Abstract
In the Rous sarcoma virus (RSV) Gag protein, the 25 amino-acid residues of the p10 domain immediately upstream of the CA domain are essential for immature particle formation. We performed systematic mutagenesis on this region and found excellent correlation between the amino-acid side chains required for in vitro assembly and those that participate in the p10–CA dimer interface in a previously described crystal structure. We introduced exogenous cysteine residues that were predicted to form disulphide bonds across the dimer interface. Upon oxidation of immature particles, a disulphide-linked Gag hexamer was formed, implying that p10 participates in and stabilizes the immature Gag hexamer. This is the first example of a critical interaction between two different Gag domains. Molecular modeling of the RSV immature hexamer indicates that the N-terminal domains of CA must expand relative to the murine leukaemia virus mature hexamer to accommodate the p10 contact; this expansion is strikingly similar to recent cryotomography results for immature human immunodeficiency virus particles.
Keywords: Gag, HIV-1, Rous sarcoma virus, virus assembly
Introduction
The major orthoretroviral structural protein Gag is necessary and sufficient for the assembly and release of virus-like particles (VLPs), which resemble actual virus particles but lack components required for infectivity. Gag is a polyprotein that is cleaved post-translationally by the viral protease PR. The major Gag cleavage products are the MA (matrix), CA (capsid) and NC (nucleocapsid) proteins, which are conserved among all retroviruses. Other cleavage products vary among retroviral families. PR cleavage is required for morphological maturation of retroviral particles, which in turn is important for infectivity. Mature particles examined by thin-section transmission electron microscopy (TEM) have a spherical outer membrane lined with MA protein and an inner core that consists of a shell of CA enclosing an NC-RNA complex. The core has a characteristic shape for each retroviral species and may be conical, cylindrical or polyhedral. In the absence of PR cleavage, full-length Gag produces non-infectious immature particles, which appear similar for all retroviruses. By TEM, immature particles are spherical with an electron-dense ring underlying the viral membrane and an electron-lucent centre. Removing the membrane yields a stable Gag-RNA shell known as the immature core. PR activation is coupled to particle release, but the precise timing and mechanism are not well understood; however, it is probable that Gag assembles into immature particles that mature during or shortly after release. Recent studies (Briggs et al, 2004, 2006) imply that the immature core dissociates upon PR cleavage and the mature core reassembles from liberated CA, rather than the immature core condensing into the mature core. Retroviral assembly is thus best understood as occurring in two separate stages, assembly of Gag into the immature core and reassembly of CA into the protein shell of the mature core.
The major protein–protein interaction domain of Gag is CA. CA contains independently folded N- and C-terminal sub-domains (NTD and CTD) connected by a short flexible linker (Khorasanizadeh et al, 1999; Campos-Olivas et al, 2000). All available retroviral NTD structures show an N-terminal β-hairpin anchored by a salt bridge between a conserved aspartate in helix 3 and the free amino terminus of the conserved N-terminal proline (Gitti et al, 1996; Jin et al, 1999; Khorasanizadeh et al, 1999; Kingston et al, 2000; Mortuza et al, 2004). Retroviral cores are highly variable in size and shape and thus not amenable to crystallization, but reconstructions of mature viral cores and in vitro-assembled CA tubes show CA in a hexagonal lattice with NTD hexamers connected by CTD dimers (Li et al, 2000; Ganser et al, 2003; Mayo et al, 2003; Ganser-Pornillos et al, 2007). For human immunodeficiency virus (HIV) at least, an intermolecular NTD–CTD interaction is also present (Lanman et al, 2003; Ganser-Pornillos et al, 2007). The NTD of murine leukaemia virus (MLV) has been crystallized as a hexamer believed to represent the NTD hexamers in the mature core (Mortuza et al, 2004). Although the CA domain of Gag drives immature assembly (Ako-Adjei et al, 2005; Lee et al, 2007), the immature lattice is strikingly different from its mature counterpart. The overall unit cell distance is smaller (Briggs et al, 2004, 2006), with the CTDs more tightly packed and the NTDs more widely spaced (Wright et al, 2007). To produce these changes, the CA–CA contacts in the mature core must be different from those in the immature core. The closer immature CTD packing may be induced by the spacer segment of Gag between CA and NC (Wright et al, 2007). The loose immature NTD hexamer, at least in part, may be a consequence of the absence of the N-terminal β-hairpin of the NTD, which forms only after PR cleavage and which appears to stabilize the mature NTD hexamer (Mortuza et al, 2004; Ganser-Pornillos et al, 2007). In summary, although both immature and mature cores contain hexagonal lattices linked by CA–CA interactions, maturation appears to involve PR-mediated changes in the CA–CA contacts.
The Rous sarcoma virus (RSV) in vitro assembly system has provided additional information about the immature NTD. RSV is a well-studied avian virus that is frequently employed as a model retrovirus. Among the advantages of RSV as a model is a robust in vitro system for immature assembly. A truncated RSV Gag termed ΔMBDΔPR (Figure 1) can be purified and assembled in vitro into spherical VLPs that are similar in shape, size and radial density profile to immature cores produced from cells (Campbell and Vogt, 1997; Yu et al, 2001). Deletion of p10, the Gag domain immediately upstream of CA, produces tubular particles (Campbell and Vogt, 1997); as purified HIV CA assembles into tubes in vitro (Ehrlich et al, 1992; Ganser-Pornillos et al, 2004), tubular morphology is considered indicative of a mature (or mature-like) CA lattice. Internal and N-terminal deletions of p10 give the same result, so the morphology change cannot be ascribed to NTD β-hairpin formation alone. The spherical assembly determinant was further localized to the C-terminal 25 amino acids of p10, both in vitro (Joshi and Vogt, 2000) and in a baculovirus expression system (Johnson et al, 2002). The same sequence also contains a CRM-1-dependent nuclear export signal (NES) required for proper intracellular trafficking of Gag and for viral genome packaging (Scheifele et al, 2002, 2005, 2007; Butterfield-Gerson et al, 2006); the significance of this dual function of p10 is unknown.
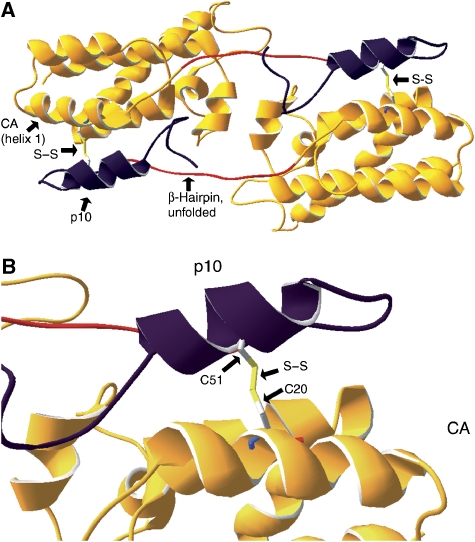
Figure 1.
(A) Single subunit of the extended NTD structure coloured by secondary structure. Those residues that form the β-hairpin in mature CA are colored red. (B) Annotated amino-acid sequence of the RSV extended NTD, including 25 amino acids of p10. Asterisks above the sequence indicate residues involved in the extended NTD dimer interface (Nandhagopal et al, 2004). Plus signs below the sequence indicate the amino acids most structurally homologous to the MLV residues involved in the MLV mature NTD hexamer interface (Mortuza et al, 2004). Secondary structural elements are indicated with colours corresponding to panel A above; note that although the helices are valid for both structures, the β-hairpin (red) is present only in mature CA. (C) Top: schematic representation of full-length RSV Gag and of ΔMBDΔPR, an N- and C-terminally truncated Gag construct used for in vitro assembly of spherical VLPs. Vertical lines represent PR cleavage sites. Bottom: C-terminal 25 amino acids of RSV p10 domain with mutagenesis results. ‘Side chain contact' indicates those amino-acid residues whose side chains contact the CA domain in the extended NTD structure. ‘Required for assembly' indicates those amino-acid residues which, when changed to alanine, abrogate assembly in vitro.
When the RSV NTD was extended N-terminally to include the 25 amino acids immediately upstream (the last 25 amino acids of p10), it crystallized as an anti-parallel dimer with an extensive p10–CA interface (Nandhagopal et al, 2004) (PDB ID: 1P7N). The extended NTD monomer (Figure 1A) is similar in structure to the mature NTD previously characterized by NMR and crystallography (Campos-Olivas et al, 2000; Kingston et al, 2000; Nandhagopal et al, 2004), but it lacks the N-terminal β-hairpin of CA (sequence marked schematically in Figure 1B), which is destabilized when proline 1 of CA is not free to form the salt bridge with aspartate 52 in helix 3. The dimer interface (Figure 1B, asterisks) includes residues in a short α-helix in the p10 segment. Owing to the anti-parallel arrangement of the NTDs, the extended NTD dimer presumably cannot be a subunit of an NTD hexamer and therefore is probably not found in the immature virus particle. Therefore, the requirement of the same 25 amino acids of p10 for both spherical virus particle morphology and the extended NTD dimer interface suggests that the p10–CA contacts in the dimer interface may have some other relevance to immature assembly.
Results
An extended NTD structure is relevant to immature assembly
To test the relevance of the extended NTD crystal structure to immature assembly, we mutated the C-terminal 25 amino acids of p10 in the context of the RSV in vitro assembly construct ΔMBD. As the results of a previous random mutagenesis of the region were inconclusive (Joshi and Vogt, 2000), we performed a systematic alanine mutagenesis. The critical 25 amino-acid region was subdivided into five segments of five amino acids, which were mutated in turn to either AAAAA or GSGSG. The resulting proteins were expressed in Escherichia coli, purified, mixed with 10% wt/wt single-stranded DNA and screened for assembly by negative staining and viewing by electron microscopy. In independently purified protein preparations, all proteins showed a consistent phenotype, either yielding abundant regular spherical particles on the EM grid or yielding no regular particles whatsoever. Segments of five amino acids that could be mutated to AAAAA or GSGSG without affecting assembly were scored as dispensable for assembly. Segments of five amino acids for which mutation to both AAAAA and GSGSG prevented assembly were subdivided into smaller alanine substitutions until all individual amino acids required for assembly were identified. The four endogenous alanine residues were not individually mutated. The results (Figure 1C) showed that the last 10 amino acids of the critical region allowed extensive substitution, whereas the first 15 amino acids of the critical region contained a mix of required and dispensable amino acids. The last 10 amino acids are extended and have no secondary structure, but they all are not functionally equivalent: the C-terminal segment (residues 58–62; PVVAM) tolerated mutation to GSGSG but not to AAAAA, whereas the preceding segment (residues 53–57; ASTGP) tolerated both mutations. These results suggest that although none of the side chains in the last 10 amino acids of p10 are required for assembly, there are some sequence constraints on the final five amino acids. Some of the first 15 amino acids of p10, however, appear to make specific side-chain contacts that are required for assembly.
Comparison of the mutagenesis results with the published amino-acid contacts in the extended NTD interface (Nandhagopal et al, 2004) revealed a striking pattern (Figure 1C). Of the six amino acids in p10 whose side chains contact the NTD in the structure, five were required for assembly and only one (E51 in the p10 helix) was dispensable. Similarly, of the 15 non-alanine amino acids whose side chains were not involved in the interface, 14 were dispensable and only 1 (D44 upstream of the p10 helix) was required. There are two amino acids in the NTD that make predicted side-chain hydrogen bonds across the dimer interface in the extended NTD structure, CA D52 and N55 in helix 3; mutation of either of these to alanine also abrogated assembly (data not shown). These results strongly support the hypothesis that the p10–CA interface in the extended NTD structure is involved in immature assembly.
p10–CA cross-linking is intermolecular and specific in VLPs
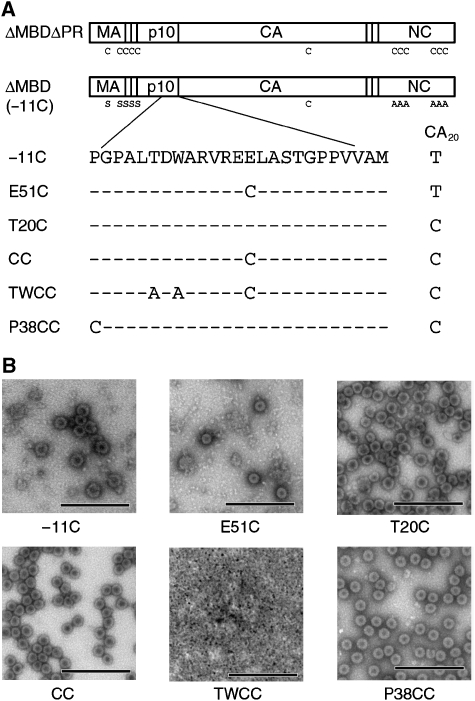
The ability of the E51A mutant to assemble despite altering a side-chain contact with the NTD offered an opportunity to use disulphide cross-linking to test for the presence of the extended NTD p10–CA interface in assembled virus particles. The Swiss-PDBViewer program (Guex, 1997) predicted a disulphide bond when E51 in the p10 helix (Figure 1B) and the residue it contacts in CA, T20 in helix 1 (Figure 1B), were mutated to cysteines (Figure 2). Therefore, the p10–CA interface depicted in the 25NTD structure could be detected in virus particles by mutating these residues to cysteine and assaying for disulphide cross-linking between p10 and CA. The ΔMBDΔPR protein has 12 endogenous cysteines that have the potential to interfere in such an assay. To reduce such interference, we mutated the cysteines in MA, p2 and NC to alanine or serine (Figure 3A). One endogenous cysteine, C192 in the C-terminal domain of CA, was known from previous studies to disrupt assembly when mutated to serine (data not shown) and was therefore left in place. The resulting construct, ΔMBD [−11C] (hereafter called −11C), was able to assemble in vitro into normal spherical particles (Figure 3B). All subsequent cysteine mutations were made in the context of −11C. Mutation of p10 E51 [E51C], CA T20 [T20C] or both [CC] to cysteines allowed spherical assembly in vitro (Figure 3B), although the E51C single mutant assembled less efficiently. Both −11C and E51C were found to assemble more efficiently at pH 6.0 than at pH 6.5, and subsequent assemblies of these proteins were performed at the lower pH. As a negative control for assembly, we combined the p10 T43A and W45A mutations, which abrogate assembly (Figure 1C), with the CC construct to produce TWCC (Figure 3A); as predicted, TWCC did not assemble (Figure 3B), confirming that the cysteine mutations cannot rescue a p10–CA interface defect.
Figure 2.
(A) High-resolution structure of the extended NTD dimer (1P7N) showing disulphide bonds predicted to form when p10 E51 and CA T20 are mutated to cysteines. The disulphide bonds (yellow) link CA (orange) and p10 (purple) domains across the dimer interface. Those residues that form the β-hairpin in mature CA are shown in red. (B) Close-up of panel A showing the predicted E51C-T20C disulphide bond.
Figure 3.
(A) Schematic representation of ΔMBDΔPR showing locations of the endogenous cysteine residues and the cysteine mutations used in this study. (B) Transmission electron microscopy images of ΔMBDΔPR cysteine mutants assembled in vitro and negative stained. −11C, top left; E51C, top middle; T20C, top right; CC, bottom left; TWCC, bottom middle; P38CC, bottom right. Scale bars, 500 nm.
Disulphide cross-linking of Gag proteins can occur in situations other than stable contacts between folded proteins in VLPs, including transient interactions between proteins in solution, protein–protein interactions in misassembled aggregates and interactions between denatured proteins in SDS–PAGE preparations. The probability of such non-specific cross-linking can be reduced by purifying the VLPs by equilibrium sedimentation to remove unassembled protein and misassembled aggregates and by quenching any remaining cysteines with an excess of a sulfhydryl-reactive reagent before denaturation. Neither of these tactics is 100% effective, so to distinguish between interface-specific and non-specific p10–CA cross-linking, we made an additional construct by mutating p10 P38 at the beginning of the critical region (Figure 1B) to cysteine and combining it with T20C to make P38CC (Figure 3A). P38 was chosen because it allows assembly when mutated to alanine (Figure 1C), but it is over 15 Å away from T20 of the other subunit in the extended NTD structure, making disulphide formation between cysteines at these sites impossible. P38CC should allow non-specific p10–CA cross-linking (during transient collisions or when proteins are denatured) but should not support cross-linking if the p10–CA interface depicted in the extended NTD structure is present. Thus, although P38CC assembled as expected (Figure 3B), it was not predicted to allow structure-specific cross-linking of p10 and CA.
To test for the presence of the p10–CA interface in particles, VLPs from assembly-competent constructs were purified, subjected to mild oxidation in the presence of stoichiometric amounts of DTT and quenched as described in the Materials and methods. To verify the identities of the cross-linked Gag domains, the oxidized particles were incubated with and without recombinant RSV PR to separate the Gag domains, as would occur during virus maturation. The products were then analysed by non-reducing and reducing SDS–PAGE (Figure 4). The parent construct −11C (lane 1) was predominantly monomeric for both full-length protein (Figure 4, top left) and CA (Figure 4, bottom left), and neither band shifted upon reduction (Figure 4, right), indicating that C192 in the CTD does not form homotypic disulphide bonds under these assay conditions. Some of the full-length E51C (lane 2) was dimeric under non-reducing conditions but reduced to a monomer, whereas the CA was monomeric even when oxidized, suggesting that the full-length dimers were due to p10–p10 disulphide bonds mediated by the exogenous cysteine. Notably, there was no evidence that C51 in p10 was able to cross-link to C192 in CA. Full-length T20C (lane 3) also formed disulphide-mediated dimers, although to a lesser extent. These must be CA–CA dimers, as there are no cysteine residues in any other Gag domain, and a faint corresponding upper band is present at approximately 50 kDa (CA is 27 kDa). It is not possible to distinguish whether these dimers are mediated by C20–C20 or C20–C192 disulphide bonds, and both species may well be present; in any case, the majority of T20C is monomeric. The results for these single mutants suggest that neither of the exogenous cysteines, C51 in p10 and C20 in CA, produces extensive cross-linking when introduced alone.
Figure 4.
Exogenous cysteines in p10 and CA form specific, intermolecular disulphide bonds in VLPs. Particles were oxidized, quenched and treated as described in the text, before SDS–PAGE. Lane 1, −11C; lane 2, E51C; lane 3, T20C; lane 4, CC; lane 5, P38CC. In the bottom panel, the samples were treated with PR before SDS–PAGE. Samples on the left were not reduced. Samples on the right were reduced before PR digestion and SDS–PAGE. The novel high molecular weight band appears in lane 4 on the top left, and the novel shifted CA band appears in lane 4 on the bottom left.
In contrast to the preceding control proteins, full-length CC protein (lane 4) formed a distinct, very high MW band with almost no monomer or dimer present, while the CA band was shifted upward by approximately 10 kDa with no 27 kDa CA remaining. Both full-length and CA bands reduced to the normal monomer position, confirming that the band shifts observed were due to disulphide bonds. The presence of both p10 and CA in the novel 37 kDa band was confirmed by immunoblotting (data not shown). For the specificity control P38CC (lane 5), the full-length protein was largely dimeric, but the CA was predominantly monomeric with a very faint upper band similar to that seen for T20C. This suggests that most of the full-length dimers were due to p10–p10 disulphide bonds mediated by C38, with a few CA–CA disulphides as for T20C; again, there was no evidence of p10–CA cross-linking, indicating that C38 in p10 was unable to form disulphide bonds with either C20 or C192 in CA. This was in marked contrast to the results seen for oxidized unassembled protein, in which either C38 or C51 in p10 was able to cross-link to C20 in CA (data not shown), and suggests that the measures taken to purify VLPs before oxidation and to quench remaining cysteines before denaturation were largely successful in eliminating non-specific p10–CA cross-linking. Together, the cross-linking results indicate that the p10–CA interface depicted in the extended NTD crystal structure is present in VLPs but that the cross-linked species is much larger than the predicted dimer.
To ensure that these results did not represent an artefact of in vitro assembly or of chemical oxidation, parallel versions of the constructs described above were made in an MLV retroviral vector expressing a mutant version of RSV Gag. As the RSV NES is contained within the region of p10 under study and includes the W45 residue, it seemed likely that the TWCC construct, and possibly others as well, would be trapped in the nucleus and therefore be unable to assemble and bud. To avoid this problem, all cell-expression constructs included an MA domain mutation called Super M, which contains two mutations in the membrane-binding domain of MA, E25K and E70K. The net +4 charge is reported to increase the affinity of Gag for the plasma membrane and thereby to bypass nuclear transit (Callahan and Wills, 2003); combining Super M MA with the constructs described in Figure 3A was predicted to ensure that all Gag proteins reached the plasma membrane. DF1 (chicken embryonic fibroblast) cell lines stably expressing these constructs were created by MLV transduction, and VLPs produced by the cell lines were collected and quenched as described in the Materials and methods and then analysed by reducing and non-reducing SDS–PAGE and immunoblotting. All proteins were expressed (data not shown) and all constructs except for TWCC were able to assemble and bud from cells (Figure 5). Gag was monomeric or dimeric and CA was the correct size in particles produced from all constructs except for CC(−11C), which produced a high molecular weight Gag complex and an upward-shifted CA, exactly as seen in vitro (Figure 5, lane 4). We conclude that the interaction between p10 and CA occurs in membrane-enclosed VLPs produced from cells as well as in particles assembled with purified protein in vitro, and that efficient disulphide bond formation can occur by air oxidation.
Figure 5.
Specific intermolecular p10–CA cross-linking occurs on air-oxidation of VLPs produced from cultured avian cells. Particles were quenched and treated as described in the text before SDS–PAGE and immunoblotting with rabbit anti-CA serum. Lane 1, −11C; lane 2, E51C; lane 3, T20C; lane 4, CC; lane 5, TWCC; lane 6, P38CC. Samples in the bottom panels were treated with PR before SDS–PAGE. Samples on the left were not reduced. Samples on the right were reduced before PR digestion and SDS–PAGE. The novel high molecular weight band appears in lane 4 on the top left, and the novel shifted CA band appears in lane 4 on the bottom left.
The p10 and CA domains of Gag become cross-linked as a hexamer
To identify the high MW band produced by undigested CC, the high molecular weight product was isolated and partly reduced to form a ladder. Owing to the amount of protein required, an additional construct, CfC(−11C), was used. This protein combines the CC construct with the p10(58–62)GSGSG mutation, which allows assembly but destroys the p10–CA cleavage site (data not shown). In the absence of PR, CfC behaves similarly to CC, but it can consistently be purified with a much higher yield. Particles were assembled, collected and oxidized as previously described (but without additional DTT) and then resuspended in 500 mM NaCl plus 2 M urea to disrupt weak non-covalent intermolecular interactions. The resulting sample was subjected to rate-zonal centrifugation in a sucrose gradient to separate protein complexes by size, and the fractions were analysed by non-reducing SDS–PAGE. Two distinct protein complexes were observed: a major complex that migrated significantly above 212 kDa and a larger minor complex that may represent a dimer of the major complex (Figure 6A). The sedimentation coefficient of the major complex was estimated at 10S compared with tetrameric β-galactosidase, which would correspond approximately to a globular protein of 300 kDa. (ΔMBD is a 52 kDa protein.) Gradient fractions that contained only the major complex were pooled, concentrated and partially reduced with 0.25% β-mercaptoethanol. The resulting mixture of species gave rise to a ladder of six bands spaced at approximately 50 kDa intervals (Figure 6B), of which the highest band is the same size as the non-reduced complex and the lowest is the correct molecular weight for a ΔMBD monomer (∼52 kDa). We conclude that the major protein complex is a hexamer linked by p10–NTD disulphide bonds.
Figure 6.
p10–CA cross-linking of VLPs produces a hexameric protein complex. (A) Analysis by rate-zonal centrifugation. Cross-linked particles were dissolved in 2 M urea and 0.5 M NaCl, mixed with E. coli β-galactosidase as in internal size standard, and then centrifuged on a sucrose gradient with the same solutes. The gradient was divided into 17 fractions and portions were submitted to SDS–PAGE. (B) Samples containing the major high MW band, such as lanes 9 and 10 in this experiment, were pooled, concentrated and partially reduced with β-mercaptoethanol. SDS–PAGE was performed on NuPAGE Tris-Acetate 3–8% gradient gels (Invitrogen), producing a ladder of six bands consistent with a ΔMBD hexamer.
Modeling of p10–NTD hexamer
The results above imply that the extended NTD structure is a dimeric version of the sixfold p10–CA hexamer observed in particles, as illustrated in Figure 7A. For the hexamer, only the NTD portion of Gag is shown. The 25 residues of p10 are drawn as a hook that acts like a flap contacting a neighbouring NTD. In the dimer, the shaft of the hook is straight and the NTDs are symmetrically arranged, pointing in opposite directions, an orientation that is inferred to be biologically irrelevant. In the hexamer, the shaft of the hook must be bent to accommodate the same p10–NTD side-chain contacts as in the dimer. As drawn, this bend would occur in the region of Thr6–Glu7 of CA.
Figure 7.
Models of the immature hexamer. (A) Schematic representation of p10–CA interaction in the extended NTD. Left: crystallographic dimer. Right: the hexamer proposed to form in immature particles. p10 is represented by the purple line, β-hairpin residues by the red line and the remainder of the NTD by solid shapes. Small yellow circles represent the N-terminus of the extended NTD. (B) Molecular models of mature and immature RSV CA NTD hexamers with approximate helix positions and interactions indicated. Top: molecular model of mature RSV NTD hexamer based on mature MLV NTD structure. The upper three subunits of the hexamer are shown as solid space-filled surfaces, whereas the lower three subunits are shown as transparent surfaces superimposed on ribbon diagrams. On the ribbon diagrams, the N-terminus is space-filled in yellow and the C-terminus in green. The β-hairpin ribbon is in dark red. Approximate helix positions are indicated by labelled circles. Dark arrows indicate direct intermolecular CA–CA interactions predicted from the MLV NTD hexamer and tan arrows indicate predicted water-mediated interactions. Bottom: molecular model of immature RSV NTD hexamer based on extended NTD structure and mature MLV NTD hexamer. On the ribbon diagrams, p10 residues are dark purple and the unfolded β-hairpin residues are dark red; the depiction is otherwise similar to the mature model. White arrows indicate intermolecular p10–CA interactions and black arrows indicate possible intramolecular interactions.
The extended NTD dimer structure does not provide any information about the relative positions of the NTD subunits in the hexamer. To elucidate the hexamer structure, we made models for both the mature and immature forms of the RSV CA NTD hexamer (Figure 7B). In this panel, the top three of the six subunits are drawn in space-filling style, with the protein–protein interactions shown by arrows between CA helices (numbers) or between the p10 segment (p) and CA helices. The bottom three subunits are drawn to show the polypeptide itself in ribbon style. The mature hexamer is based on the MLV CA NTD hexamer as determined by crystallography, which is stabilized primarily by interactions between the N-terminal β-hairpins of adjacent subunits and between the first helix of adjacent subunits, and secondarily by interactions between helix 2 and helix 3 of adjacent subunits (Mortuza et al, 2004). Although RSV lacks sufficient sequence similarity to MLV to allow the creation of a detailed model of the RSV NTD–NTD interface, a structural alignment of the MLV and RSV NTD sequences (Mortuza et al, 2004) has allowed us to speculate which amino acids may be involved; these are shown in Figure 1B (plus symbols). Nonetheless, differences between the RSV and MLV monomer structures mean that the relative helix positions within the hexamer are somewhat different as well, and thus the interactions inferred from the MLV structure must be considered approximate for RSV.
The compact nature of the mature NTD hexamer seems to leave little space for p10, and the crystallographic contacts between p10 and helices 1 and 3 in the extended RSV NTD structure and the putative NTD–NTD contacts in the mature RSV CA NTD hexamer are very similar and thus unlikely to form at the same time (Figure 1B, compare asterisk and plus symbols). This implies that the immature CA NTD hexamer must be significantly different from the mature NTD hexamer. The addition of p10 in the immature model expands the hexamer (Figure 7B, bottom), opening up a much larger hole in the centre. Although the nature of the model does not allow for exact measurements, the immature hole is approximately 3–4 times larger than the mature hole (Mortuza et al, 2004), with respective diameters of about 42 and 12 Å. A similarly large hole in the NTD hexamer of the immature HIV-1 Gag lattice was recently observed in immature HIV-1 particles analysed by electron cryotomography (Wright et al, 2007).
Discussion
Retroviral assembly is a multistep process in which the structural protein Gag assembles into immature particles and then is cleaved by the viral protease PR into products that reassemble to form the internal structure of the mature infectious virus. The CA domain of Gag forms a hexameric lattice in both immature and mature particles, but differences in the lattice suggest that the Gag-embedded CA and the liberated CA make different protein–protein contacts. The nature of these differences is not well understood. The N-terminal β-hairpin of free CA is highly conserved and is important for correct mature assembly of at least some (von Schwedler et al, 1998) retroviruses. In addition, RSV has a specific sequence requirement for correct immature assembly, namely the C-terminal 25 amino acids of the p10 domain that adjoin CA (Joshi and Vogt, 2000). A high-resolution structure of an RSV NTD extended to include this critical region of p10 showed a dimer with an extensive p10–NTD interface (Nandhagopal et al, 2004), but the relevance of this structure to immature assembly was uncertain. We have now provided evidence that the p10–NTD interface identified in the structure is relevant to immature assembly, but that in immature virus particles the interface forms a hexamer rather than a dimer. As the NTD is believed to form a hexamer in both the immature and the mature CA lattices, we conclude that p10 participates in the immature NTD hexamer. This is the first example of a critical heterologous interdomain Gag-Gag contact in retroviral assembly.
The interaction between p10 and the NTD in the extended NTD structure is incompatible with the NTD–NTD interactions seen in the MLV mature NTD hexamer structure (Mortuza et al, 2004). Our molecular modelling of an immature RSV NTD hexamer, based on the RSV extended NTD dimer and the MLV hexamer structures, shows that the p10–NTD interaction forces the NTD subunits further apart, opening a central cavity. The final NTD arrangement is strikingly similar to that seen for immature HIV-1 particles examined by electron cryotomography (Wright et al, 2007). This similarity suggests that immature retroviral lattices are structurally homologous, just as are their mature CA proteins. However, it is unknown by what mechanism the HIV-1 NTD lattice is expanded in the immature lattice as compared with the mature lattice.
In vitro studies of HIV-1 assembly show that assembly pH acts as a morphological switch for immature-like assembly: an HIV Gag protein lacking the C-terminal p6 domain and amino acids 16–99 of MA assembles only into spheres at pH 8.0 but assembles into both spheres and tubes at pH 6.5 (Gross et al, 2000). The assembly morphology switch is paralleled by changes in epitope availability in the NTD of monomeric Gag, with epitopes in helix 3 and helix 6 available at pH 8.0 but masked at pH 6.5 (Gross et al, 2000). These results suggest that for HIV-1 a pH-mediated conformational change in Gag—which may reprise changes caused by PR cleavage in vivo—is responsible for the switch between immature-like spheres and mature-like tubes. The molecular nature of this switch remains to be unraveled.
Electron cryotomography has recently provided the first data on the retroviral immature lattice (Wright et al, 2007). This study shows that, in contrast to the mature lattice, the immature lattice has the NTD hexamers more widely spaced, with almost no NTD–NTD contact, and the CTDs are more tightly packed. The hexamer appears to be stabilized by interactions among the spacer domains C-terminal to the CTD, a region that is also important for RSV assembly (Keller and V.M.V, unpublished data.) In addition, a novel ‘domain-swapped' structure has been proposed for the HIV CTD (Ivanov et al, 2007); this dimer is much tighter than the traditional dimer structure seen for both purified protein (Gamble et al, 1997) and the reconstructed mature lattice (Ganser-Pornillos et al, 2007), and it is tempting to speculate that the immature lattice, with its greater stability and tighter CTD spacing, incorporates the domain-swapped CTD dimer instead.
Our model of the RSV immature NTD hexamer is similar to that observed for HIV, with widely spaced monomers and a large central cavity. However, in contrast to the immature HIV lattice, in which neither the NTD nor MA appears to stabilize the hexamer (Wright et al, 2007), the RSV immature hexamer is stabilized, at least in part, by p10–NTD interactions. This difference is not surprising; previous studies with chimeric RSV–HIV Gag constructs have shown that constructs with RSV CA require the critical p10 sequence from RSV upstream for assembly, whereas constructs with HIV CA tolerate RSV sequence upstream (Ako-Adjei et al, 2005), suggesting that the HIV CA domain does not interact with upstream Gag sequences. Similar studies with chimeric MLV and spleen necrosis virus Gag constructs show that assembly of these distantly related γ-retroviruses also requires that the domain upstream of CA (p12) be homologous to the CA domain. We therefore speculate that the γ-retrovirus immature hexamer may be stabilized by interactions between CA and p12.
The incompatibility of the immature p10–CA interaction with the mature MLV hexamer supports the idea that retroviruses have distinct immature and mature NTD–NTD interfaces, with PR cleavage favouring the mature over the immature interface. The participation of the upstream p10 domain in the immature lattice is intriguing because it suggests a mechanism by which proteolysis leads to maturation. The RSV p10 domain both stabilizes the immature hexamer and prevents mature NTD–NTD contacts from forming. The maturation process can thus be imagined as follows: upon PR activation, cleavage between p10 and CA destabilizes the immature hexamer and allows p10 to diffuse away from CA, which in turn unmasks mature contact surfaces and allows the mature NTD hexamer to form.
The nature of the link that couples proteolysis to maturation is likely to differ among retroviral families. There are three broad categories of mechanisms by which PR cleavage can change the CA–CA contacts: by initiating refolding of CA, by removing sequences that promote immature contacts or by removing sequences that inhibit mature contacts. The first mechanism, exemplified by the refolding of the β-hairpin, is well documented. Disrupting the β-hairpin of Mason–Pfizer monkey virus CANC by removing the N-terminal proline converts the in vitro assembly products from disorganized aggregates into spherical particles (Ulbrich et al, 2006), suggesting that proteolytic refolding is incompatible with the immature lattice. HIV-1 appears to use both refolding (based on pH-dependent epitope availability) and removing required sequences; as the immature hexamer is stabilized by interactions between downstream SP1 domains, proteolysis should dissociate the immature hexamer. For RSV, the contact between p10 and CA both stabilizes the putative immature NTD hexamer and interferes with the inferred mature NTD–NTD contacts, so PR cleavage between p10 and CA both destabilizes the immature particle and promotes mature assembly. Thus, although the structural properties of both immature and mature retroviral particles appear to be similar across families, the mechanics of maturation may prove to be quite different.
Materials and methods
DNA constructs
The pET3xc.ΔMBDΔPR plasmid has been described previously (Campbell and Vogt, 1997). pET3xc.ΔMBDΔPR (ΔBlpI) was generated by partially digesting ΔMBDΔPR with BlpI, gel-purifying single-cut plasmid, blunting the ends with Taq polymerase, and ligating to destroy the site; clones were screened by XhoI/BlpI double digestion to identify one with a unique BlpI site in CA. pET3xc.ΔMBD(−6C) is an unpublished plasmid in which the six cysteines in NC were replaced by alanine by two-step PCR. ΔMBD(−11C) was made by changing the five cysteines upstream of p10 to serine by two rounds of two-step PCR using the XhoI and FseI sites. Mutations involving P38, G39 or P40 of p10 were made by replacing the XhoI/BlpI fragment of ΔMBDΔPR(ΔBlpI) with two-step PCR products containing the desired mutation; the remainder of the alanine and GSGSG substitutions, and E51C, T20C and E51C/T20C, were made by replacing the FseI/BlpI fragments of ΔMBDΔPR(ΔBlpI) with two-step PCR products containing the desired mutation. E51C(−11C), T20C(−11C) and E51C/T20C(−11C) were made by replacing the FseI/SacII fragment of ΔMBD(−11C) with the analogous fragment from the analogous ΔMBDΔPR(ΔBlpI) constructs. TWCC(−11C) was made by replacing the FseI/SacII fragment from ΔMBD(−11C) with a two-step PCR product that used E51C/T20C(−11C) as a template. P38CC(−11C) was made by replacing the XhoI/FseI fragment of T20C(−11C) with a two-step PCR fragment that used ΔMBD(−11C) as a template. E51Cf was made by replacing the FseI/BlpI fragment of ΔMBDΔPR(ΔBlpI) with a two-step PCR product using p10(58–62)f as a template. E51C-f-T20C was made by replacing the FseI/BlpI fragment of ΔMBDΔPR(ΔBlpI) with a 2-step PCR product using E51Cf as a template to make E51C-f-T20C. Finally, E51C-f-T20C(−11C) was made by replacing the FseI/SacII fragment of ΔMBD(−11C) with that from E51C-f-T20C. The protease ePR C113S was made by replacing the AvrII fragment of pET11c.his-ePR (Schatz et al, 2001) with two annealed oligonucleotides. pQCXIP.SMGagΔPR(−11C) was made by replacing the XhoI-PacI fragment of pQCXIP.SMGag (Dalton and Vogt, unpublished data) with a PCR product amplified from ΔMBD(−11C). The other cell expression constructs were then made by replacing the XhoI/BlpI fragment of SMGagΔPR(−11C) with that from the analogous E. coli expression vector. All PCR-derived fragments and cloning sites were confirmed by sequencing.
Protein purification, in vitro assembly and electron microscopy
ΔMBDΔPR and derivatives were expressed and purified as previously described (Yu et al, 2001) with the following changes: after ammonium sulphate precipitation, the protein was bound to an Amersham SP FPLC column in 20 mM Tris pH 8.0, 100 mM NaCl and eluted in 20 mM Tris pH 8.0 and 500 mM NaCl. Dialysis steps were omitted. Protein was stored at 5 mg/ml and assembled by diluting to 1 mg/ml in 50 mM MES pH 6.0 or 6.5 with or without 50 μM DTT, adding 10% wt/wt ssDNA (GT50, an oligonucleotide with 25 GT repeats (Yu et al, 2001)), mixing gently and incubating at room temperature for 15 min.
Assembly reactions were spotted onto carbon-Formvar-coated grids and stained with 2% uranyl acetate (pH 5.0). The particles were visualized on a Morgagni 268 transmission electron microscope. Proteins scored as assembly competent yielded many particles visible in a single field, as exemplified by the micrographs in Figure 3B. Proteins scored as assembly incompetent yielded no regular particles in at least two independent protein preparations.
Centrifugation
Equilibrium centrifugation was performed on 10–60% (wt/wt) sucrose gradients in assembly buffer (50 mM MES pH 6.5 and 100 mM NaCl). Gradients were spun in an SW60 rotor at 50 000 r.p.m. for 4 h at 4°C. Fractions containing VLPs were collected from the gradient and their densities measured with a refractometer. The VLP fractions were diluted at the ratio of 1:5 with assembly buffer and the VLPs were pelleted by spinning in a microfuge at 16 000 g for 30 min at 4°C. Rate-zonal centrifugation was performed on 10–30% wt/wt sucrose gradients in 50 mM MES pH 6.5, 500 mM NaCl, 2 M urea. Gradients were spun in an SW60 rotor at 50 000 r.p.m. for 24 h at 4°C.
Oxidation and PR digestion
Particles were resuspended in assembly buffer with or without 10 μM DTT. Resuspended particles were oxidized with copper phenanthroline (60 μM CuSO4, 267 μM O-phenanthroline), mixed by vortexing for approximately 5 s and immediately quenched with 20 mM iodoacetamide and 3.7 mM Neocuproine (Sigma). In vitro PR digestion was carried out as previously described (Schatz et al, 2001). In brief, reactions were adjusted to 100 mM MES pH 6.5, 700 mM NaCl, 0.7 mM EDTA and 5.3% glycerol by adding 5 × PR buffer. Before digestion, DTT to 15 mM was added to reduced samples and fresh iodoacetamide was added to oxidized samples. A cysteine-lacking mutant of RSV PR (PR C113S) purified from E. coli was added at 2 μg PR/10 μg ΔMBDΔPR. (Wild-type PR is inactive in the presence of iodoacetamide.) Reactions were incubated at 37°C for 16 h. Fresh iodoacetamide was added to oxidized samples immediately before SDS–PAGE.
Cell culture and immunoblotting
DF1 (chicken embryonic fibroblast) cells were maintained in Dulbecco's modified Eagle's medium supplemented with 5% foetal bovine serum, 5% NuSerum, 1% heat-inactivated chick serum, standard vitamins, L-glutamine, penicillin and streptomycin. Stable cell lines expressing Gag proteins were created by transduction with virus derived from Phoenix cells transfected with pQCXIP.SMGagΔPR(−11C) and related constructs using FuGene 6 followed by selection with 2 μg/ml puromycin. Medium was harvested after 3 days and pelleted through a 15% sucrose cushion by centrifugation for 20 min at 70K in a TLA100.4 rotor. Pellets were resuspended in VSB (20 mM Tris pH 7.5, 100 mM NaCl, 1 mM EDTA) plus 20 mM iodoacetamide and permeabilized with 0.01% Triton X-100 in the same buffer. PR digestion was carried out as previously described. Immunoblotting was performed using a polyclonal rabbit anti-RSV CA antibody.
Modelling
The structures for the MuLV CA NTD hexamer (PDB code 1U7K, (Mortuza et al, 2004)) and the RSV-extended NTD dimer (PDB code 1P7N (Nandhagopal et al, 2004)) were used to model the mature RSV CA NTD hexamer. The protein secondary structure prediction programs, PSIPRED (McGuffin et al, 2000), 3D-PSSM (Kelley et al, 2000) and Scratch (Pollastri et al, 2002) predict a β-hairpin in residues 1–15 of RSV CA NTD, as seen in the mature RSV CA NTD structure (PDB code 1EM9 (Kingston et al, 2000)). Consequently, we used the modelling program Nest (Petrey et al, 2003) to construct a composite model of RSV CA NTD in which residues 1–21 were modelled after 1U7K and 22–144 were modelled after 1P7N. The resulting mature monomer model was then structurally superimposed, using the combinatorial extension (CE) server (Shindyalov and Bourne, 1998), on each subunit (A–F) of 1U7K, the mature MuLV CA NTD hexamer, thus yielding a mature model of the RSV CA NTD hexamer (Figure 7B).
The same structures (1U7K and 1P7N) were used to construct an immature model of the RSV CA NTD hexamer. As opposed to the mature CA, immature CA is not cleaved between Met0 and Pro1; that is, the upstream p10 helix is joined to and, as depicted in Figure 2, free to interact with CA. To model the immature hexamer, we excised the a–B intermolecular pair of p10–CA NTD from 1P7N. The p10 half, ‘a', was cut after CA Thr6; therefore, p10 comprises Pro-24 to Thr6 of ‘a'. The CA NTD half, ‘B', was cut before Leu14; therefore, CA NTD comprises Leu14 to Ala144 of ‘B'. The ‘a–B' pair was then structurally superimposed, using CE, on each subunit of 1U7K. With the ‘p10' region added in, there was significant structural overlap between the subunits of the immature hexamer. To relieve this overlap, each subunit was radially moved away from the centre of the hexamer until there was no structural overlap. The program Loopy (Petrey et al, 2003) was used to model in the missing segment between residues Thr6 and Leu14. This segment was modelled such that each p10 region is joined to the next CA NTD of the hexamer, moving clockwise (Figure 7B).
Acknowledgments
We are indebted to Peter Prevelige and Chi-Yu Fu for carrying out initial experiments to investigate the multimeric status of the ΔMBDΔPR protein by sedimentation equilibrium and velocity and to Marc Johnson and Richard Kingston for reviewing the manuscript. The cartoon in Figure 7A is based on a sketch by Richard Kingston.
References
- Ako-Adjei D, Johnson MC, Vogt VM (2005) The retroviral capsid domain dictates virion size, morphology, and coassembly of gag into virus-like particles. J Virol 79: 13463–13472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JA, Johnson MC, Simon MN, Fuller SD, Vogt VM (2006) Cryo-electron microscopy reveals conserved and divergent features of gag packing in immature particles of Rous sarcoma virus and human immunodeficiency virus. J Mol Biol 355: 157–168 [DOI] [PubMed] [Google Scholar]
- Briggs JA, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC (2004) The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol 11: 672–675 [DOI] [PubMed] [Google Scholar]
- Butterfield-Gerson KL, Scheifele LZ, Ryan EP, Hopper AK, Parent LJ (2006) Importin-beta family members mediate alpharetrovirus gag nuclear entry via interactions with matrix and nucleocapsid. J Virol 80: 1798–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan EM, Wills JW (2003) Link between genome packaging and rate of budding for Rous sarcoma virus. J Virol 77: 9388–9398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Vogt VM (1997) In vitro assembly of virus-like particles with Rous sarcoma virus gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J Virol 71: 4425–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Olivas R, Newman JL, Summers MF (2000) Solution structure and dynamics of the Rous sarcoma virus capsid protein and comparison with capsid proteins of other retroviruses. J Mol Biol 296: 633–649 [DOI] [PubMed] [Google Scholar]
- Ehrlich LS, Agresta BE, Carter CA (1992) Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J Virol 66: 4874–4883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble TR, Yoo S, Vajdos FF, von Schwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP (1997) Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278: 849–853 [DOI] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, Cheng A, Yeager M (2007) Structure of full-length HIV-1 CA: A model for the mature capsid lattice. Cell 131: 70–79 [DOI] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI (2004) Assembly properties of the human immunodeficiency virus type 1 CA protein. J Virol 78: 2545–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganser BK, Cheng A, Sundquist WI, Yeager M (2003) Three-dimensional structure of the M-MuLV CA protein on a lipid monolayer: a general model for retroviral capsid assembly. EMBO J 22: 2886–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, Sundquist WI (1996) Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273: 231–235 [DOI] [PubMed] [Google Scholar]
- Gross I, Hohenberg H, Wilk T, Wiegers K, Grättinger M, Müller B, Fuller S, Kräusslich H-G (2000) A conformational switch controlling HIV-1 morphogenesis. EMBO J 19: 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- Ivanov D, Tsodikov OV, Kasanov J, Ellenberger T, Wagner G, Collins T (2007) Domain-swapped dimerization of the HIV-1 capsid C-terminal domain. Proc Natl Acad Sci USA 104: 4353–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Jin L, Peterson DL, Lawson CL (1999) Model for lentivirus capsid core assembly based on crystal dimers of EIAV p26. J Mol Biol 286: 83–93 [DOI] [PubMed] [Google Scholar]
- Johnson MC, Scobie HM, Ma YM, Vogt VM (2002) Nucleic acid-independent retrovirus assembly can be driven by dimerization. J Virol 76: 11177–11185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi SM, Vogt VM (2000) Role of the Rous sarcoma virus p10 domain in shape determination of Gag virus-like particles assembled in vitro and within Escherichia coli. J Virol 74: 10260–10268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, MacCallum RM, Sternberg MJ (2000) Enhanced genome annotation using structural profiles in the program 3D-PSSM. J Mol Biol 299: 499–520 [DOI] [PubMed] [Google Scholar]
- Khorasanizadeh S, Campos-Olivas R, Summers MF (1999) Solution structure of the capsid protein from the human T-cell leukemia virus type-I. J Mol Biol 291: 491–505. [DOI] [PubMed] [Google Scholar]
- Kingston RL, Fitzon-Ostendorp T, Eisenmesser EZ, Schatz GW, Vogt VM, Post CB, Rossmann MG (2000) Structure and self-association of the Rous sarcoma virus capsid protein. Structure 8: 617–628 [DOI] [PubMed] [Google Scholar]
- Lanman J, Lam TT, Barnes S, Sakalian M, Emmett MR, Marshall AG, Prevelige PE Jr (2003) Identification of novel interactions in HIV-1 capsid protein assembly by high-resolution mass spectrometry. J Mol Biol 325: 759–772 [DOI] [PubMed] [Google Scholar]
- Lee SK, Boyko V, Hu WS (2007) Capsid is an important determinant for functional complementation of murine leukemia virus and spleen necrosis virus Gag proteins. Virology 360: 388–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Hill CP, Sundquist WI, Finch JT (2000) Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407: 409–413 [DOI] [PubMed] [Google Scholar]
- Mayo K, Huseby D, McDermott J, Arvidson B, Finlay L, Barklis E (2003) Retrovirus capsid protein assembly arrangements. J Mol Biol 325: 225–237 [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16: 404–405 [DOI] [PubMed] [Google Scholar]
- Mortuza GB, Haire LF, Stevens A, Smerdon SJ, Stoye JP, Taylor IA (2004) High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature 431: 481–485 [DOI] [PubMed] [Google Scholar]
- Nandhagopal N, Simpson AA, Johnson MC, Francisco AB, Schatz GW, Rossmann MG, Vogt VM (2004) Dimeric Rous sarcoma virus capsid protein structure relevant to immature Gag assembly. J Mol Biol 335: 275–282 [DOI] [PubMed] [Google Scholar]
- Petrey D, Xiang Z, Tang CL, Xie L, Gimpelev M, Mitros T, Soto CS, Goldsmith-Fischman S, Kernytsky A, Schlessinger A, Koh IY, Alexov E, Honig B (2003) Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling. Proteins 53 (Suppl 6): 430–435 [DOI] [PubMed] [Google Scholar]
- Pollastri G, Przybylski D, Rost B, Baldi P (2002) Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins 47: 228–235 [DOI] [PubMed] [Google Scholar]
- Schatz GW, Reinking J, Zippin J, Nicholson LK, Vogt VM (2001) Importance of the N terminus of Rous sarcoma virus protease for structure and enzymatic function. J Virol 75: 4761–4770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheifele LZ, Garbitt RA, Rhoads JD, Parent LJ (2002) Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc Natl Acad Sci USA 99: 3944–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheifele LZ, Kenney SP, Cairns TM, Craven RC, Parent LJ (2007) Overlapping roles of the Rous sarcoma virus Gag p10 domain in nuclear export and virion core morphology. J Virol 81: 10718–10728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheifele LZ, Ryan EP, Parent LJ (2005) Detailed mapping of the nuclear export signal in the Rous sarcoma virus Gag protein. J Virol 79: 8732–8741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindyalov IN, Bourne PE (1998) Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng 11: 739–747 [DOI] [PubMed] [Google Scholar]
- Ulbrich P, Haubova S, Nermut MV, Hunter E, Rumlova M, Ruml T (2006) Distinct roles for nucleic acid in in vitro assembly of purified Mason-Pfizer monkey virus CANC proteins. J Virol 80: 7089–7099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler UK, Stemmler TL, Klishko VY, Li S, Albertine KH, Davis DR, Sundquist WI (1998) Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J 17: 1555–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ (2007) Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J 26: 2218–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Joshi SM, Ma YM, Kingston RL, Simon MN, Vogt VM (2001) Characterization of Rous sarcoma virus gag particles assembled in vitro. J Virol 75: 2753–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]