |
| 30 mM EMS3B | cpr1 | c4 | 51 | 3 | 0 |
| 30 mM EMS3C | Chimeric | c5* | 42 | 0 | 43 |
| 30 mM EMS | Chimeric | c6 | 21 | 0 | 18 |
| Experiment #2 | |||||
| Treatment | |||||
| Mock | cpr1 | c7 | 85 | 0 | 0 |
| Mock | cpr1 | c8 | 96 | 0 | 0 |
| 30 mM EMS | cpr1 | c9 | 50 | 3 | 0 |
| 30 mM EMS | cpr1 | c10 | 28 | 0 | 0 |
| 30 mM EMS | Chimeric | c11 | 4 | 0 | 31 |
| 30 mM EMS | Chimeric | c12 | 31 | 0 | 74 |
Plants with slightly curled leaves that were wider than cpr1 were called intermediate in phenotypic categorization. Chimeric M1 plants were those that developed chimeric sectors of wild type-looking leaves and stems. 3A, 3B, and 3C: Lines for which representative M2 plants are shown in Figure 3A to 3C.
*: P < 0.05 by chi square test with expected ratio of 5:3 or 1:3 and degree of freedom = 1.
bal, cpr1, and snc1 haplotypes show synergistic interactions in F1 hybrids
Previously, we found that phenotypes in bal × cpr1 F1 hybrids are more severe than those in heterozygous bal plants (bal × CPR1) [8]. This result suggests that the phenotypic interaction between bal and cpr1 haplotypes is synergistic as the cpr1 allele is recessive relative to the wild-type allele with regards to morphological phenotypes and activation of defense signaling. We hypothesized that this synergistic interaction is the result of transcriptional activation of the RPP5 locus in the cpr1 haplotype by the semidominant bal allele [6]. We tested this idea by combining the cpr1 haplotype with the snc1 haplotype or a SNC1 transgenic background. The SNC1 transgene under the control of the constitutive 35S promoter (35S::SNC1) and the snc1 allele affect SNC1 activity at the transcript and protein level, respectively, leading to the activation of defense signaling in a dominant manner [4,6,9,10]. F1 hybrids carrying the cpr1 haplotype showed stronger phenotypes than those with the wild-type (CPR1) haplotype, consistent with our hypothesis (Figure 4A and Figure 4B). Previously, we demonstrated that the steady-state expression level of SNC1 is significantly lower in the cpr1 mutant compared to that in the bal variant [6]. Therefore, a stronger induction of SNC1 expression in the 35S:SNC1 × cpr1 F1 hybrid compared to the 35S:SNC1 × bal hybrid was unexpected, suggesting that the SNC1 expression level in 35S:SNC1 × cpr1 is not additive (Figure 4B, Figure 4C, and Additional File 2) [6]. In addition, bal × snc1 F1 hybrids also exhibited dwarfism and a curled leaf phenotype comparable to their parents (Figure 4D). Our results show that phenotypes characteristic of bal, cpr1, and snc1 alleles are enhanced in hybrid contexts.
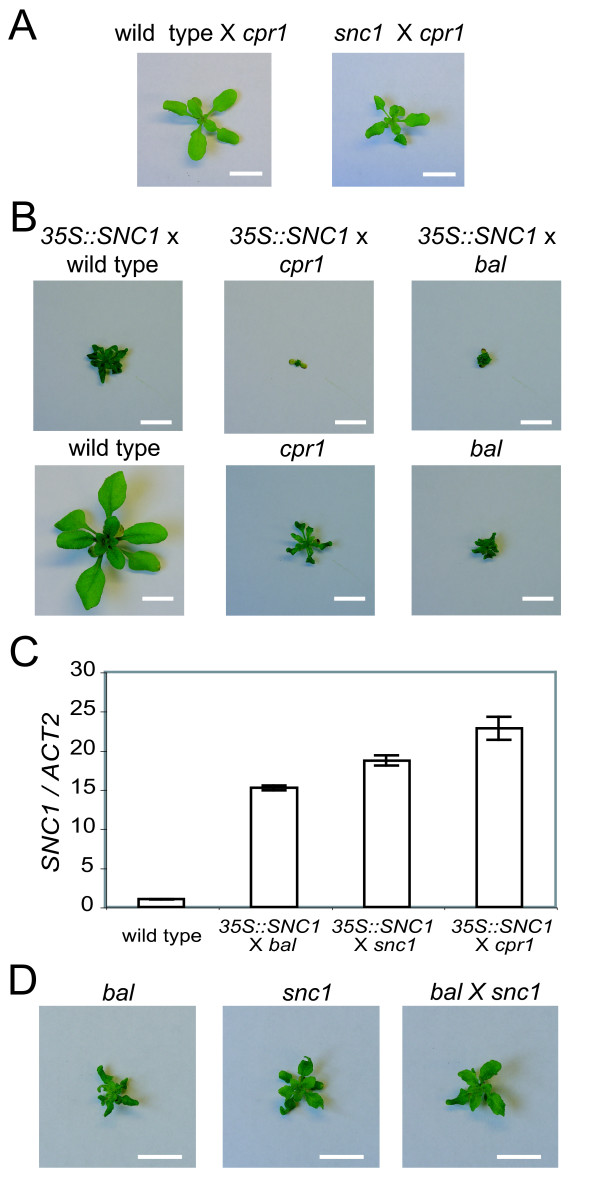
Figure 4.
Combining the cpr1 haplotype with the snc1 haplotype or SNC1 transgene enhances the phenotypes. (A) Phenotypes of F1 plants from wild type (SNC1) × cpr1 and snc1 × cpr1 crosses. (B) Phenotypes of F1 hybrids carrying a hemizygous 35S:SNC1 transgene. (C) Steady-state expression levels of SNC1 determined by quantitative real-time PCR. SNC1 transcript level was compared to that of the Actin 2 (ACT2) gene. (D) Phenotypes of bal or snc1 homozygotes and their F1 hybrid. Scale bar: 1 cm.
Induced instability of the cpr1 allele after pairing with the snc1 allele in F1 hybrids
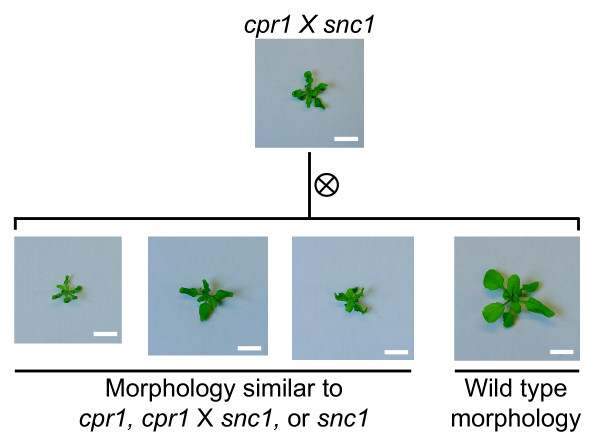
In addition to EMS treatment, interaction between the bal and cpr1 haplotype in a hybrid context can cause phenotypic instability and produce F2 plants with intermediate and wild-type morphologies [8,9]. We tested whether bal × snc1 and cpr1 × snc1 F1 hybrids can also generate phenotypically normal F2 progeny. Although bal × snc1 F1 hybrids exhibit a similar dwarf morphology compared to bal × cpr1 F1 hybrids, all 342 F2 progeny examined from bal × snc1 F1 hybrids showed dwarf phenotypes regardless of the direction of the cross (Table 3, Additional File 3). In contrast, F2 plants with normal morphology were frequently identified among cpr1 × snc1 F2 progeny (Figure 5 and Table 3). We genotyped 35 wild-type F2 plants generated from cpr1 × snc1 hybrids to determine which allele was associated with phenotypic suppression. In 17 of 35 plants, both the cpr1- and snc1-linked markers were detected while in the remainder of the plants only the cpr1-linked marker was detected. The lack of snc1/snc1 genotypes among the wild-type F2 progeny suggested that the destabilized cpr1 allele is associated with wild-type morphology in the F2 generation. Our F3 progeny test of four phenotypically normal F2 plants supported the idea that these plants contain a destabilized cpr1 allele (CPR1F1) that no longer induces cpr1 mutant phenotypes after hybridization (Table 4). snc1 and cpr1 mutants in the F3 generation with characteristic phenotypes were produced exclusively from F2 parents whose presumed genotypes were CPR1F1/snc1 and CPR1F1/cpr1, respectively.
Table 3.
Phenotypes of F2 progeny from F1 hybrids carrying different alleles in RPP5 locus.
| Crosses | F1 phenotype | F2 phenotypes | Total number of F2 plants | ||
| Wild-type | Intermediate | Dwarf | |||
| snc1 × bal | Dwarf | 0 | 0 | 244 | 244 |
| bal × snc | Dwarf | 0 | 0 | 218 | 218 |
| Cpr1 × snc1 #1 | Dwarf | 20 | 0 | 212a | 232 |
| Cpr1 × snc1 #2 | Dwarf | 22 | 0 | 211b | 234 |
| snc1 × cpr1 | Dwarf | 22 | 0 | 215c | 215 |
| bal × cpr1 | Dwarf | 13 | 14 | 241d | 268 |
a This number includes 18 plants with severe dwarfism in which cotyledons were not covered by later developing very small true leaves
b This number includes 13 plants with severe dwarfism. Two plants with severe dwarfism in this family are shown in Additional File 3.
c This number includes 17 plants with severe dwarfism.
d This number includes 24 plants with severe dwarfism.
Figure 5.
Wild-type morphology is observed in some F2 plants from cpr1 × snc1 F1 hybrids. Both dwarf and wild-type morphologies are observed among F2 progeny generated by self-pollination of cpr1 × snc1 F1 hybrids. Note that segregation ratios of F2 siblings are included in Table 3 as cpr1 × snc1 #2. Scale bar: 1 cm.
Table 4.
Segregation of cpr1 and snc1 phenotypes in F3 generation.
| Genotypes in F2 parents | Phenotypes of F2 parents | F3 phenotypes | Total number of F3 plants | ||
| Wild-type | cpr1 | snc1 | |||
| cpr1/cpr1 | Wild-type | 62 | 23 | 0 | 85 |
| cpr1/cpr1 | Wild-type | 74 | 22 | 0 | 96 |
| cpr1/snc1 | Wild-type | 74 | 0 | 23 | 97 |
| cpr1/snc1 | Wild-type | 65 | 0 | 24 | 89 |
Note that cpr1/cpr1 and snc1/snc1 genotypes were confirmed from sample genotyping of F3 plants with cpr1 and snc1 phenotypes, respectively.
Discussion
Here we report the genetic behavior of three different A. thaliana mutations that map to the RPP5 locus R gene cluster in strain Columbia. All three mutations condition a similar, but not identical, dwarfing phenotype accompanied by leaf-curling. Interestingly, only the snc1 allele behaves like a conventional mutation in terms of its phenotypic stability, as expected from this allele's known molecular nature. In contrast, the bal and cpr1 alleles show a high degree of phenotypic instability after exposure to EMS and F1 hybrid contexts.
Phenotypic suppression is induced by EMS treatment during the M1 generation in bal and cpr1 mutants
One demonstration of the instability of both the bal and cpr1 alleles was the high incidence of non-parental phenotypes in the M1 generation (Figure 2). More than 1/3 of EMS-treated bal plants and 10% of EMS-treated cpr1 plants carried at least one suppressor or revertant allele in the M1 generation (Figure 2, Table 1, and Table 2). The frequency of phenotypic suppression observed in our EMS-treated bal and cpr1 M1 plants is at least an order of magnitude higher than the expected M1 mutation frequency of ~10-2 to 10-3 (Additional File 4) [22,23]. We previously reported a high frequency of phenotypic suppression in EMS-treated M2 populations of the bal variant [9]. Now, we extend these findings by demonstrating that the frequency of reversion or suppressor mutations recovered in the M2 generation in the cpr1 mutant was also much higher than the expected frequency of recessive loss-of-function mutations after EMS mutagenesis (~0.1% of the M2 population) (Table 2).
The following observations suggest that EMS treatment destabilizes the bal allele and produces revertant BAL or suppressed BALEMS alleles, which no longer induce dwarf and curled leaf phenotypes. First, mutations that suppress bal phenotypes in M1 plants show tight linkage to the RPP5 locus [9], suggesting that the strain carries a revertant BAL, an intragenic suppressor BALEMS allele, or a linked dominant suppressor. However, F1 progeny of stable true-breeding phenotypic revertants and bal variants display the morphology of heterozygous BAL/bal plants, arguing against the possibility that any suppressor mutations are dominant mutations tightly linked to RPP5 locus (Data Not Shown). Second, Li and colleagues recovered less than 0.1% (50 out of ~150,000) of snc1 M2 plants carrying recessive mutations that suppressed snc1-dependent phenotypes, while more than 10% of bal M2 plants showed non-parental morphology in our experiments [9,14]. This large difference is not expected if the primary mechanism for recovery of phenotypically suppressed plants is extragenic suppression, assuming that the spectrum of possible extragenic suppressor mutations is similar for the bal and snc1 mutations.
We consider two different mechanisms that might explain why bal and cpr1 phenotypes are suppressed at a high frequency after EMS mutagenesis. One possibility is that bal and cpr1 are conditionally metastable epialleles. Specifically, the erasure and resetting of epigenetic alterations through DNA repair initiated by DNA damage (e.g., via EMS treatment) might suppress the phenotypes. The RPP5 locus is a complex mixture of transposable elements and tandemly arrayed paralogous genes that are targets of small RNA species. Genomic regions with this type of organization are frequent targets of epigenetic regulation [24,25]. Nonetheless, the sectoring of phenotypes observed in many M1 plants suggests that phenotypic suppression by RNA silencing, which acts systemically, cannot easily explain the phenotypic instability in EMS-treated bal and cpr1 mutants [26]. An alternative hypothesis is that the mutation rate in the RPP5 locus in the cpr1 and bal mutants is elevated. This scenario might occur if the constitutively active state or possible aberrant epigenetic modification(s) in the RPP5 locus contributes to the high frequency of phenotypic suppression, perhaps by making the locus more susceptible to mutagenesis when treated with EMS. We note that the snc1 mutation induces transcription of RPP5 locus R genes through a positive feedback mechanism [6], which might be distinct from the mechanism(s) operating in the bal and cpr1 mutants.
In both EMS-treated bal and cpr1 plants (M1), non-parental phenotypes were more frequently observed in later developmental stages. This trend can be explained in three different ways. First, the number of meristematic initial cells that generate organs emerging later in development gradually decreases in Arabidopsis [27,28]. The effect of phenotypic suppression in one variant cell among 8–9 initial cells can be easily masked by more abundant unaffected initial cells in the leaf development stage. However, suppression will be readily observable at the later inflorescence development stage since the inflorescence is produced from only one or two initial cells. Second, phenotypically-suppressed cells might have a selective advantage in the stem cell niche in the meristem and over time outcompete those without suppressor mutations. This competition-selection model is consistent with the finding that constitutive activation of defense signaling in the cpr1 mutant has a fitness cost in vegetative growth [29]. Third, suppression of bal-like phenotypes by RNA silencing becomes obvious only two weeks after germination in transgenic plants over-expressing SNC1 [6]. Regardless of the mechanism responsible for high frequency of phenotypic suppression in bal and cpr1 mutants, a gradient of phenotypes in body size and leaf curliness observed in EMS-treated M2 populations might parallel the generation of novel genetic or epigenetic variation in the RPP5 locus in natural contexts.
Destabilization of the cpr1 allele is induced between late F1 generation and early F2 generation when paired with the bal or snc1 allele
In F1 hybrids, all possible allelic combinations of bal, cpr1, and snc1 cause dwarf phenotypes similar to their parents (Figure 4) [8]. Stronger phenotypes were observed in 35S:SNC1 × cpr1 hybrids compared to 35S:SNC1 × bal or 35S:SNC1 × wild type hybrids. The enhanced phenotypes are unlikely to be caused by additive interaction of the alleles because the bal allele is semidominant and shows stronger phenotypes than the recessive cpr1 allele. Therefore, we propose that the enhanced phenotypes are caused by synergistic interactions between alleles of the RPP5 locus, and that RPP5 locus-wide transcriptional activation by SNC1 plays an important role in these interactions.
Unexpected non-dwarf phenotypes segregate among F2 progeny from the cpr1 × bal and cpr1 × snc1 crosses (Figure 5 and Table 3) [8]. Genotyping results in the F2 generation showed that, in most cases, the cpr1 allele is associated with these unexpected phenotypes (Table 3) [8]. Given the frequency of non-dwarf F2 plants recovered (~1/10) and specific instability of the cpr1 allele, it is unlikely that a recessive suppressor allele for the cpr1 phenotype was introduced from the bal or snc1 background. Instead, the data are most consistent with a paramutation-like mechanism whereby pairing of the cpr1 allele and the other two alleles of the RPP5 locus occasionally induces the formation of CPR1F1, a derivative of the cpr1 allele that no longer causes dwarf and curled-leaf phenotypes. In contrast to the phenotypic reversion noted in our EMS experiments, no sign of chimeric development was observed in cpr1 × bal or cpr1 × snc1 hybrid plants during the F1 generation.
Two models, which are not mutually exclusive, can explain the hybridization-induced phenotypic instability. In the first model, RNA silencing of SNC1 and possibly other R genes in cis causes the phenotypic suppression. Previously, we showed that 21–24 nucleotide small RNA species complementary to many paralogous R genes in the RPP5 locus are produced and demonstrated that over-expression of SNC1 can induce coordinate suppression of these R genes [6]. In this model, preferential destabilization of the cpr1 allele is consistent with the observation that the steady-state expression level of SNC1 is higher in 35S:SNC1 × cpr1 hybrids compared to 35S:SNC1 × bal or 35S:SNC1 × snc1 hybrids (Figure 4C). Over-expression of SNC1 and the possible presence of unpaired DNA, which can be formed by out-of-register meiotic pairing among tandem repeats of the paralogous RPP5 locus R genes, are potential triggers of RNA silencing. Silencing of unpaired DNA during meiosis leading to a stable epigenetic state can be inherited in the progeny was reported in Arabidopsis [30]. In the second model, homologous recombination (e.g., gene conversion or unequal crossing over) during meiosis disrupts the cpr1 allele or produces haplotypes without any mutant alleles. Consistent with an unequal crossing over mechanism, we recovered extreme dwarf and seedling lethal plants in F2 populations, such as would be predicted from RPP5 locus R gene amplification, along with wild-type plants (Additional File 3). There is precedence for similar mechanisms involving R genes in other plant species. Cf-4 and Cf-9 are two homologous R genes located at the same locus in different Lycopersicon species [31,32]. Although both genes are meiotically stable in homozygotes, haplotypes carrying neither Cf-4 nor Cf-9 were produced at a frequency of ~1/2000 through meiotic recombination in a trans-heterozygote (Cf-4 × Cf-9). In addition, meiosis-specific, intrachromatidal, homologous recombination that preferentially eliminates DNA between homologous sequences was recently reported for the human male germ line [33]. We also cannot rule out the possibility that DNA repair processes accompanying recombination may remove epigenetic alterations responsible for the up-regulation of RPP5 locus R genes or deposit silencing marks such as cytosine methylation [34]. Meiotic recombination between cpr1 and other haplotypes may be facilitated by a constitutively active transcriptional state or by aberrant epigenetic alterations in RPP5 locus. These results demonstrate that the stability of the cpr1 allele can be affected by the genetic interaction of the cpr1 haplotype with the bal or snc1 haplotype, although the mechanism involved remains to be determined.
Conclusion
Our results showed that EMS treatment induces phenotypic instability in the cpr1 mutant, as well as in the bal variant. Phenotypic suppression was observed in the M1 generation in more than 10% of the EMS-treated bal and cpr1 mutants. Moreover, exceptions to a simple Mendelian inheritance from the M1 to M2 generation were observed for both mutants. We also found that phenotypic instability of the cpr1 allele was induced in cpr1 × snc1 F1 hybrids, in addition to cpr1 × bal F1 hybrids as previously reported. However, no phenotypic instability was observed among F2 progeny from bal × snc1 F1 hybrids. We conclude that bal, cpr1, and snc1 alleles with similar phenotypes can be differentiated in terms of phenotypic stability after EMS mutagenesis and hybrid formation. A high degree phenotypic instability in bal and cpr1 mutants suggests that metastable states, which are associated with constitutive over-expression of RPP5 locus R genes in these mutants, can facilitate genetic or epigenetic variation in RPP5 locus.
Methods
Plants and Growth Conditions
We previously described the bal variant [8,9]. cpr1 and snc1 were kindly provided by Drs. Xinnian Dong and Xin Li [10,11]. The point mutation in the snc1 allele disrupts an XbaI restriction enzyme site in the SNC1 coding region [4]. XbaI cleavage of the PCR product amplified with 5'-GTGGAGTTCCCATCTGAACATC-3' and 5'-CCCATTTTGATTGCTGGAAAG-3' allowed us to differentiate the snc1 allele from other alleles (Xin Li, Personal Communication). All plants were grown on soil in growth chambers under long day conditions (16 hours light) as described previously, except those shown in Additional File 2, which were grown under short day conditions (8 hours light) [9]. EMS mutagenesis was performed as described previously using an 8 hour 30 mM EMS treatment [9].
Nucleic Acid Isolation
Total RNA for construction of 1st strand cDNA libraries was isolated from aerial parts of 2 week-old plants using the TRIzol reagent (Invitrogen). Genomic DNA for genotyping was isolated using the urea lysis miniprep protocol [35].
Expression Analysis
Steady-state expression levels of SNC1 were determined by quantitative real-time PCR as described previously using 2-week old plants [6]. Information on primers and Taqman MGB probe (Applied Biosystems) used in PCR reactions are shown below. 5'-TCGGTGGTTCCATTCTTGCT-3', 5'-GCTTTTTAAGCCTTTGATCTTGAGAG-3', and 5'-NED-AGCACATTCCAGCAGATGTGGATCTCCAA-3' for Actin2. 5'-GCCGGATATGATCTTCGGAA-3', 5'-CGGCAAGCTCTTCAATCATG-3', and 5'-6FAM-TGGCCTAGTGAAGCA-3' for SNC1.
Authors' contributions
HY and EJR conceived the study. HY performed the molecular genetic studies and the expression analysis. HY and EJR drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Steady-state expression levels of many RPP5 locus R genes are increased in the bal variant. RT-PCR was used to compare the steady-state transcript levels of different R genes in the RPP5 locus. Two genes located outside of the RPP5 locus, Actin 2 (ACT2) and phosphofructokinase β subunit (PFKβ), were used as loading controls. -RT and +RT: 1st strand cDNA was constructed without or with reverse transcriptase (RT). BAL: wild-type plants; bal: bal variant.
Phenotypic interaction of the SNC1 transgene with the cpr1 and bal allele in short day. Note that the bal and cpr1 alleles display milder phenotypes in short day conditions compared to long day conditions.
Extremely severe dwarfism is observed in some F2 progeny from cpr1 × snc1 cross. Plants show representative phenotypes of plants with severe dwarfism. These plants are siblings of the plants shown in Figure 4. Scale bar: 1 cm.
Values used to calculate the frequency of loss-of-function mutation by EMS treatment.
Acknowledgments
Acknowledgements
We thank Hye Ryun Woo, Travis Dittmer, and Ashley Galant for helpful comments on the manuscript, and Mike Dyer and the greenhouse staff for plant care. Seeds were generous gifts from Xinnian Dong, Barbara Hohn, and Xin Li. This work was supported by grants from the National Science Foundation to E.J.R. (MCB-0321990 and MCB-0548597). Additional support was provided by the Danforth Foundation.
Contributor Information
Hankuil Yi, Email: hyi@artsci.wustl.edu.
Eric J Richards, Email: richards@biology.wustl.edu.
References
- Noel L, Moores TL, van Der Biezen EA, Parniske M, Daniels MJ, Parker JE, Jones JD. Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell. 1999;11:2099–2112. doi: 10.1105/tpc.11.11.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen EA, Freddie CT, Kahn K, Parker JE, Jones JD. Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 2002;29:439–451. doi: 10.1046/j.0960-7412.2001.01229.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Goritschnig S, Dong X, Li X. A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell. 2003;15:2636–2646. doi: 10.1105/tpc.015842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JE, Coleman MJ, Szabo V, Frost LN, Schmidt R, van der Biezen EA, Moores T, Dean C, Daniels MJ, Jones JD. The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. Plant Cell. 1997;9:879–894. doi: 10.1105/tpc.9.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Richards EJ. A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell. 2007;19:2929–2939. doi: 10.1105/tpc.107.051821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yang S, Yang H, Hua J. The TIR-NB-LRR gene SNC1 is regulated at the transcript level by multiple factors. Mol Plant Microbe Interact. 2007;20:1449–1456. doi: 10.1094/MPMI-20-11-1449. [DOI] [PubMed] [Google Scholar]
- Stokes TL, Richards EJ. Induced instability of two Arabidopsis constitutive pathogen-response alleles. Proc Natl Acad Sci USA. 2002;99:7792–7796. doi: 10.1073/pnas.112040999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes TL, Kunkel BN, Richards EJ. Epigenetic variation in Arabidopsis disease resistance. Genes Dev. 2002;16:171–182. doi: 10.1101/gad.952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Clarke JD, Zhang Y, Dong X. Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol Plant Microbe Interact. 2001;14:1131–1139. doi: 10.1094/MPMI.2001.14.10.1131. [DOI] [PubMed] [Google Scholar]
- Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X. A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell. 1994;6:1845–1857. doi: 10.1105/tpc.6.12.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JD, Aarts N, Feys BJ, Dong X, Parker JE. Constitutive disease resistance requires EDS1 in the Arabidopsis mutants cpr1 and cpr6 and is partially EDS1-dependent in cpr5. Plant J. 2001;26:409–420. doi: 10.1046/j.1365-313X.2001.2641041.x. [DOI] [PubMed] [Google Scholar]
- Kakutani T, Jeddeloh JA, Flowers SK, Munakata K, Richards EJ. Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc Natl Acad Sci USA. 1996;93:12406–12411. doi: 10.1073/pnas.93.22.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li X. A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1, constitutive 1. Plant Cell. 2005;17:1306–1316. doi: 10.1105/tpc.104.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cheng YT, Bi D, Palma K, Li X. MOS2, a protein containing G-patch and KOW motifs, is essential for innate immunity in Arabidopsis thaliana. Curr Biol. 2005;15:1936–1942. doi: 10.1016/j.cub.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Goritschnig S, Zhang Y, Li X. The ubiquitin pathway is required for innate immunity in Arabidopsis. Plant J. 2007;49:540–551. doi: 10.1111/j.1365-313X.2006.02978.x. [DOI] [PubMed] [Google Scholar]
- Palma K, Zhang Y, Li X. An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Curr Biol. 2005;15:1129–1135. doi: 10.1016/j.cub.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Palma K, Zhao Q, Cheng YT, Bi D, Monaghan J, Cheng W, Zhang Y, Li X. Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev. 2007;21:1484–1493. doi: 10.1101/gad.1559607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J, Ries G, Bonhoeffer S, Hohn B. Interchromatid and interhomolog recombination in Arabidopsis thaliana. Plant Cell. 2004;16:342–352. doi: 10.1105/tpc.019042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page DR, Grossniklaus U. The art and design of genetic screens: Arabidopsis thaliana. Nat Rev Genet. 2002;3:124–136. doi: 10.1038/nrg730. [DOI] [PubMed] [Google Scholar]
- Li SL, Rédei GP. Estimation of mutation rates in autogamous diploids. Radiation Botany. 1969;9:125–131. doi: 10.1016/S0033-7560(69)80079-7. [DOI] [Google Scholar]
- Kovalchuk I, Kovalchuk O, Hohn B. Genome-wide variation of the somatic mutation frequency in transgenic plants. Embo J. 2000;19:4431–4438. doi: 10.1093/emboj/19.17.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera G, Baute J, Bauwens M, Peck I, Piette D, Pycke M, Asselman P, Depicker A. Development and Application of Novel Constructs to Score C:G-to-T:A Transitions and Homologous Recombination in Arabidopsis. Plant Physiol. 2008;146:22–31. doi: 10.1104/pp.107.105213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja JH, Clough SJ, Chan WC, Vodkin LO. Tissue-specific gene silencing mediated by a naturally occurring chalcone synthase gene cluster in Glycine max. Plant Cell. 2004;16:819–835. doi: 10.1105/tpc.021352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Vedova CB, Lorbiecke R, Kirsch H, Schulte MB, Scheets K, Borchert LM, Scheffler BE, Wienand U, Cone KC, Birchler JA. The dominant inhibitory chalcone synthase allele C2-Idf (inhibitor diffuse) from Zea mays (L.) acts via an endogenous RNA silencing mechanism. Genetics. 2005;170:1989–2002. doi: 10.1534/genetics.105.043406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. Non-cell autonomous RNA silencing. FEBS Lett. 2005;579:5858–5871. doi: 10.1016/j.febslet.2005.09.039. [DOI] [PubMed] [Google Scholar]
- Furner IJ, Pumfrey JE. Cell fate in the shoot apical meristem of Arabidopsis thaliana. Development. 1992;115:755–764. [Google Scholar]
- Irish VF, Sussex IM. A fate map of the Arabidopsis embryonic shoot apical meristem. Development. 1992;115:745–753. [Google Scholar]
- Heidel AJ, Clarke JD, Antonovics J, Dong X. Fitness costs of mutations affecting the systemic acquired resistance pathway in Arabidopsis thaliana. Genetics. 2004;168:2197–2206. doi: 10.1534/genetics.104.032193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux FG, Pont-Lezica R, Galaud JP. Relationship between allelic state of T-DNA and DNA methylation of chromosomal integration region in transformed Arabidopsis thaliana plants. Plant Mol Biol. 2005;58:295–303. doi: 10.1007/s11103-005-4808-0. [DOI] [PubMed] [Google Scholar]
- Wulff BB, Thomas CM, Parniske M, Jones JD. Genetic variation at the tomato Cf-4/Cf-9 locus induced by EMS mutagenesis and intralocus recombination. Genetics. 2004;167:459–470. doi: 10.1534/genetics.167.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske M, Wulff BB, Bonnema G, Thomas CM, Jones DA, Jones JD. Homologues of the Cf-9 disease resistance gene (Hcr9s) are present at multiple loci on the short arm of tomato chromosome 1. Mol Plant Microbe Interact. 1999;12:93–102. doi: 10.1094/MPMI.1999.12.2.93. [DOI] [PubMed] [Google Scholar]
- Turner DJ, Miretti M, Rajan D, Fiegler H, Carter NP, Blayney ML, Beck S, Hurles ME. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat Genet. 2008;40:90–95. doi: 10.1038/ng.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuozzo C, Porcellini A, Angrisano T, Morano A, Lee B, Pardo AD, Messina S, Iuliano R, Fusco A, Santillo MR, Muller MT, Chiariotti L, Gottesman ME, Avvedimento EV. DNA Damage, Homology-Directed Repair, and DNA Methylation. PLoS Genet. 2007;3:e110. doi: 10.1371/journal.pgen.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cocciolone SM, Cone KC. Pl-Bh, an anthocyanin regulatory gene of maize that leads to variegated pigmentation. Genetics. 1993;135:575–588. doi: 10.1093/genetics/135.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Steady-state expression levels of many RPP5 locus R genes are increased in the bal variant. RT-PCR was used to compare the steady-state transcript levels of different R genes in the RPP5 locus. Two genes located outside of the RPP5 locus, Actin 2 (ACT2) and phosphofructokinase β subunit (PFKβ), were used as loading controls. -RT and +RT: 1st strand cDNA was constructed without or with reverse transcriptase (RT). BAL: wild-type plants; bal: bal variant.
Phenotypic interaction of the SNC1 transgene with the cpr1 and bal allele in short day. Note that the bal and cpr1 alleles display milder phenotypes in short day conditions compared to long day conditions.
Extremely severe dwarfism is observed in some F2 progeny from cpr1 × snc1 cross. Plants show representative phenotypes of plants with severe dwarfism. These plants are siblings of the plants shown in Figure 4. Scale bar: 1 cm.
Values used to calculate the frequency of loss-of-function mutation by EMS treatment.