Abstract
Developing autoreactive B cells edit their B cell antigen receptor (BCR) in the bone marrow and are clonally deleted when they fail to reexpress an innocent BCR. Here, inducible Cre-loxP-mediated gene inversion is used to change the specificity of the BCR on mature IgM+ IgD+ B cells in vivo to address the fate of lymphocytes encountering self-antigens at this developmental stage. Expression of an autoreactive BCR on mature B cells leads to their rapid elimination from the periphery, a process that is inhibited by constitutive bcl-2 transgene expression in an antigen dose-dependent manner. Thus, selection of mature B cells into the long-lived peripheral pool does not prevent their deletion upon encounter of self-antigens.
Tolerance to self-antigens is critical in preventing autoimmunity in the organism. In the B cell lineage, central tolerance occurs in the bone marrow where immature self-reactive B cells edit their autoreactive B cell antigen receptors (BCRs) (1, 2) and may undergo anergy (3) or clonal deletion (4, 5) when they failed in this process. This prevents autoreactive B cells from gaining functional competence and entering the long-lived peripheral pool. In addition, peripheral tolerance is thought to exist to control B lymphocytes that recognize tissue- or organ-specific self-antigens.
Peripheral B cell tolerance previously had been studied by mating Ig-transgenic mice with mice that expressed the corresponding autoantigen in a peripheral organ (6). In these studies, Ig-transgenic B cells were found in the bone marrow and not in the spleen and lymph nodes of the offspring harboring the organ-specific autoantigen, suggesting that these cells were eliminated at a post-bone-marrow stage of differentiation. However, it is not known whether under these experimental conditions clonal deletion of self-reactive B cells occurs at the mature or, indeed more likely, the “transitional” (7) immature-to-mature B cell stage of differentiation. Other studies have shown that peripheral B cells in the germinal centers undergo apoptosis in the presence of saturating amounts of antigen (8, 9). This also can be applied to germinal center B cells expressing self-reactive somatic antibody mutants. However, germinal center B cells are thought to be programmed to die unless rescued by antigen-dependent and costimulatory signals (10), so that their behavior may not reflect that of the bulk of the mature B cells in the peripheral lymphoid organs upon encounter of autoantigens. There is evidence that BCR engagement shortens the life span of the latter cells under certain conditions (11, 12). The present study addresses this issue further by studying in situ the fate of IgM+ IgD+ mature cells that recognize a self antigen after their selection into the long-lived peripheral B cell pool.
Cre-mediated conditional gene targeting can be used to inactivate genes in vivo in a tissue-specific and temporal manner (13). This often is accomplished by using Cre to excise a gene segment flanked by two loxP sites in the same orientation. However, Cre is also capable of inverting a gene segment if the two flanking loxP sites are placed in opposite orientation to each other. We have adapted this property of the Cre-loxP recombination system to change gene expression in vivo, specifically, to switch BCR specificity on mature B cells to study peripheral B cell tolerance.
MATERIALS AND METHODS
Construction of the 3–83B1–8f Targeting Vector.
The targeting vector was a modification of pIVhL2neor used by Sonoda et al. (14). The VH3–83 gene was cloned in a head-to-head orientation upstream of the VHB1–8 gene in pBluescript. A third loxP site, inverted with respect to the two flanking the neor gene, was placed downstream of the VHB1–8 VDJ. Finally, the restriction fragment comprising the two VH genes and the third loxP site was cloned into the unique ClaI-SalI site of pIVhL2neor to generate the final targeting vector.
Transfection of Embryonic Stem (ES) Cells and Generation of 3–83B1–8f Mice.
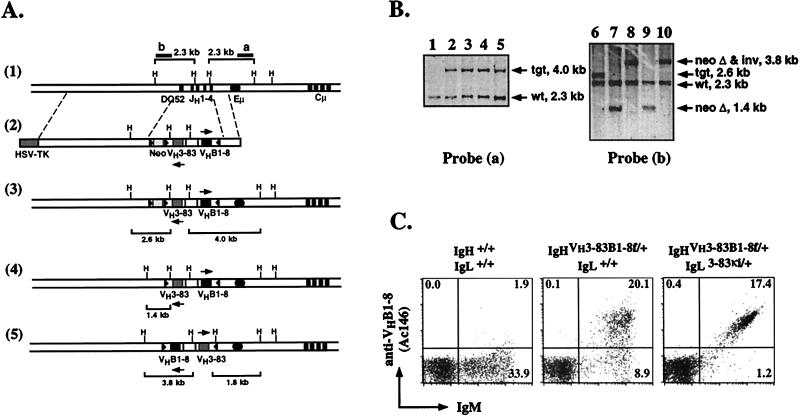
E14.1 ES cells (107) were electroporated with 10 μg of NotI-linearized targeting vector as described previously (14). Transfected ES cells were selected with G418 (350 μg/ml) and gancyclovir (2 μM). Homologous recombinants first were identified by PCR by using a primer that anneals to the JH2 element in the B1–8 VDJ (5′-CAAGGCACCACTCTCACAGTCT-3′) and a primer that is located external to the targeting construct (5′-GGAAACTAGAACTACTCAAGCTA-3′). The expected sizes of the PCR products for the wild-type and targeted alleles are 2.2 and 1.4 kb, respectively. Homologous recombinants were later verified by Southern blotting by using an external probe as shown in Fig. 2. The frequency of homologous recombinants was 1 out of 20 of double-drug-resistant clones. For the deletion of the neor gene, homologous recombinants were transfected with pIC-Cre (30 μg/107 cells) and G418-sensitive clones were selected. For the identification of clones that had deleted neor only or that had deleted neor and undergone an inversion event we used an internal probe as shown in Fig. 2. Approximately 12% of the G418-sensitive clones had undergone an inversion event. ES cell clones with the correct configuration of the targeted allele were injected into blastocysts to generate chimeric mice for germ-line transmission.
Figure 2.
Generation of mice allowing inducible, cre-mediated change in BCR specificity. (A) Targeting of the VH3–83 and VHB1–8 genes into the endogenous IgH locus. (1) Partial restriction endonuclease map of the wild-type IgH locus. The solid circle and rectangles represent the IgH enhancer and the JH and Cμ exons, respectively. (2) The targeting vector used for the insertion of the 3–83 and B1–8 VDJs. The structures of the locus after homologous recombination in ES cells (3); removal of the neor gene (4); and removal of the neor gene and inversion of the loxP-flanked gene segment (5) are shown. The solid triangles represent the loxP sites, one of which is inverted. The sizes of the restriction fragments (H, HindIII) as revealed by the probes after homologous recombination and each subsequent deletion or inversion event are shown. Maps are not drawn to scale. (B) Southern Blot analysis of ES cell clones. DNA from parental E14 (lane 1) and targeted subclones (lanes 2–5) were digested with HindIII and hybridized with probe (a) to verify the targeting event. After transient transfection with a cre-expressing plasmid, DNA from neo-sensitive subclones was digested with HindIII and hybridized with probe (b) to distinguish clones that had deleted the neor gene only (lanes 7 and 9) or that had deleted the neor gene and undergone an inversion event (lanes 8 and 10). Targeted clone before neo-deletion is shown as a control (lane 6). (C) Flow cytometry analysis of peripheral blood lymphocytes from wild-type, 3–83B1–8f/+, and 3–83B1–8f/+, 3–83κi/+ (Dtg) mice. Cells were stained with anti-IgM and anti-VHB1–8 (Ac146) mAbs. Numbers indicate percentage of total lymphocytes.
Mouse Strains.
The Mx-cre (15), 3–83κi (16), and B1–8f (17) mice used in this study were bred and maintained in our animal facilities. The Eμ-bcl-2–22 transgenic mouse strain (18) was obtained from The Jackson Laboratory.
Induced Inversion of the loxP-flanked VH Genes.
The 3–83B1–8f/+ mice were crossed with the 3–83κi/+ and the Mx-cre mice to generate 3–83B1–8f/+, 3–83κi/+, Mx-cre mice. These mice were given three doses of 1 mg anti-IL7R (A7R34) mAb i.v. every other day for a week (19). Fluorescence-activated cell sorter (FACS) analyses revealed that the three doses of anti-IL7R mAb were sufficient to inhibit B lymphopoiesis in the bone marrow. To induce Cre-recombinase expression and subsequent inversion of the loxP-flanked gene segment, mice were given one dose (2 × 106 units) of recombinant α/β interferon (IFN) 1 week after the anti-IL7R mAb treatment. DNA was isolated from the livers of these mice to assess the efficiency of inversion by Southern blotting.
Flow Cytometry.
Cells obtained from the various lymphoid organs of mice were stained with fluorochrome (fluorescein and phycoerythrin) or biotin-conjugated mAbs for flow cytometric analyses on a FACScan (Becton Dickinson) as described previously (20). Biotin-conjugated mAbs were revealed with streptavidin-Cychrome. The following mAbs were prepared in our laboratory: R33–24.12 (anti-IgM), RA3–6B2 (anti-B220), Ac146 (anti-VHB1–8), 54.1 (anti-VH3–83/Vκ3–83), H141–29 (anti-H-2b), and M5–114 (anti-MHC class II). mAbs to the following cell surface antigens were purchased from PharMingen: anti-H-2d, CD23, CD40, CD44, CD69, CD86 (B7.2), and CD95 (Fas).
RESULTS
Experimental System for Changing Antigen Receptors on Mature B Cells in Vivo.
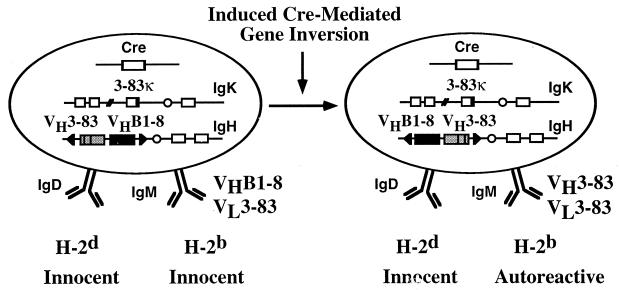
The strategy for changing BCR specificity on mature B cells is depicted in Fig. 1. In our experimental system, developing B lymphocytes in the bone marrow first express a BCR of nonautoreactive but unknown specificity (21). Its variable (V) regions are encoded by the heavy (H) chain V region gene VHB1–8 (22, 23) and the light (L) chain V region gene Vκ3–83 (6, 8). At the mature B cell stage of differentiation and after the lymphocytes have seeded the peripheral organs, Cre recombinase is used to switch BCR specificity from that of VHB1–8, Vκ3–83 to that of VH3–83, Vκ3–83. This second BCR recognizes MHC class I molecules of the b and k haplotype with high affinity but does not bind or binds weakly those MHC class I molecules of the d haplotype (4, 6). Thus, it is autoreactive in mice of the H-2b but not H-2d haplotype.
Figure 1.
Model for the analysis of peripheral B cell tolerance by changing BCR specificity in vivo. Schematic representation of changing the expression of an innocent BCR (VHB1–8, Vκ3–83) to an autoreactive (VH3–83, Vκ3–83) one on mature B cells by using cre-mediated inducible gene targeting. The autoreactive BCR recognizes MHC class I molecules of the b but not d haplotype.
Generation of Mice Allowing Inducible, Cre-Mediated Change in B Cell Antigen Receptor Specificity.
For the generation of mice that allow inducible switching of BCR specificity, we positioned two rearranged variable (V) region genes, VHB1–8 and VH3–83, into the IgH locus (24). The gene-targeting strategy and characterization of recombinant ES cells by Southern blotting are depicted in Fig. 2. The two V region genes, flanked by loxP sites (floxed) in opposite orientation, are placed head to head in opposite transcriptional orientation such that VHB1–8 will be expressed first (Fig. 2A). Transient transfection of targeted ES cell clones with a Cre-expressing plasmid yielded subclones that had deleted the neo-resistance (neor) gene or had deleted the neor gene and undergone a gene-inversion event (Fig. 2B). The occurrence of the latter in the rapidly dividing ES cells demonstrates that Cre-mediated gene inversion is not necessarily detrimental to mammalian cells in vivo, contrasting a previous report (25). We designate the targeted IgH allele, from which the B1–8 H chain is first expressed, as 3–83B1–8f. The second targeted allele, generated by inversion and allowing expression of the 3–83 H chain, is designated B1–83–83f. It is evident in the present strategy that one can obtain two strains of Ig insertion mice with a single gene-targeting event. This can be achieved either by injecting ES cells bearing the two different targeted alleles separately into blastocysts to derive the two strains of mice, or by crossing mice bearing one of the targeted alleles (e.g., 3–83B1–8f) with the deleter mice (26) to derive the second strain (e.g., B1–83–83f).
B lymphopoiesis in 3–83B1–8f/+ mice is normal and the size of the peripheral pool is comparable to that of wild-type animals (data not shown). Heterozygous mice carrying the 3–83B1–8f allele, which is of the a allotype, and a wild-type IgH b allele exhibit perfect allelic exclusion of the latter (data not shown). The anti-idiotypic antibody (Ab), Ac146 (22, 23), recognizes the V region encoded by VHB1–8 in combination with λ and most κ Ig L chains. Flow cytometric analysis of 3–83B1–8f/+ mice indicated that >70% of the B cells are Ac146+ and essentially all B cells express this idiotype (Id) when the 3–83κ L chain is introduced as a transgene (16, 21) into the mice (Fig. 2C). Together, these data indicate that essentially all B cells in 3–83B1–8f/+ mice first express the VHB1–8 gene only. Furthermore, comparable levels of surface Ig expression are found on B cells of 3–83B1–8f/+ and B1–8f/+ mice (17), which carry VHB1–8 as the sole transgene (data not shown). Thus, the positioning of two rearranged V region genes in tandem but opposite transcriptional orientation into the IgH locus neither perturbs B cell development in vivo nor affects the level of Ig expression on the B cells.
Comparison of the Efficiency of Induced Cre-Mediated Gene Inversion Versus Deletion.
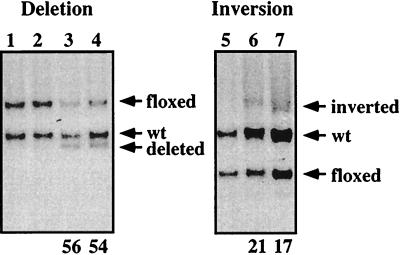
The Cre-mediated inversion reaction should result in an equal ratio of inverted and noninverted alleles. To directly compare the in vivo efficiency of Cre-mediated gene inversion with that of deletion, we injected type I IFN into 3–83B1–8f/+, Mx-cre and B1–8f/+, Mx-cre mice, derived, respectively, from crosses of 3–83B1–8f/+ and B1–8f/+ mice with the Mx-cre mice (15). The B1–8f/+, Mx-cre mice harbor a floxed VHB1–8 gene that can be inducibly deleted (17). In both mice, the floxed gene segments are of equivalent size (3 kb) and are targeted into the same locus, thus ruling out any size and positional discordance in the comparison. To avoid biasing of the results through cellular selection, we assayed the efficiencies of gene deletion and inversion in liver cells where Ig V region genes are not expressed. As shown in Fig. 3, the level of Cre-mediated gene deletion after a single, nonsaturating dose of IFN is approximately 55% whereas that of inversion is 20%. Thus, the efficiency of cre-mediated gene inversion is 40% of that of deletion, i.e., close to the theoretical expectation of 50%.
Figure 3.
Efficiency of induced cre-mediated gene inversion versus deletion. Southern blot analysis of DNA from liver of B1–8f/+, Mx-cre (lanes 1–4) and 3–83B1–8f/+, Mx-cre (lanes 5–7) mice that received a single dose of PBS (lanes 1, 2, and 5) or IFN (lanes 3, 4, 6, and 7). Genomic DNA were obtained from cells 3 days after treatment, digested with HindIII, and hybridized with probe (b) as shown in Fig. 2. Numbers indicate the amount of deletion or inversion of the floxed gene segment.
Rapid Elimination of Mature B Cells upon Expression of an Autoreactive Antigen Receptor.
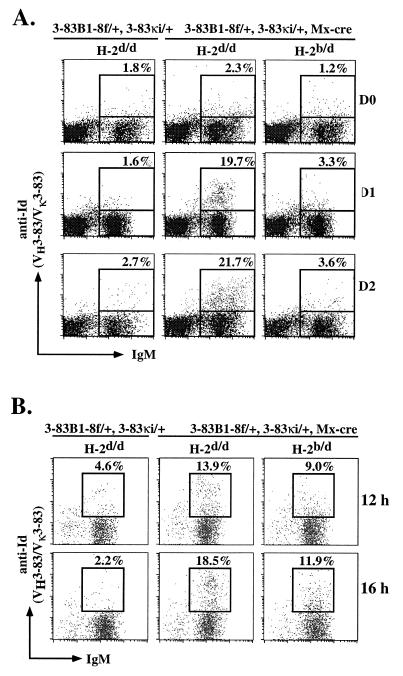
For the analysis of peripheral B cell tolerance, we crossed 3–83B1–8f/+, Mx-cre mice with 3–83κi/+ mice (16) and mice of the H-2b or H-2d haplotypes to obtain mice of the following genotypes: 3–83B1–8f/+, 3–83κi/+ (abbreviated Dtg), H-2d/d; Dtg, Mx-cre, H-2d/d; and Dtg, Mx-cre, H-2b/d. Before the induction of the BCR specificity switch on B cells by IFN injection, these mice were treated with the anti-interleukin-7 receptor (IL-7R) mAb to block B lymphopoiesis in the bone marrow (19). This is to ensure that the subsequent analysis of peripheral B cell tolerance is restricted to the mature IgM+ IgD+ B cell compartment in the peripheral tissues.
Expression of VH3–83 in association with Vκ3–83 can be monitored by the anti-idiotypic antibody, 54.1 (4, 6). As shown in Fig. 4A, 54.1 Id+ B cells are not detected in control Dtg, H-2d/d mice in the absence of the Mx-cre transgene or in its presence without IFN treatment, again suggesting that the VH3–83 gene is not expressed as protein when positioned in the wrong transcriptional orientation. IFN induction of BCR specificity switch is rather rapid, as 24 hr after treatment approximately 20% of all B cells in the spleens (Fig. 4A) and 15% of the B cells in the lymph nodes (data not shown) of the nonautoreactive Dtg, Mx-cre, H-2d/d mice express the 54.1 Id. These are cells that have switched BCR specificity in vivo from that of VHB1–8, Vκ3–83 to that of VH3–83, Vκ3–83. The frequency of switched cells is consistent with the Southern blotting data in Fig. 3. In line with our previous work (17), the initial BCR expressed by the cells before switching was gradually lost from the cell surface, and this is detected as a population of Ac146low cells in the spleen at early time points after IFN treatment (data not shown). The loss of the Ac146 Id from the switched cells was not studied in detail because of technical problems in the concomitant staining of cells with the Ac146 and 54.1 mAbs.
Figure 4.
Induced change in BCR specificity on mature B cells in vivo. Analysis of splenic B cells from 3–83B1–8f/+, 3–83κi/+, Mx-cre, H-2b/d and 3–83B1–8f/+, 3–83κi/+, Mx-cre, H-2d/d mice at various days (A) and at early time points (B) after IFN treatment. Control mice were of the H-2d/d haplotype and without the Mx-cre transgene. Cells were stained with fluorescein isothiocyanate (FITC)-anti-IgM and biotin-anti-Id (54.1) mAbs in A and with FITC-anti-IgM, phycoerythrin (PE)-anti-B220, and biotin-anti-Id (54.1) mAbs in B. Biotin-mAb is revealed with streptavidin-Cychrome. Numbers indicate percentage of total B cells. Only B220+ B cells are shown in B. Figures shown are representative of three separate experiments.
In contrast to the situation in the Dtg, Mx-cre, H-2d/d mice, 54.1 Id+ cells are not found in the spleens and lymph nodes of Dtg, Mx-cre mice of the autoreactive H-2b/d background (Fig. 4A) 1 day after IFN treatment. Analyses of mice 2–5 days after IFN treatment yielded similar results (Fig. 4A and data not shown). In addition, the population of Ac146low cells also is not detected (data not shown).
To ensure that the VH3–83, Vκ3–83 receptors are indeed expressed on the cell surface of H-2b/d B cells, we examined these cells at very early time points after IFN injection. Indeed, as shown in Fig. 4B, 54.1 Id+ B cells can be seen in Dtg, Mx-cre, H-2b/d mice 12–16 hr after IFN treatment. However, these cells express lower levels of the autoreactive BCR on their cell surfaces than control Dtg, Mx-cre, H-2d/d mice, perhaps reflecting the fact that these receptors continuously are being internalized upon engagement of the self-antigen. Since cells expressing the 54.1 Id are no longer detected in mice of the autoreactive background 1–2 days after IFN treatment, mature B lymphocytes that recognize a membrane-bound self-antigen with high affinity appear to be eliminated rapidly in the periphery.
Surprisingly, comparison of 54.1 Id+ cells from Dtg, Mx-cre mice of the autoreactive H-2b or nonautoreactive H-2d background at 12–16 hr after IFN treatment revealed no changes in the cell surface expression of MHC class II, CD23, CD40, CD44, CD69, CD86 (B7.2), and CD95 (Fas). Similarly, no differences in the expression of these molecules were detected between splenic 54.1 Id+ and 54.1 Id− cells in Dtg, Mx-cre, H-2b/d mice (data not shown). Thus, the cellular activation that could be expected to precede the deletion of mature B cells upon encountering a membrane self-antigen may be inefficient or abortive, perhaps because of a lack of T cell help.
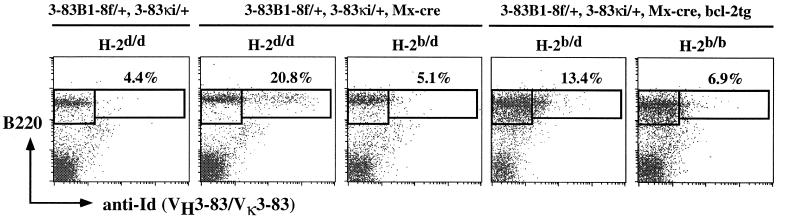
Constitutive bcl-2 Transgene Expression Prevents the Elimination of Mature Autoreactive B Cells.
Bcl-2 transgene (tg) expression has been shown to delay the elimination of immature autoreactive (27) and rescue peripheral (and likely, transitional) autoreactive B cells (28). To determine the effect of bcl-2 tg expression on mature autoreactive B cells, we crossed Dtg, Mx-cre mice of the H-2d/d, H-2b/d, or H-2b/b background with Eμ-bcl-2–22 mice (18). As shown in Fig. 5, 1 week after IFN treatment, a population of 54.1 Id+ B cells is found in Dtg, Mx-cre, bcl-2tg, H-2b/d mice, suggesting that constitutive bcl-2 tg expression indeed can rescue mature autoreactive B cells from elimination. This population of cells, however, expresses lower levels of cell surface Ig as compared with control mice of the nonautoreactive background. Surprisingly, 54.1 Id+ cells are barely detectable in Dtg, Mx-cre, bcl-2tg, H-2b/b mice that express twice as much self-antigens as H-2b/d mice. This suggests that the bcl-2tg can rescue mature autoreactive B cells in an antigen dose-dependent manner.
Figure 5.
Effect of constitutive bcl-2 expression on the survival of mature autoreactive B cells. Analysis of splenic B cells from Dtg, Mx-cre, bcl-2tg, H-2b/d, and H-2b/b mice and control Dtg, H-2d/d; Dtg, Mx-cre, H-2d/d; and Dtg, Mx-cre, H-2b/d mice 7 days after IFN treatment. Cells were stained with FITC-IgM, PE-B220, and biotin-anti-Id (54.1) mAb. Numbers indicate percentage of total B cells. Figures shown are representative of four sets of experiments.
DISCUSSION
The experimental strategy described here has allowed us to establish unequivocally that mature autoreactive B cells that recognize a membrane antigen with high affinity are eliminated by what appears to be clonal deletion, as indicated by the partial rescue of these autoreactive cells with a bcl-2 transgene.
Could our results be influenced by the IFN given to the animals to induce expression of the autoreactive BCR, given that type I IFN seems to increase the sensitivity of B cells to BCR ligation (29)? While this possibility can be excluded formally only in an experimental system that allows efficient Cre induction by a different inducer, we consider it unlikely. The high affinity of the 3–83 antibody for its target epitope together with the multivalent interaction of the 3–83 antibody-expressing B cells with their cellular target should guarantee potent B cell triggering also in the absence of IFN, which does not have a potentiating effect under the condition of saturating stimulation through the BCR (29).
Thus, selection of the mature IgM+ IgD+ B cells into the peripheral pool does not appear to prevent their rapid elimination once they encounter self-antigens in the periphery that bind to their BCR with high affinity. This rapid clonal deletion may reflect the physiological need to establish peripheral tolerance quickly to avoid autoimmunity. Given this rapid elimination process, it is unlikely that the mature autoreactive B cells that recognize membrane antigens with high affinity have the opportunity to edit their receptors even though RAG genes have been reported to be expressed in the periphery (30, 31). Indeed, recent studies have indicated that editing of high-affinity receptors may be confined to developing B cells (32), whereas mature B cells initiate receptor editing only upon low-affinity antigen binding (33). Presently, it is not known whether the lack of persistence of these mature autoreactive B cells results from a lack of T cell help after BCR engagement by membrane self-antigens, and whether the elimination process requires the presence of T cells and occurs via Fas–Fas ligand interaction (34).
The current experimental system now will allow us to address the mechanism(s) of peripheral tolerance that acts on Ig class-switched memory B cells whose BCR cross-reacts with self-antigens. This is of particular interest as memory B cells are selected and, perhaps, maintained on the basis of their high-affinity recognition of antigens.
Finally, the Cre-mediated gene inversion strategy allows one to reversibly or irreversibly (35) change gene expression in vivo. It thus can be used as a molecular “on/off” switch to study the reversibility of a mutant phenotype.
Acknowledgments
We thank Y.-R. Zou for drawing our attention to the utility of Cre-mediated inversion; R. Pelanda for the 3–83κi mice and VH3–83 gene; C. Weissmann for recombinant α/β IFN; S.-I. Nishikawa for anti-IL7R mAb (A7R34); H. von Boehmer for critical reading of the manuscript; and A. Egert and C. Königs for technical help. This work was supported by fellowships from the European Molecular Biology Organization and the Human Frontier Science Program to K.-P.L. and by grants from the Volkswagen Foundation, the Deutsche Forschungsgemeinschaft through SFB 243, the European Union (Bio4-CT96-0077), and the Land Nordrhein-Westfalen.
ABBREVIATIONS
- BCR
B cell antigen receptor
- H
heavy
- L
light
- V
variable
- Dtg
3–83B1–8f/+, 3–83ki/+
- IFN
interferon
- tg
transgene
- Id
idiotype
References
- 1. Tiegs S L, Russell D M, Nemazee D. J Exp Med. 1993;177:1009–1020. doi: 10.1084/jem.177.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gay D, Saunders T, Camper S, Weigert M. J Exp Med. 1993;177:999–1008. doi: 10.1084/jem.177.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodnow C C, Crosbie J, Adelstein S, Lavoie T B, Smith-Gill S J, Brink R A, Pritchard-Briscoe H, Wotherspoon J S, Loblay R H, Raphael K, et al. Nature (London) 1988;334:676–682. doi: 10.1038/334676a0. [DOI] [PubMed] [Google Scholar]
- 4.Nemazee D A, Burki K. Nature (London) 1989;337:562–566. doi: 10.1038/337562a0. [DOI] [PubMed] [Google Scholar]
- 5.Hartley S B, Crosbie J, Brink R, Kantor A B, Basten A, Goodnow C C. Nature (London) 1991;353:765–769. doi: 10.1038/353765a0. [DOI] [PubMed] [Google Scholar]
- 6.Russell D M, Dembic Z, Morahan G, Miller J F A P, Burki K, Nemazee D. Nature (London) 1991;354:308–311. doi: 10.1038/354308a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carsetti R, Kàhler G, Lamers M C. J Exp Med. 1995;181:2129–2140. doi: 10.1084/jem.181.6.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pulendran B, Kannourakis G, Nouri S, Smith K G, Nossal G J. Nature (London) 1995;375:331–334. doi: 10.1038/375331a0. [DOI] [PubMed] [Google Scholar]
- 9.Shokat K M, Goodnow C C. Nature (London) 1995;375:334–338. doi: 10.1038/375334a0. [DOI] [PubMed] [Google Scholar]
- 10.Lebecque S, de Bouteiller O, Arpin C, Banchereau J, Liu Y-J. J Exp Med. 1997;185:563–571. doi: 10.1084/jem.185.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkelman F D, Holmes J M, Dukhanina O I, Morris S C. J Exp Med. 1995;181:515–525. doi: 10.1084/jem.181.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fulcher D A, Lyons A B, Korn S L, Cook M C, Koleda C, Parish C, Fazekas de St. Groth B, Basten A. J Exp Med. 1996;183:2313–2328. doi: 10.1084/jem.183.5.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajewsky K, Gu H, Kühn R, Betz U A K, Müller W, Jürgen R, Schwenk F. J Clin Invest. 1996;98:600–603. doi: 10.1172/JCI118828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sonoda E, Pewzner-Jung Y, Schwers S, Taki S, Jung S, Eilat D, Rajewsky K. Immunity. 1997;6:225–233. doi: 10.1016/s1074-7613(00)80325-8. [DOI] [PubMed] [Google Scholar]
- 15.Kühn R, Schwenk F, Aguet M, Rajewsky K. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 16.Pelanda R, Schaal S, Torres R M, Rajewsky K. Immunity. 1996;5:229–239. doi: 10.1016/s1074-7613(00)80318-0. [DOI] [PubMed] [Google Scholar]
- 17.Lam K-P, Kühn R, Rajewsky K. Cell. 1997;90:1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- 18.Strasser A, Whittingham S, Vaux D L, Bath M L, Adams J M, Cory S, Harris A W. Proc Natl Acad Sci USA. 1991;88:8661–8665. doi: 10.1073/pnas.88.19.8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sudo T, Nishikawa S, Ohno N, Akiyama N, Tamakoshi M, Yoshida H, Nishikawa S-I. Proc Natl Acad Sci USA. 1993;90:9125–9129. doi: 10.1073/pnas.90.19.9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam K-P, Stall A M. J Exp Med. 1994;180:507–516. doi: 10.1084/jem.180.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelanda R, Schwers S, Sonoda E, Torres R M, Nemazee D, Rajewsky K. Immunity. 1997;7:765–775. doi: 10.1016/s1074-7613(00)80395-7. [DOI] [PubMed] [Google Scholar]
- 22.Reth M, Hämmerling G J, Rajewsky K. Eur J Immunol. 1978;8:393–400. doi: 10.1002/eji.1830080605. [DOI] [PubMed] [Google Scholar]
- 23.Reth M, Imanishi-Kari T, Rajewsky K. Eur J Immunol. 1979;9:1004–1013. doi: 10.1002/eji.1830091216. [DOI] [PubMed] [Google Scholar]
- 24.Taki S, Meiering M, Rajewsky K. Science. 1993;262:1268–1271. doi: 10.1126/science.8235657. [DOI] [PubMed] [Google Scholar]
- 25.Lewandoski M, Martin G. Nat Genet. 1997;17:223–225. doi: 10.1038/ng1097-223. [DOI] [PubMed] [Google Scholar]
- 26.Schwenk F, Baron U, Rajewsky K. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartley S B, Cooke M P, Fulcher D A, Harris A W, Cory S, Basten A, Goodnow C C. Cell. 1993;72:325–335. doi: 10.1016/0092-8674(93)90111-3. [DOI] [PubMed] [Google Scholar]
- 28.Lang J, Arnold B, Hämmerling G, Harris A W, Korsmeyer S, Russell D, Strasser A, Nemazee D. J Exp Med. 1997;186:1513–1522. doi: 10.1084/jem.186.9.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demengeot J, Vasconcellos R, Modigliani Y, Grandien A, Coutinho A. Int Immunol. 1997;9:1677–1685. doi: 10.1093/intimm/9.11.1677. [DOI] [PubMed] [Google Scholar]
- 30.Han S, Zheng B, Schatz D G, Spanopoulou E, Kelsoe G. Science. 1996;274:2094–2097. doi: 10.1126/science.274.5295.2094. [DOI] [PubMed] [Google Scholar]
- 31.Hikida M, Mori M, Takai T, Tomochika K, Hamatani K, Ohmori H. Science. 1996;274:2092–2094. doi: 10.1126/science.274.5295.2092. [DOI] [PubMed] [Google Scholar]
- 32.Melamed D, Benschop R J, Cambier J C, Nemazee D. Cell. 1998;92:173–182. doi: 10.1016/s0092-8674(00)80912-5. [DOI] [PubMed] [Google Scholar]
- 33.Hertz M, Kouskoff V, Nakamura T, Nemazee D. Nature (London) 1998;394:292–295. doi: 10.1038/28419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagata S, Golstein P. Science. 1995;267:1449–1456. doi: 10.1126/science.7533326. [DOI] [PubMed] [Google Scholar]
- 35.Araki K, Araki M, Yamamura K. Nucleic Acids Res. 1997;25:868–872. doi: 10.1093/nar/25.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]