Abstract
Inhibitors of poly (ADP-ribose)-polymerase-1 (PARP) are highly lethal to cells with deficiencies in BRCA1, BRCA2 or other components of the homologous recombination pathway. This has led to PARP inhibitors entering clinical trials as a potential therapy for cancer in carriers of BRCA1 and BRCA2 mutations. To discover new determinants of sensitivity to these drugs, we performed a PARP-inhibitor synthetic lethal short interfering RNA (siRNA) screen. We identified a number of kinases whose silencing strongly sensitised to PARP inhibitor, including cyclin-dependent kinase 5 (CDK5), MAPK12, PLK3, PNKP, STK22c and STK36. How CDK5 silencing mediates sensitivity was investigated. Previously, CDK5 has been suggested to be active only in a neuronal context, but here we show that CDK5 is required in non-neuronal cells for the DNA-damage response and, in particular, intra-S and G2/M cell-cycle checkpoints. These results highlight the potential of synthetic lethal siRNA screens with chemical inhibitors to define new determinants of sensitivity and potential therapeutic targets.
Keywords: CDK5, cell cycle, DNA repair, poly(ADP)ribose polymerase, RNAi screen
Introduction
Poly (ADP-ribose)-polymerase-1 (PARP) is a highly abundant nuclear enzyme involved in the repair of single-strand breaks (SSBs) (Hoeijmakers, 2001). Inhibition of PARP induces accumulation of large numbers of unrepaired SSBs, leading to the collapse of replication forks during S-phase and the consequent generation of double-strand breaks (DSBs). Cells deficient in DNA DSB repair, in particular homologous recombination (HR) by gene conversion, are highly sensitive to chemical inhibitors of PARP (Bryant et al, 2005; Farmer et al, 2005; McCabe et al, 2006). In contrast, cells with intact DNA DSB-response pathways repair damage with high fidelity and accordingly show very little sensitivity to PARP inhibitors. The breast and ovarian cancer predisposition genes, BRCA1 and BRCA2, encode proteins that are required for efficient HR (Moynahan et al, 1999; Tutt et al, 2001). Tumours arising in the carriers of heterozygous germline BRCA mutations have generally lost the wild-type BRCA allele, resulting in defective HR, which may be targeted in a synthetic lethal approach (Farmer et al, 2005). PARP inhibitors have now entered clinical trials and initial results are promising, with frequent sustained responses in BRCA mutation carriers (Yap et al, 2007).
Despite the clinical promise of PARP inhibitors in the treatment of BRCA-related cancer, extending the utility of these agents to other cancers is challenging. Little is known about the determinants of PARP-inhibitor sensitivity, other than the profound sensitivity of cells with defects in HR (McCabe et al, 2006). The identification of novel mediators of cellular response to PARP inhibitors may highlight additional patient populations that might benefit form this therapeutic approach. Furthermore, mechanisms of drug resistance and potential combination therapies may also be uncovered. RNA interference (RNAi) screens have the potential to identify novel determinants of drug response and hence enhance the application of novel and existing drugs (Iorns et al, 2007), and have already proven highly effective in the unbiased identification of novel genes involved in biological processes (Aza-Blanc et al, 2003; Mukherji et al, 2006). These screens exploit the naturally occurring mechanism of RNAi that controls gene expression at the post-transcriptional level by mediating degradation of mRNA transcripts in a sequence-specific fashion (Meister and Tuschl, 2004). With the development of RNAi libraries composed of reagents that allow targeting a wide range of transcripts, it is now possible to conduct high-throughput screens (HTS) that simultaneously interrogate phenotypes associated with the loss of function of many genes (Iorns et al, 2007). Here, we have used a high-throughput RNAi screen to identify new determinants of sensitivity to a PARP inhibitor.
Results
siRNA screen for kinases sensitising to a PARP inhibitor
RNAi screens that have previously examined sensitivity to DNA-damaging chemotherapy drugs have been limited by the small relative sensitivity, or therapeutic window, that exists between cells that are sensitive and resistant, limiting screens to identification of genes that cause profound effects when silenced (Bartz et al, 2006). DNA DSB repair-deficient cells are potentially more than a thousandfold more sensitive than resistant cells to PARP inhibitor (Bryant et al, 2005; Farmer et al, 2005; McCabe et al, 2006), probably due to the specificity of the DNA damage induced, increasing the ability of a screen to detect significant but less sensitising effects. We performed a PARP inhibitor synthetic lethal screen with a short interfering RNA (siRNA) library targeting 779 human protein kinase and kinase-associated genes. We selected kinases as they represent drugable targets. CAL51 cells were used for the screen, which are a diploid, TP53 wild-type breast cancer cell line. The HTS assay involved transfecting CAL51 cells with siRNA in a 96-well plate format and dividing the cells the day after transfection into replica plates, treating half with the PARP inhibitor KU0058948 and half with the vehicle (Figure 1A). The screen was optimised to detect modestly sensitising effects by using a dose of KU0058948 sufficient to inhibit the repair of SSBs (data not shown) and equivalent to the SF80 (80% survival after KU0058948 administration). Furthermore, cells were exposed to drug continuously for 5 days to allow multiple cell cycles to occur, allowing effects of PARP inhibition to accumulate and modelling chronic exposure to these drugs in the clinic.
Figure 1.
PARP-inhibitor synthetic lethality screen with protein kinase siRNA library. (A) HTS method. CAL51 cells plated in 96-well plates were transfected with siRNA. Each transfection plate contained 80 experimental siRNAs (SMARTPools of four different siRNA targeting the same gene) supplemented with four wells of non-targeting siCON, and two wells of siRNA directed against BRCA1 (positive control). Transfected cells were divided into six replica plates, half treated with DMSO vehicle alone and half with PARP inhibitor KU0058948 at 1 μM, the SF80 of CAL51. Cell viability was assessed after 5 days of KU0058948 exposure using CellTiter-Glo Luminescent Cell Viability Assay (Promega). (B) Reproducibility of HTS method. Correlation of the effect of siRNA on cell growth in vehicle-treated plates from two replicates of the entire screen. Spearman correlation coefficient, r=0.83. (C) Correlation of KU0058948 sensitivity Z-scores from two replicates of the entire screen. r=0.54. (D) Scatter plot of averaged Z-scores from PARP inhibitor sensitivity screen carried out in duplicate with KU0058948. Red line indicates −3 averaged Z-score significance threshold. Black, siRNA (SMARTPools) targeting 779 protein kinase genes; red, siCON and blue, siBRCA1. Reflecting the reproducibility and sensitivity of the screen, siCON and siBRCA1 Z-scores were widely separated, with a screen Z′-factor (Zhang et al, 1999) of 0.34.
The screen was completed in duplicate. Comparison of the two duplicates revealed the screen to be highly reproducible (Figure 1B and C). The duplicates of the screen were combined in the final results of the screen displayed in Figure 1D. A robust significance or ‘hit' threshold of a combined Z-score of −3 or less was selected, with 24 gene-specific siRNA pools (SMARTPools®; Dharmacon) fulfilling this criterion (Table I). Full results of the screen are supplied as Supplementary Table 1. Internal validation of the high sensitivity of the screen was provided by the demonstration that siRNAs targeting the key DNA-damage response genes, ataxia–telangiectasia related (ATR), ataxia–telangiectasia mutated (ATM) and CHK1, significantly sensitised to the PARP inhibitor (Table I), as we have previously reported (McCabe et al, 2006), emphasising the importance of intact DNA DSB-response pathways in tolerance to PARP inhibitors.
Table 1.
Results of PARP inhibitor synthetic lethal siRNA screen
| siRNA SMARTpool® | KU0058948-sensitivity Z-score | Growth % siCONTROL |
|---|---|---|
| IMPK | −8.33 | 96 |
| PNKP | −8.21 | 54 |
| ATR | −7.27 | 106 |
| TTBK1 | −6.31 | 60 |
| STK36 | −5.50 | 65 |
| PLK1 | −5.15 | 6 |
| STK35 | −5.09 | 77 |
| CDC2 | −4.17 | 55 |
| ADRA1A | −4.15 | 56 |
| PIP5K2B | −4.10 | 48 |
| PLK3 | −3.99 | 73 |
| ITGB1BP1 | −3.94 | 86 |
| STK22C | −3.85 | 87 |
| AKAP1 | −3.64 | 48 |
| FLJ34389 | −3.35 | 63 |
| GSK3A | −3.32 | 78 |
| CHEK1 | −3.26 | 42 |
| CDK5 | −3.18 | 106 |
| ATM | −3.12 | 92 |
| MAPK12 | −3.11 | 59 |
| PMVK | −3.07 | 83 |
| DAPK1 | −3.07 | 66 |
| MGC5601 | −3.05 | 52 |
| PRKD2 | −3.00 | 97 |
| Results of siRNA screen displayed in Z-score order with siRNA SMARTpool target gene, KU0058948 PARP-inhibitor sensitivity Z-score and the effect of siRNA on cell growth in vehicle-alone plates. The first 24 siRNAs are statistically significant, with a Z-score of less than −3. Hits in bold italic have well-documented roles in the DNA-damage response, and have previously been shown to sensitise to PARP inhibitors. Hits in boldface are revalidated in subsequent experiments as being on-target. Statistically significant hits 21–24 were not further revalidated. |
Validation of siRNA screen hits
In addition to silencing a target gene, siRNAs potentially suppress the expression of a large number of other genes through off-target effects. Therefore, we repeated the HTS assay separately with each of the four different siRNA species included in the original SMARTPool®. The HTS results were likely to be ‘on-target' when two or more individual siRNA targeting the same gene sensitised to KU0058948 (Echeverri et al, 2006). The top 20 hits from the screen were re-examined (Table I), excluding ATR, ATM and CHK1 as we have established previously that these are determinants of KU0058948 sensitivity (McCabe et al, 2006). Of the remaining 17 hits, 11 were potentially shown to be due to off-target effects of single-siRNA species (data not shown).
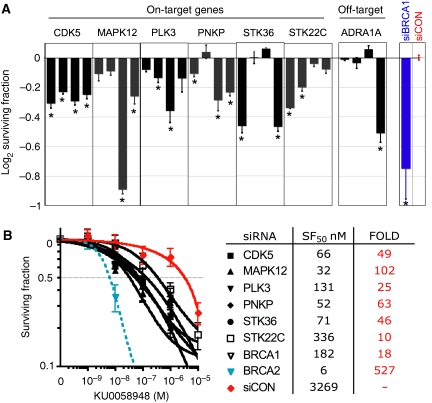
The six on-target hits (Figure 2A) were cyclin-dependent kinase 5 (CDK5, Entrez GeneID 1020), which revalidated with all four individual siRNAs; polynucleotide kinase 3′-phosphatase (PNKP, aka PNK, GeneID 11284) positive with three siRNAs and mitogen-activated protein kinase 12 (MAPK12, aka P38γ, GeneID 6300), Polo-like kinase 3 (PLK3, GeneID 1263), serine/threonine kinase 36 (STK36, fused homologue, GeneID 27148) and serine/threonine kinase 22C (STK22C, aka TSSK3, GeneID 81629), all of which revalidated with two siRNAs. We repeated the PARP sensitivity assay in HeLa cells, a cervical carcinoma cell line. Silencing of CDK5, MAPK12, PNPK and STK22C significantly sensitised to PARP inhibitor in this cell line (Supplementary Figure 3), confirming the effects we had seen in CAL51 cells. However, PLK3 silencing had no effect on PARP-inhibitor sensitivity in HeLa cells, suggesting the mechanism of sensitivity following PLK3 silencing might be restricted to specific cell lines. To establish the sensitivity of the revalidated on-target hits to PARP inhibitor, dose–response relationships were determined by clonogenic assay following SMARTpool® siRNA silencing (Figure 2B). All six hits dramatically sensitised cells to KU0058948 (10- to 102-fold sensitivity). Confirmation of gene silencing by siRNA was performed by quantitative real-time polymerase chain reaction (PCR) (Supplementary Figure 1).
Figure 2.
Validation of PARP-inhibitor synthetic lethality screen. (A) Validation of hits from the PARP inhibitor HTS. PARP inhibitor sensitivity assay repeated in triplicate with the four different siRNA originally included in each SMARTPool. The surviving fractions following PARP inhibition are shown, including those after transfection with siBRCA1 (blue) and siCON (red). CDK5 revalidated with all four siRNA; PNKP revalidated with three siRNAs; and MAPK12, PLK3, STK36 and STK22C revalidated with two siRNA. As an example of a probable off-target hit, ADRA1A silencing sensitises to PARP inhibitor due to an off-target effect of the fourth siRNA. *P<0.0227 compared with siCON (Student's t-test). Error bars represent the s.e.m. (B) PARP-inhibitor sensitivity assessed by clonogenic assay in the six novel validated hits from the screen. Error bars represent s.e.m. of three independent experiments.
CDK5 functions in the DNA-damage response
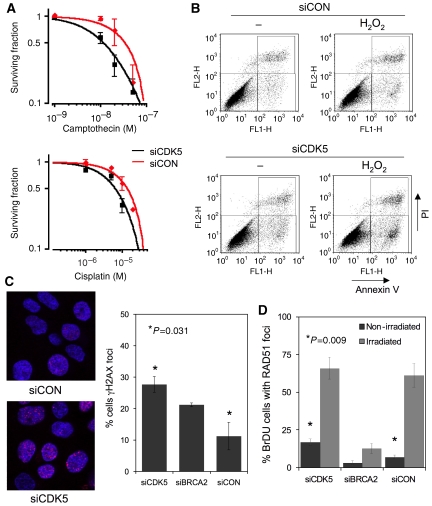
CDK5 is an unusual CDK, previously thought to be active only in post-mitotic neurones due to the perceived neuronal-specific expression of its activators, CDK5R1 (p35) and CDK5R2 (p39) (Dhavan and Tsai, 2001). In non-neuronal cells, no role for CDK5 in the DNA-damage response has previously been suggested and we, therefore, chose to examine the biological significance of CDK5 silencing and the mechanism of sensitivity to PARP inhibitor in more detail. Having confirmed CDK5 silencing by siRNA (Supplementary Figure 1), we initially investigated whether CDK5 silencing sensitised to other DNA-damaging agents. CDK5-silenced cells were also more sensitive than control cells to camptothecin and cisplatin (Figure 3A), but not to the microtubule poison docetaxel (data not shown), suggesting that the effect of CDK5 silencing was not restricted to DNA damage by PARP inhibition. This was accompanied by induction of an apoptotic response following DNA damage in CDK5-silenced cells (Figure 3B). An early marker of the presence of DNA DSBs is the phosphorylation of the highly conserved histone H2AX. Following CDK5 knockdown, phosphorylated H2AX (γH2AX) foci were increased in the absence of exogenous DNA damage (Figure 3C). We examined whether the observed increase represented induction or persistence of γH2AX foci. While the basal level of γH2AX foci was increased in siCDK5-transfected (CDK5 SiRNA) cells, the resolution of ionising radiation-induced foci was similar to that in siCONTROL (siCON)-transfected cells (Supplementary Figure 2). This suggested that the increase in γH2AX foci following CDK5 silencing was due to increased induction of γH2AX and not persistence. This increase in γH2AX foci was also associated with a small, but significant, increase in RAD51 foci, a hallmark of repair by HR (Figure 3D).
Figure 3.
CDK5 silencing sensitises cells to DNA-damaging agents and induces a DNA-damage response. (A) Clonogenic survival assays in CAL51 cells transfected with siCDK5 or siCON and treated with camptothecin (SF50 siCON 33.5 nM, siCDK5 11.3 nM, threefold more sensitive) or cisplatin (SF50 siCON 13.8 μM, siCDK5 6.4 μM, 2.2-fold more sensitive). (B) Sensitivity to DNA damage in CDK5-silenced cells is characterised by an increase in apoptosis. FACS plots showing Annexin V and propidium iodide (PI) staining after treatment with hydrogen peroxide and either siCON or siCDK5 transfection. Percentages of Annexin V-positive/PI-negative cells were as follows: siCON-transfected cells, 3.4% (−H2O2) and 4.5% (+H2O2); siCDK5-transfected cells, 6.2% (−H2O2) and 10.4% (+H2O2). (C) CDK5 silencing induces γH2AX foci formation. The panels indicate confocal microscopic images from siCON- and siCDK5-transfected cells, with red indicating γH2AX and blue indicating ToPro3 DNA staining. Quantification of cells with ⩾5 γH2AX foci from three independent experiments. Error bars represent s.e.m.; silencing of CDK5 P=0.031 relative to siCON (Student's t-test). Silencing of BRCA2 acted as a control, P>0.05 relative to siCON. (D) CDK5 silencing induces basal RAD51 foci formation. Quantification of basal RAD51 foci, and RAD51 foci 5 h after 8-Gy irradiation (non-irradiated, siCDK5 transfected 17% versus siCON 7%). Error bars represent s.e.m. of three independent experiments; *P=0.009 relative to siCON (Student's t-test). Silencing of BRCA2 acted as a control for impairment of the radiation-induced foci.
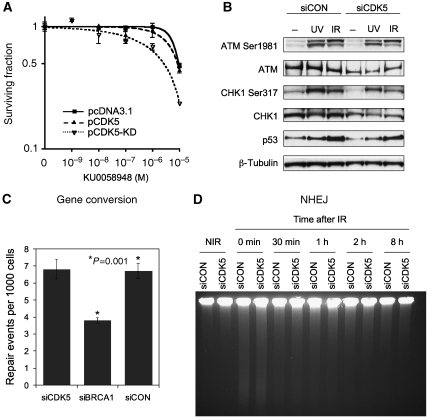
CDK5 silencing, therefore, induced spontaneous formation of DNA DSB and induced markers of DNA DSB repair. We assessed the relevance of CDK5 kinase activity to PARP-inhibitor sensitivity by transiently expressing a dominant-negative, kinase-dead D145N CDK5 mutant (van den Heuvel and Harlow, 1993). This mutant differs from wild-type CDK5 in only a single amino-acid change that abrogates the kinase activity of CDK5 (van den Heuvel and Harlow, 1993). Expression of dominant-negative CDK5 sensitised to PARP inhibitor, whereas expression of exogenous wild-type CDK5 did not (Figure 4A). This suggested that kinase activity of CDK5 was required for tolerance of PARP inhibitors. CDK5 activity was examined by IP kinase assay. CDK5 kinase activity increased after irradiation in CAL51 cells (Supplementary Figure 2A). To address the possibility that ATM and CDK5 act in a common pathway, we performed epistasis experiments. We observed no significant difference in PARP-inhibitor sensitivity between ATM inhibition and ATM inhibition with additional CDK5 silencing, suggesting that CDK5 and ATM act in a common pathway that sensitises to PARP inhibitor (Supplementary Figure 2B).
Figure 4.
CDK5 is not required for early DNA DSB signalling or DNA DSB repair. (A) Expression of dominant-negative CDK5 sensitises to PARP inhibitor. CAL51 cells transfected with empty vector (pcDNA3.1empty), CDK5 expression vector (pCDK5) or kinase-dead D145N CDK5 expression vector, and 24 h following transfection plated for clonogenic assay with KU0058948. (B) CDK5 is not required for ATM and ATR activation. CAL51 cells transfected 72 h earlier with siRNA where treated with 10-Gy irradiation, 50 J/m2 ultraviolet light or not treated. Lystates where made 1 h following treatment, and a western blot was probed with antibodies against ATM Ser1981 autophosphorylation site and CHK1 Ser317 ATM/ATR phosphorylation site, with total ATM, CHK1, TP53 and a β-Tubulin loading control. (C) CDK5 does not affect homologous recombination by gene conversion. Gene conversion was measured using an adapted single-copy, chromosomally integrated HR reporter construct in a 293 human embryonic kidney cell line (Tutt et al, 2001). Silencing of BRCA1 acted as a positive control for reduced gene conversion. Details of this assay are outlined in Supplementary Figure 4. (D) CDK5 silencing does not affect DNA DSB end joining. FIGE assay in CAL51 cells transfected with siCDK5 or siCON. Time points represent minutes after 30 Gy irradiation and non-irradiated control (NIR).
The potential mechanism of sensitivity to PARP inhibitor following CDK5 silencing was also investigated. We first examined the integrity of early DNA DSB-damage signalling. Following CDK5 silencing there was normal autophosphorylation of ATM on serine 1981 following irradiation, and normal phosphorylation of CHK1 on serine 317 after ultraviolet light exposure, indicating retained ATR signalling (Jazayeri et al, 2006; Figure 4B). Induction of TP53 expression following irradiation was also normal following CDK5 silencing. CDK5 was therefore not required for activation of ATM or ATR following DNA damage, and most likely functioned downstream of initial DNA-damage signalling.
The integrity of DNA DSB repair pathways, one determinant of PARP-inhibitor sensitivity (Farmer et al, 2005; McCabe et al, 2006), was investigated in CDK5-silenced cells. Two main DNA DSB repair pathways predominate, HR by gene conversion and non-homologous end joining (NHEJ) (Hoeijmakers, 2001). We measured gene conversion using an adapted single-copy, chromosomally integrated HR reporter construct present in a 293 cell line (Tutt et al,2001; described in Supplementary Figure 4). Transfection of cells with BRCA1 siRNA significantly reduced HR in this assay (Figure 4C), as expected, of a well-defined HR gene. Silencing of CDK5 had no detectable effect on HR (Figure 4C), in keeping with the normal formation of irradiation-induced RAD51 foci (Figure 3D). We also confirmed this observation using a green fluorescent protein reporter of HR (Pierce et al, 1999; Supplementary Figure 5). NHEJ was assessed using field-inversion gel electrophoresis (FIGE) to measure gross DNA fragmentation following irradiation. NHEJ was also found to be normal in siCDK5-transfected cells (Figure 4D). We conclude that, although CDK5 is required for response to DNA damage, and activated after DNA damage, it is not directly required for DNA DSB repair.
CDK5 is required for DNA-damage checkpoint activation
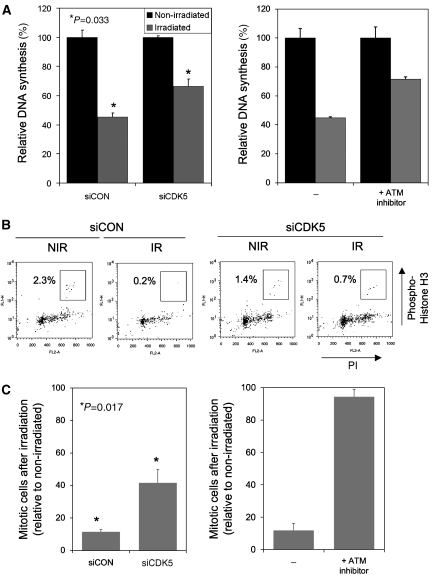
A number of partially overlapping DNA-damage response pathways regulate the cell cycle following DNA damage. The intra-S-phase checkpoint inhibits firing of new replication origins after DNA damage, causing a relative decrease in DNA synthesis after irradiation (Bartek et al, 2004). Following CDK5 silencing, radiation-resistant DNA synthesis (RDS) was assayed by 3H-labelled thymidine DNA incorporation and was shown to increase, suggesting a defect in the intra-S-phase checkpoint (Figure 5A). The magnitude of RDS after CDK5 silencing was comparable to that previously observed with silencing of MDC1, a protein crucial to activation of the intra-S-phase checkpoint (Goldberg et al, 2003). We compared the intra-S-phase checkpoint defect in siCDK5-transfected cells with that in cells with ATM-kinase activity inhibited. There was no significant difference in the magnitude of RDS between siCDK5-transfected cells and those treated with a concentration of the small-molecule inhibitor KU0055933 known to effectively abolish ATM-kinase activity (Hickson et al, 2004; Figure 5A, relative DNA incorporation siCDK5 66% versus ATM inhibitor 72%, P=0.48). A defect in the intra-S-checkpoint was also observed in cells transfected with a series of different siRNAs targeting CDK5 (Supplementary Figure 2), and in HeLa cells (Supplementary Figure 3), suggesting this phenotype was unlikely to be the result of off-target effects nor specific to CAL51 cells.
Figure 5.
CDK5 is required for cell-cycle DNA-damage checkpoints. (A) CDK5 is required for the intra-S-phase checkpoint. 3H-labelled thymidine incorporation was assessed by scintillation counting in an assay of radiation resistant DNA synthesis. DNA synthesis in cells irradiated 1 hour earlier with 10-Gy expressed relative to non-irradiated cells. Error bars represent s.e.m. of three independent experiments. Relative DNA synthesis after irradiation 45% siCON versus 66% siCDK5, *P=0.033 (Student's t-test). Right panel, DNA synthesis in untransfected CAL51 cells and CAL51 cells exposed to 10 μM KU0055933 (ATM inhibitor), relative DNA synthesis 45 versus 72%, respectively P<0.01. (B, C) CDK5 is required for G2/M checkpoint function. (B) FACS plots of cells stained with propidium iodide and an antibody directed against the mitosis marker phospho-histone H3. Mitotic cells are highlighted in the box. (C) Quantification of three independent experiments, with proportion of mitotic cells after irradiation expressed relative to non-irradiated cells. Error bars represent s.e.m. of three independent experiments. Relative mitotic entry after irradiation 12% siCON versus 42% siCDK5; *P=0.017 (Student's t-test). Right panel, mitotic entry in untransfected CAL51 cells and CAL51 cells exposed to 10 μM KU0055933 (ATM inhibitor), relative mitotic entry 11 versus 97%, respectively; P<0.001.
Genes involved in the intra-S-phase checkpoint are frequently involved in the G2/M checkpoint that prevents cells with unrepaired DNA damage from entering mitosis by arresting the cell cycle at the G2/M transition (Mailand et al, 2002). Following CDK5 silencing, an abnormally high percentage of cells remained in mitosis after irradiation, suggesting significant defect in the G2/M checkpoint (Figure 5B and C), although it should be noted that the siCDK5-mediated effect was modest compared with that of ATM inhibition. A modest G2/M defect was also observed in HeLa cells transfected with siCDK5. This supports our contention of a generally applicable non-neuronal role for CDK5 in the DNA-damage response.
Discussion
Our screen has identified new determinants of sensitivity to PARP inhibitors and highlights how the functional profiling of new cancer drugs may become valuable in the drug development process (Iorns et al, 2007). PARP inhibitors are showing considerable promise as cancer drugs in early clinical trials (Yap et al, 2007), and the work described here identifies new avenues of research to extend the utility of these agents. We have demonstrated that sensitivity to PARP inhibitors can result from defective DNA-damage cell-cycle checkpoints, identifying a novel mechanism of sensitivity to PARP inhibitors. We envisage that tumours with reduced or no expression of the genes identified by us might be selectively sensitive to PARP inhibitors, as has been shown for BRCA1- and BRCA2-deficient cells. In addition, this screen has identified therapeutic targets whose inhibition would potentially synergise with PARP inhibitors in the clinic.
Of the novel determinants of PARP sensitivity identified in our screen, some have been previously linked to DNA-damage response pathways. PNKP is a DNA kinase/phosphatase enzyme involved in the processing of damaged DNA ends prior to ligation. PNKP has previously been shown to be involved the repair of SSBs (Jilani et al, 1999) and also in DNA DSBs repair by NHEJ (Chappell et al, 2002). It will be interesting to elucidate which of these potential mechanisms underlie the sensitivity to PARP inhibitors on PNKP silencing. PLK3 has previously been suggested to have a role in the G2/M checkpoint following irradiation (Bahassi el et al, 2004), and in G1/S-phase progression (Zimmerman and Erikson, 2007). MAPK12/P38γ has also been shown previously to be required for the G2/M checkpoint (Wang et al, 2000). These previous observations provide further evidence of the importance of cell-cycle checkpoints in the cellular response to PARP inhibitors. Interestingly the magnitude of sensitivity to PARP inhibitor after silencing of these genes was lower than that observed with BRCA2 silencing (Figure 2). This may suggest different levels of sensitivity to PARP inhibitor, depending on the underlying defect in the DNA-damage response. It could be that defects in the core HR proteins may lead to profound sensitivity, whereas defects in cell-cycle control may lead to significant, although less marked, sensitivity. STK36, the fused homologue, has not been shown previously to have a role in DNA-damage responses and neither had CDK5.
The CDK5 gene is located at the telomeric region of chromosome 7q, distal to the fragile site FRA7I (Ciullo et al, 2002). A re-analysis of previously published data from breast cancers (Chin et al, 2006) revealed that genomic loss of CDK5 occurred in 5.5% (8/145) of breast cancers, with evidence of homozygous loss in one cancer (data not shown). Loss of CDK5 was associated with significant reduction in gene expression. Furthermore, CDK5 expression data in Oncomine (http://www.oncomine.org) reveal that variations in the expression of CDK5 are common during tumour progression (Chen et al, 2002; Graudens et al, 2006; Sanchez-Carbayo et al, 2006). Therefore, a significant population group may exist that could benefit from PARP-inhibitor treatment because of reduced, tumour-specific, CDK5 expression.
A number of previous studies have demonstrated that CDK5 is activated in neuronal cells after DNA damage, but its precise role in DNA-damage responses is unclear (Strocchi et al, 2003; Lee and Kim, 2007). Our results demonstrate that CDK5 plays a key role in DNA-damage response and in cell-cycle checkpoint activation in non-neuronal cells. We show that CDK5 is required for intra-S-phase checkpoint, suggesting that the mechanism of sensitivity to PARP inhibitor in CDK5-silenced cells is a failure of this checkpoint. It is possible that in the presence of greatly increased SSBs, failure of this checkpoint leads to increased replication fork collapse and subsequent cell death. The observed induction of γH2AX foci after CDK5 silencing is also likely to result from an impaired intra-S-phase checkpoint, perhaps arising from increased replication fork collapse at sites of endogenous DNA damage. The specific role of CDK5 in cell-cycle checkpoints remains to be determined. CDK5 silencing does not grossly modify the stability of the CDC25A phosphatase that partially determines S-phase delay (NC Turner and A Ashworth, unpublished observations). However, we have identified elements of the SCF (scf, Cullin, F-box containing) ubiquitin ligase complex as CDK5-interacting proteins (R Elliott and A Ashworth, unpublished observations). This is intriguing given the role of SCF ubiquitin ligase components in cell-cycle control (Bai et al, 1996). We provide evidence that kinase activity of CDK5 is required for PARP-inhibitor sensitivity; expression of a dominant-negative, kinase-dead CDK5 mutant sensitises to KU0058948 and CDK5 kinase activity increases after DNA damage. However, we cannot exclude the possibility that the kinase activity identified in the IP kinase assays is due to a CDK5-associated protein and not CDK5 itself. Non-catalytic functions have previously been reported for cyclin A–cdk2 in cell-cycle control through interaction with the SCF complex (Zhu et al, 2004). Potentially, CDK5 could mediate checkpoint activation through a non-catalytic interaction with DNA-damage kinases or complexes such as SCF.
CDK5 has also been implicated in Alzheimer's disease pathogenesis through aberrant phosphorylation of TAU and the resultant formation of neuro-fibrillary tangles (Dhavan and Tsai, 2001; Cruz and Tsai, 2004). Neurodegenerative disorders, including Alzheimer's disease, are characterised by reactivation of the cell-cycle machinery in previously quiescent, post-mitotic, neurones (Woods et al, 2007). It is, therefore, possible that the role we have identified for CDK5 in cell-cycle checkpoint regulation is relevant to reactivation of the cell cycle in neurodegeneration. Finally, as germline mutations in other DNA-damage checkpoint genes are linked to development of breast and other cancers (Renwick et al, 2006) our results suggest that it will be important to examine the role of CDK5, and other genes we have identified, in cancer predisposition.
Materials and methods
Cell lines, compounds and siRNA
CAL51 and HeLa cells were obtained from ATCC (USA) and maintained in Dulbecco's modified Eagle's medium (Sigma, Poole, UK) supplemented with 10% fetal calf serum (10% vol/vol) glutamine and antibiotics. The inhibitors of PARP (KU0058684, IC50 3.2 nM) (Farmer et al, 2005) and ATM (KU0055933, IC50 13 nM) (Hickson et al, 2004) have been described previously. Unless otherwise stated, siCDK5 was a pool of four different siRNA all targeting CDK5 (CDK5 SMARTPool). Sequences of all siRNAs are supplied in Supplementary Table 2. The protein kinase siRNA library (siARRAY, targeting 779 known and putative human protein kinase genes) was obtained in 10, 96-well plates from Dharmacon (USA). Each well in this library contained a SMARTPool of four distinct siRNA species targeting different sequences of the target transcript.
Antibodies
Antibodies targeting the following epitopes were used: ATM (ab2631; Abcam, UK), phospho-Ser1981–ATM (17168; Rockland, USA), BRCA1 (8F7; GeneTex, USA), BRCA2 (Ab-1; Calbiochem, USA), CDK5 (C-8/sc-173; Santa-Cruz, USA), CDK5 (DC17/ab3226; Abcam, UK), CHK1 (Ab2845; Abcam), phospho-S317–CHK1 (BL229; Bethyl, USA), phospho-histone–Ser10-H3 (06-570; Upstate, USA), phospho-Ser139–H2AX (05-636, γH2AX; Upstate, USA), RAD51 (sc-8349; Santa-Cruz, USA), P53 (Ab8; Neomarkers, USA), β-tubulin (T4026; Sigma, UK).
HTS screen method
CAL51 cells plated in 96-well plates were transfected 24 h later with siRNA (final concentration 100 nM), using Oligofectamine (Invitrogen, USA) as per manufacturer's instructions. Twenty-four hours following transfection, cells were trypsinised and divided into six identical replica plates. At 48 h after transfection, three replica plates were treated with 0.01% (vol/vol) dimethylsulphoxide (DMSO) vehicle in media and three replica plates with 1 μM KU0058948 (PARP inhibitor) in media. Media containing KU0058948 or vehicle was replenished after 48 h, and cell viability was assessed after 5 days of KU0058948 exposure using CellTiter-Glo® Luminescent Cell Viability Assay (Promega, USA) as per manufacturer's instructions. The luminescence reading for each well on a plate was expressed relative to the median luminescence value of all wells on the plate. The screen was completed in duplicate after rejecting plates from the screen if mean growth in siCON wells was less than 60% of untransfected control wells. For each transfection, the following were calculated:
Cell growth. The effect of each individual siRNA SMARTPool on cell growth alone was calculated by dividing mean luminescence in the three replica wells treated with DMSO by the mean luminescence of the replica wells transfected with siCON, and expressed as a percentage. Cell growth effect of siRNA (%)=mean (three replica wells with siRNA)/mean (12 replica wells with treated siCON) × 100.
PARP-inhibitor sensitivity. Sensitivity to PARP inhibitor for each siRNA SMARTPool was assessed by calculating the surviving fraction following PARP inhibitor. Surviving fraction=log2mean (three replica wells with KU0058948)−log2mean (three replica wells with DMSO).
The surviving fractions were centred on the median surviving fraction of all 80 SMARTPools from one 96-well plate transfection, the results from all ten siRNA plates combined and results expressed as a Z-score. For the Z-score the standard deviation of the screen was estimated from the median absolute deviation of all 779 SMARTPools adjusted by a factor of 1.4826 for equivalence with an asymptotically normal distribution. A robust significance threshold of 3 Z-scores was selected to reduce the identification of screen false positives. The Z′-factor was calculated using the siCON and siBRCA1 control wells, as described elsewhere (Zhang et al, 1999).
Validation of HTS screen
Four distinct siRNA species targeting each gene were used to revalidate hits from the screen. A significance threshold of P<0.0227 was used for each siRNA, to adjust for multiple comparisons, yielding a combined P<0.003 that two or more siRNA sensitise to KU0058948 for any one gene. Following 17 comparisons, P<0.00301 would be considered statistically significant (Sidak's adjustment). Validation of RNAi gene silencing was by real-time reverse transcriptase–PCR, or western blotting, as described previously (McCabe et al, 2006).
Clonogenic survival assays to measure drug sensitivity
CAL51 cells were transfected with siRNA using Oligofectamine (Invitrogen, UK) as per manufacturer's instructions, divided 48 h following transfection into six-well plates and exposed to various doses of drug from 60 h post transfection. Colonies were fixed and counted at 10–14 days post transfection, and the surviving fraction for each dose of drug was assessed. Survival curves were generated as described previously (Farmer et al, 2005). Drug treatments consisted of either continuous exposure to KU0058948, 24-h exposure to camptothecin and 1 h exposure to cisplatin at 72 h post transfection. For assessment of PARP sensitivity following exogenous CDK5 expression, CAL51 cells were transfected with Fugene HD (Roche Applied Science, USA) as per manufacturer's instructions, with pcDNA3.1 empty vector (Invitrogen), CDK5-HA and CDK5-DN-HA (van den Heuvel and Harlow, 1993). The transfected cells were selected with G418 (Invitrogen) for the initial 4 days of the clonogenic assay.
Western blotting and IP/kinase assay
Western blots were carried out with precast TA or Bis-Tris gels (Invitrogen) as described previously (Farmer et al, 2005). The IP kinase assay was performed essentially as described previously (Patrick et al, 1999).
Immunofluorescence and FACS analysis
Formation and quantification of DNA-damage-induced foci and Annexin V fluorescence-activated cell sorting (FACS) were performed as described previously (Farmer et al, 2005). For RAD51 foci, cells were pulsed with 10 μM bromodeoxyuridine (BrDU) for 30 min before irradiation to identify cells in S-phase at time of irradiation. The percentage of BrDU-incorporating cells with ⩾5 RAD51 foci was assessed in irradiated and non-irradiated cells. FACS analysis of histone H3 phosphorylation was carried out on CAL51 cells transfected 72 h earlier. CAL51 cells were irradiated (3 Gy) and then fixed 1 h later with 75% (vol/vol) ethanol, permeabilised with 0.25% (vol/vol) Triton X-100 in phosphate-buffered saline (PBS), incubated with 1 μg/ml anti-phospho-Histone H3 antibody for 3 h and incubated with secondary anti-rabbit-Alexa-555 antibody conjugate (1:1000) for 1 h at room temperature. DNA was stained with propidium iodide in the presence of RNAse A. The proportion of cells in mitosis after irradiation was expressed relative to the proportion of cells in mitosis in a non-irradiated sample. For ATM inhibitor controls, CAL51 cells were exposed to 10 μM KU0055933 for 30 min prior to irradiation and throughout the assay.
Radiation resistant DNA synthesis
RDS was assessed 72 h post transfection of siRNA. Cells were irradiated (10 Gy), or not, and 1 h post-irradiation pulsed with 10 μM 3H-labelled thymidine (Amersham, UK) in media for 1 h. Cells were washed twice with PBS, followed by a 30 min chase with media lacking 3H-labelled thymidine. Cells were lysed with 0.25 M NaOH, lysates transferred to scintillation vials and counts per minute were measured in a scintillation counter. DNA incorporation after irradiation was expressed relative to DNA incorporation in non-irradiated wells. For ATM-inhibitor controls, CAL51 cells were exposed to 10 μM KU0055933 for 30 min prior to irradiation and throughout the assay.
Field-inversion gel electrophoresis
FIGE was performed as described previously (Wong et al, 2000).
Assay of HR by gene conversion
This procedure is described in Supplementary Figure 4.
Supplementary Material
Supplementary Figures Legends
Supplementary Table 1 and Table 2
Supplementary Figures 1–5
Acknowledgments
We thank Jorge Reis-Filho of The Institute of Cancer Research for helpful discussions, G Smith at Kudos Pharmaceuticals for provision of inhibitors and A Smith of The Institute of Cancer Research for technical assistance. Grant support was obtained from Breakthrough Breast Cancer Research and Cancer Research UK.
References
- Aza-Blanc P, Cooper CL, Wagner K, Batalov S, Deveraux QL, Cooke MP (2003) Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol Cell 12: 627–637 [DOI] [PubMed] [Google Scholar]
- Bahassi el M, Hennigan RF, Myer DL, Stambrook PJ (2004) Cdc25C phosphorylation on serine 191 by Plk3 promotes its nuclear translocation. Oncogene 23: 2658–2663 [DOI] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ (1996) SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86: 263–274 [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas C, Lukas J (2004) Checking on DNA damage in S phase. Nat Rev Mol Cell Biol 5: 792–804 [DOI] [PubMed] [Google Scholar]
- Bartz SR, Zhang Z, Burchard J, Imakura M, Martin M, Palmieri A, Needham R, Guo J, Gordon M, Chung N, Warrener P, Jackson AL, Carleton M, Oatley M, Locco L, Santini F, Smith T, Kunapuli P, Ferrer M, Strulovici B et al. (2006) Small interfering RNA screens reveal enhanced cisplatin cytotoxicity in tumor cells having both BRCA network and TP53 disruptions. Mol Cell Biol 26: 9377–9386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T (2005) Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434: 913–917 [DOI] [PubMed] [Google Scholar]
- Chappell C, Hanakahi LA, Karimi-Busheri F, Weinfeld M, West SC (2002) Involvement of human polynucleotide kinase in double-strand break repair by non-homologous end joining. EMBO J 21: 2827–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Cheung ST, So S, Fan ST, Barry C, Higgins J, Lai KM, Ji J, Dudoit S, Ng IO, Van De Rijn M, Botstein D, Brown PO (2002) Gene expression patterns in human liver cancers. Mol Biol Cell 13: 1929–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, Chen F, Feiler H, Tokuyasu T, Kingsley C, Dairkee S, Meng Z, Chew K, Pinkel D, Jain A, Ljung BM et al. (2006) Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 10: 529–541 [DOI] [PubMed] [Google Scholar]
- Ciullo M, Debily MA, Rozier L, Autiero M, Billault A, Mayau V, El Marhomy S, Guardiola J, Bernheim A, Coullin P, Piatier-Tonneau D, Debatisse M (2002) Initiation of the breakage–fusion–bridge mechanism through common fragile site activation in human breast cancer cells: the model of PIP gene duplication from a break at FRA7I. Hum Mol Genet 11: 2887–2894 [DOI] [PubMed] [Google Scholar]
- Cruz JC, Tsai LH (2004) Cdk5 deregulation in the pathogenesis of Alzheimer's disease. Trends Mol Med 10: 452–458 [DOI] [PubMed] [Google Scholar]
- Dhavan R, Tsai LH (2001) A decade of CDK5. Nat Rev Mol Cell Biol 2: 749–759 [DOI] [PubMed] [Google Scholar]
- Echeverri CJ, Beachy PA, Baum B, Boutros M, Buchholz F, Chanda SK, Downward J, Ellenberg J, Fraser AG, Hacohen N, Hahn WC, Jackson AL, Kiger A, Linsley PS, Lum L, Ma Y, Mathey-Prevot B, Root DE, Sabatini DM, Taipale J et al. (2006) Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods 3: 777–779 [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434: 917–921 [DOI] [PubMed] [Google Scholar]
- Goldberg M, Stucki M, Falck J, D'Amours D, Rahman D, Pappin D, Bartek J, Jackson SP (2003) MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 421: 952–956 [DOI] [PubMed] [Google Scholar]
- Graudens E, Boulanger V, Mollard C, Mariage-Samson R, Barlet X, Gremy G, Couillault C, Lajemi M, Piatier-Tonneau D, Zaborski P, Eveno E, Auffray C, Imbeaud S (2006) Deciphering cellular states of innate tumor drug responses. Genome Biol 7: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC (2004) Identification and characterization of a novel and specific inhibitor of the ataxia–telangiectasia mutated kinase ATM. Cancer Res 64: 9152–9159 [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH (2001) Genome maintenance mechanisms for preventing cancer. Nature 411: 366–374 [DOI] [PubMed] [Google Scholar]
- Iorns E, Lord CJ, Turner N, Ashworth A (2007) Utilizing RNA interference to enhance cancer drug discovery. Nat Rev Drug Discov 6: 556–568 [DOI] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP (2006) ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 8: 37–45 [DOI] [PubMed] [Google Scholar]
- Jilani A, Ramotar D, Slack C, Ong C, Yang XM, Scherer SW, Lasko DD (1999) Molecular cloning of the human gene, PNKP, encoding a polynucleotide kinase 3′-phosphatase and evidence for its role in repair of DNA strand breaks caused by oxidative damage. J Biol Chem 274: 24176–24186 [DOI] [PubMed] [Google Scholar]
- Lee JH, Kim KT (2007) Regulation of cyclin-dependent kinase 5 and p53 by ERK1/2 pathway in the DNA damage-induced neuronal death. J Cell Physiol 210: 784–797 [DOI] [PubMed] [Google Scholar]
- Mailand N, Podtelejnikov AV, Groth A, Mann M, Bartek J, Lukas J (2002) Regulation of G(2)/M events by Cdc25A through phosphorylation-dependent modulation of its stability. EMBO J 21: 5911–5920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, Giavara S, O'Connor MJ, Tutt AN, Zdzienicka MZ, Smith GC, Ashworth A (2006) Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res 66: 8109–8115 [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T (2004) Mechanisms of gene silencing by double-stranded RNA. Nature 431: 343–349 [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Chiu JW, Koller BH, Jasin M (1999) Brca1 controls homology-directed DNA repair. Mol Cell 4: 511–518 [DOI] [PubMed] [Google Scholar]
- Mukherji M, Bell R, Supekova L, Wang Y, Orth AP, Batalov S, Miraglia L, Huesken D, Lange J, Martin C, Sahasrabudhe S, Reinhardt M, Natt F, Hall J, Mickanin C, Labow M, Chanda SK, Cho CY, Schultz PG (2006) Genome-wide functional analysis of human cell-cycle regulators. Proc Natl Acad Sci USA 103: 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick GN, Zukerberg L, Nikolic M, de la Monte S, Dikkes P, Tsai LH (1999) Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature 402: 615–622 [DOI] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M (1999) XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 13: 2633–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renwick A, Thompson D, Seal S, Kelly P, Chagtai T, Ahmed M, North B, Jayatilake H, Barfoot R, Spanova K, McGuffog L, Evans DG, Eccles D, Easton DF, Stratton MR, Rahman N (2006) ATM mutations that cause ataxia–telangiectasia are breast cancer susceptibility alleles. Nat Genet 38: 873–875 [DOI] [PubMed] [Google Scholar]
- Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C (2006) Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol 24: 778–789 [DOI] [PubMed] [Google Scholar]
- Strocchi P, Pession A, Dozza B (2003) Upregulation of cDK5/p35 by oxidative stress in human neuroblastoma IMR-32 cells. J Cell Biochem 88: 758–765 [DOI] [PubMed] [Google Scholar]
- Tutt A, Bertwistle D, Valentine J, Gabriel A, Swift S, Ross G, Griffin C, Thacker J, Ashworth A (2001) Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J 20: 4704–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S, Harlow E (1993) Distinct roles for cyclin-dependent kinases in cell cycle control. Science 262: 2050–2054 [DOI] [PubMed] [Google Scholar]
- Wang X, McGowan CH, Zhao M, He L, Downey JS, Fearns C, Wang Y, Huang S, Han J (2000) Involvement of the MKK6-p38gamma cascade in gamma-radiation-induced cell cycle arrest. Mol Cell Biol 20: 4543–4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KK, Chang S, Weiler SR, Ganesan S, Chaudhuri J, Zhu C, Artandi SE, Rudolph KL, Gottlieb GJ, Chin L, Alt FW, DePinho RA (2000) Telomere dysfunction impairs DNA repair and enhances sensitivity to ionizing radiation. Nat Genet 26: 85–88 [DOI] [PubMed] [Google Scholar]
- Woods J, Snape M, Smith MA (2007) The cell cycle hypothesis of Alzheimer's disease: suggestions for drug development. Biochim Biophys Acta 1772: 503–508 [DOI] [PubMed] [Google Scholar]
- Yap TA, Boss DS, Fong PC, Roelvink M, Tutt A, Carmichael J, O'Connor MJ, Kaye SB, Schellens JH, de Bono J (2007) First in human phase I pharmacokinetic (PK) and pharmacodynamic (PD) study of KU-0059436 (Ku), a small molecule inhibitor of poly ADP-ribose polymerase (PARP) in cancer patients (p), including BRCA1/2 mutation carriers. J Clin Oncol 25: A3529 [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 4: 67–73 [DOI] [PubMed] [Google Scholar]
- Zhu XH, Nguyen H, Halicka HD, Traganos F, Koff A (2004) Noncatalytic requirement for cyclin A–cdk2 in p27 turnover. Mol Cell Biol 24: 6058–6066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman WC, Erikson RL (2007) Polo-like kinase 3 is required for entry into S phase. Proc Natl Acad Sci USA 104: 1847–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures Legends
Supplementary Table 1 and Table 2
Supplementary Figures 1–5