Abstract
In human the brain represents only about 2% of the body’s mass but contains about one quarter of the body’s free cholesterol. Cholesterol is synthesised de novo in brain, and removed by metabolism to oxysterols. 24S-Hydoxycholesterol represents the major metabolic product of cholesterol in brain, being formed via the cytochrome P450 (CYP) enzyme CYP46A1. CYP46A1 is expressed exclusively in brain, normally by neurons. In this study we investigated the effect of 24S-hydroxycholesterol on the proteome of rat cortical neurons. By using two dimensional liquid chromatography linked to nano-electrospray tandem mass spectrometry over 1040 proteins were identified including members of the cholesterol, isoprenoid and fatty acid synthesis pathways. By using stable isotope labelling technology, the protein expression patterns of enzymes in these pathways were investigated. 24S-Hydroxycholesterol was found to down-regulate the expression of members of the cholesterol/isoprenoid synthesis pathways including 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 (EC 2.3.3.10), diphosphomevalonate decarboxylase (EC 4.1.1.33), isopentenyl-diphosphate delta isomerase (EC 5.3.3.2), farnesyl-diphosphate synthase (Geranyltranstransferase, EC 2.5.1.10), and dedicated sterol synthesis enzymes, farnesyl-diphosphate farnesyltransferase 1 (squalene synthase, EC 2.5.1.21), and methylsterol monooxygenase (EC 1.14.13.72). The expression of many enzymes in the cholesterol/isoprenoid and fatty acid synthesis pathways are regulated by the membrane-bound transcription factors named sterol regulatory element-binding proteins (SREBPs), which themselves are both transcriptionally and posttranscriptionally regulated. The current proteomic data indicates that 24S-hydroxycholesterol down-regulates cholesterol synthesis in neurons, possibly, in a posttranscriptional manner through SREBP-2. In contrast to cholesterol metabolism, enzymes responsible for the synthesis of fatty acids were not found to be down-regulated in neurons treated with 24S-hydroxycholesterol, while apolipoprotein E (apo E), a cholesterol trafficking protein, was found to be up-regulated. Taken together, this data leads to the hypothesis that in times of cholesterol excess, 24S-hydroxycholesterols signals down-regulation of cholesterol synthesis enzymes through SREBP-2, but up-regulates apo E synthesis (through the liver X receptor) leading to cholesterol storage and restoration of cholesterol balance.
Keywords: cholesterol, oxysterol, neuron, quantitative proteomics, SREBP, LXR, iTRAQ
Introduction
Most recent studies of cholesterol homeostasis pathways have been performed at the mRNA level, on transgenic animals, or using Western blot analysis for specified protein1-7. However, with the maturation of proteomics technology, alternative methodology is now available for the study of signalling, synthesis and metabolism8,9. Using proteomics methods the identification and relative quantification of proteins does not require their pre-selection (as with antibodies) and is made at the protein level (cf. mRNA). However, proteomic identifications are restricted to the more abundant proteins and for deep mining of the proteome some form of enrichment, pre-fractionation or subcellular fractionation may be desirable10-12. Alternatively, deep mining can be achieved by concentrating on one particular cell type e.g. neuron13, astrocyte14, rather than a tissue consisting of many different cell types.
In the current study, our interest is focussed on primary cortical neurons derived from embryonic rats. In the embryonic stage, before astrocytes differentiate, neurons meet their cholesterol requirements by de novo synthesis. Using proteomic methods we wish to shed light on the hypothesis that metabolites of cholesterol provide a feedback control on the cell content of free cholesterol.
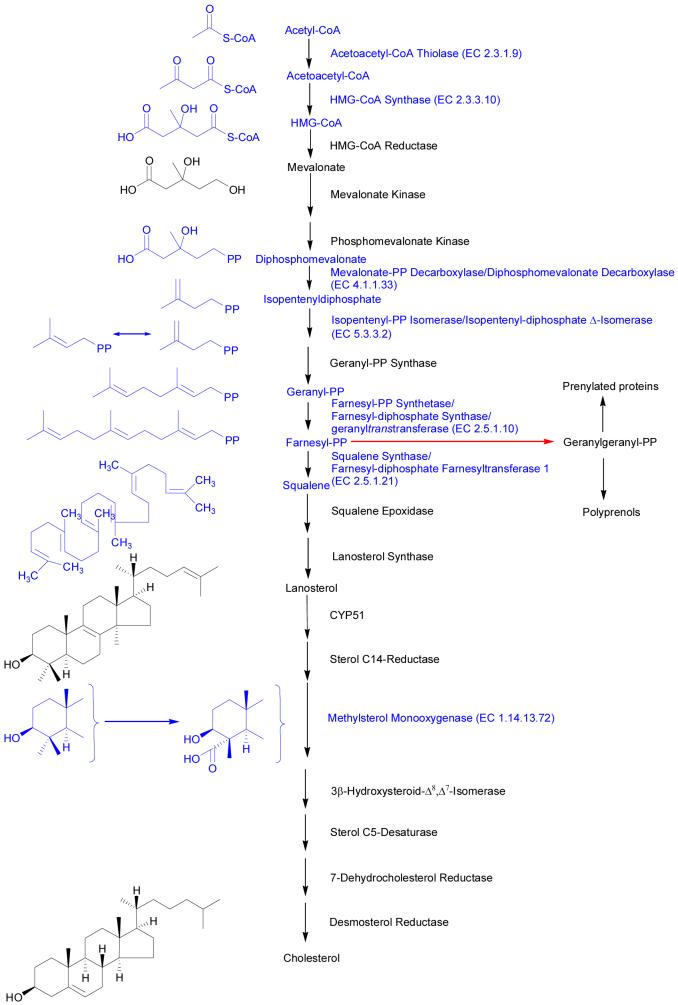
Lipid homeostasis in vertebrate cells is regulated by a family of membrane bound transcription factors called sterol regulatory element-binding proteins (SREBPs)1,2. SREBPs activate the expression of over 30 genes involved in the synthesis and uptake of cholesterol, fatty acids, triglycerides and phospholipids as well as NADPH (Figure 1)2. SREBPs are synthesised as inactive precursors bound to the endoplasmic reticulum (ER) and in order to reach the nucleus and act as transcription factors, the NH2-terminal domain must be released. SREBPs in the ER form a complex with an escort protein SREBP cleavage-activating protein (SCAP), which in times of cholesterol deprivation escorts SREBPs from the ER to the Golgi where SREBPs are proteolytically processed to their nuclear forms (nSREBP) which activate genes for cholesterol synthesis and uptake. In times of cholesterol wealth, cholesterol binds to the sterol sensing domain (SSD) of SCAP and triggers a conformational change that causes SCAP to bind to membrane bound proteins called INSIGs (1 and 2). The abbreviation INSIG signifying “insulin-induced gene”. When SCAP binds to INSIGs, SCAP and the attached SREBP are retained in the ER, preventing transport to the Golgi and further processing of SREBPs to the active form (see Figure 3 in reference 1). SREBPs exist in three forms, SREBP-2 which is primarily involved in cholesterol synthesis, SREBP-1c that primarily stimulates fatty acid synthesis and SREBP-1a which preferably activates cholesterol synthesis. As well as cholesterol, the oxysterol, 25-hydroxycholesterol can trigger binding between SCAP and INSIGs, but in this case by sterol binding to INSIGs, with the resultant inhibition of cholesterol synthesis6,7,15. Cholesterol synthesis is further regulated by a mechanism involving sterols, INSIGs and HMG-CoA (3-hydroxy-3-methylglutaryl-Coenzyme A) reductase, the first committed enzyme in the cholesterol synthesis pathway. When cholesterol and its precursor lanosterol accumulate in ER membranes HMG-CoA reductase is induced to bind to INSIGs, probably by direct binding of sterol to the SSD of HMG-CoA reductase, initiating ubiquitin-mediated proteosomal degradation of the reductase, the postubiquination step is stimulated by geranylgeraniol through an unidentified mechanism that likely involves a geranylgeranylated protein, such as one of the Rab proteins (see Figure 5 in reference 1). It is postulated that an oxysterol binding protein (OBP) mediates similar oxysterol effects on the reductase1.
Figure 1.
Enzymes of the cholesterol synthesis pathway regulated by SREBP-2. Enzymes written in blue were found to be down-regulated in cortical neurons by treatment with 24S-hydroxycholesterol. The EC number is given for these enzymes. Some enzymes have multiple names as indicated in the figure. Farnesyl-PP represents a branch point between cholesterol synthesis and that of non-steroidal isoprenoids. Formation of geranylgeranyl-PP (indicated by the red arrow) by farnesyltranstransferase (EC 2.5.1.29) leads to the further formation of geranylgeraniol, polyprenols, dolichols and ubiquinone.
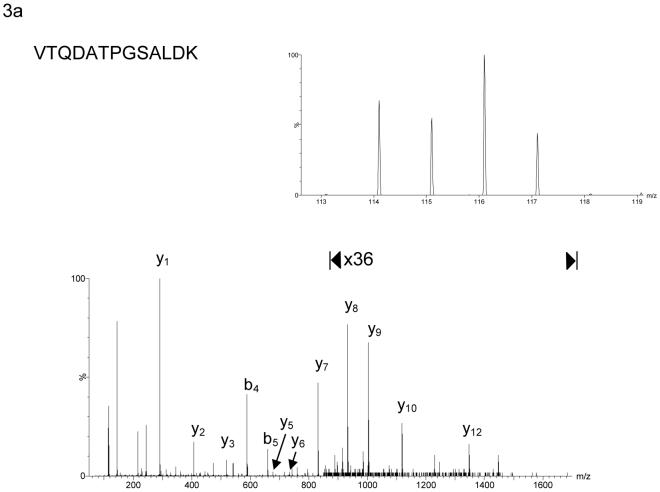
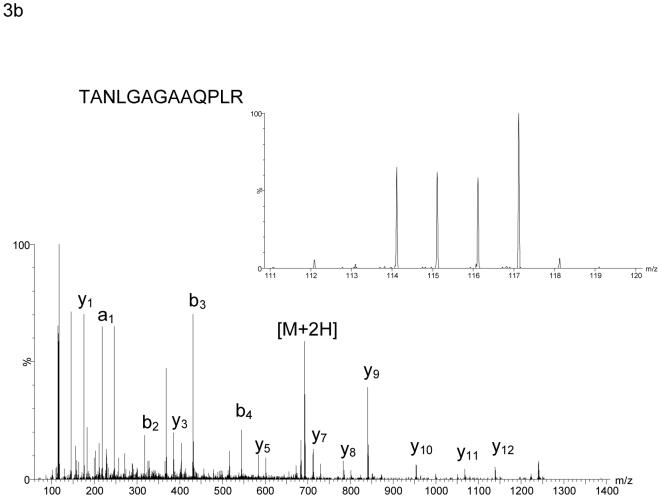
Figure 3.
MS/MS spectra of iTRAQ labelled peptides derived from (a) HMG-CoA synthase 1 (EC 2.3.3.10) and (b) apo E precursor. The insets in the right-hand corner show the iTRAQ reporter ions m/z 114.1 - 117.1 on an expanded scale. The reporter ions at m/z 116.1 and 117.1 are derived from peptides originating from proteins isolated from cortical neurons treated with HPBCD vehicle alone and with 24S-hydroxycholesterol in HPBCD vehicle, respectively. In (a) HMG-CoA synthase 1 is demonstrated to be down-regulated by treatment with 24S-hydroxycholesterol, while in (b) apo E precursor is up-regulated. The reporter ions at m/z 114.1 and 115.1 in (a) and (b) are derived from peptides originating from proteins isolated from neurons treated with ethanol as vehicle, and 24S-hydroxycholesterol in ethanol vehicle, respectively. In the absence of HPBCD, which enhances the solubility of sterol in aqueous environment and its penetration into cells, the effect of 24S-hydroxycholesterol on protein expression is greatly reduced.
Oxysterols are naturally occurring ligands for the liver X receptors (LXRs), which bind to DNA as obligate heterodimers with the retinoid X receptor (RXR)16. LXRs serve as sterol sensors to regulate transcription of gene products that control intracellular cholesterol homeostasis. LXRs target genes include cytochrome P450 7A1 (CYP7A1), the rate-limiting enzyme in the classic pathway of bile acid biosynthesis17, apoliporotein E (apo E) and ATP-binding cassette transporters A1 (ABCA1) and G1 (ABCG1)18, SREBP-1c, but not SREBP-1a or SREBP-219. Thus, oxysterols regulate genes which modulate intacellular free cholesterol through increased elimination as bile acids, efflux via ABC1 transporters and storage as fatty acid esters via apo E.
Although, in man, the brain makes up only about 2% of body mass, it contains about 25% of the body’s cholesterol20, essentially all of which is synthesised de novo on account of the impermeability of the blood brain barrier (BBB) to cholesterol. Excess cholesterol is exported from brain in the form of oxysterols which can cross the BBB. It appears that this mechanism accounts for about two thirds of the elimination of cholesterol in mouse, rat and man20. The major oxysterol in brain is 24S-hydroxycholesterol, which is present at a level of about 10 ng/mg in man21 and 20 ng/mg in rat22. In brain, cholesterol is mostly located in glial cells20, however, the enzyme responsible for cholesterol oxidation, CYP46A1, is normally expressed in neurons23. Perhaps, significantly LXRβ, the LXR isoform found in brain, has a similar distribution pattern to CYP46A124. As neuronal cells contain only a small proportion of total brain cholesterol (∼2%), and as CYP46A1 is only normally expressed in neurons, it is evident that only a small pool of total brain cholesterol is available for CYP46A1 mediated turnover, however this pool of cholesterol has a high turnover rate in contrast to the bulk of cholesterol which is turned over much slower i.e. half life of bulk cholesterol is 5 years in human adult brain20,25. This leads to the questions, what factors control cholesterol homeostasis in neurons and does 24S-hydroxycholesterol, the product of CYP46A1 mediated oxidation of cholesterol, have a role to play in this process in brain? To answer these questions we have taken a quantitative proteomic approach where we have treated rat cortical neurons with 24S-hydroxycholesterol using (2-hydroxypropyl)-β-cyclodextrin (HPBCD) as a delivery vehicle (HPBCD enhances the solubility of sterol in aqueous environment and its penetration into cells) and monitored the resultant proteomic changes using a stable-isotope tagging approach using iTRAQ labelling26.
Experimental
Cortical neuron preparation and 24S-hydroxycholesterol treatment
Primary cortical neurons were prepared from rat embryonic pups day 19, cultured on poly-L-lysine coated plates using 108 cells/plate in neurobasal medium (Invitrogen, UK), containing B27 supplement, glutamine, penicillin/streptomycin and glucose27. The purity of cortical neuron preparations was verified by staining with anti-glial fibrillary acidic protein (Sigma, UK) which showed the cultures contained less than 3% glial cells. 24S-Hydroxycholesterol (Steraloids, London, UK) stock solution was prepared by dissolving the sterol in ethanol to 67 mM. Before each cell treatment, 24S-hydroxycholesterol was first diluted with either saline or 45% HPBCD (w/v) in saline, and then added to culture medium to a final concentration of 10 μM, containing 0.014% ethanol (v/v) with or without 0.13% HPBCD (w/v). It should be noted that in the absence of HPBCD 24S-hydroxycholesterol is only partially soluble in culture medium (and saline solution) giving a white insoluble precipitate. Similar solubility problems have been noted by others6,28,29. At day 7, medium was replaced with fresh medium containing either: (a) ethanol vehicle, (b) 10 μM 24S-hydroxycholesterol in ethanol, (c) HPBCD vehicle, or (d) 10 μM 24S-hydroxycholesterol in HPBCD vehicle, and incubation continued for a further 24 hrs. Experiments were repeated three times.
Protein digestion and iTRAQ reagent labelling
After 24 hr, cells were lysed in 0.5 M triethylammonium bicarbonate (Sigma, UK), containing 0.1% sodium dodecyl sulphate (SDS). The supernatant was obtained by centrifugation at 16000 g for 20 min at 4 °C. Equal amounts of protein (80 μg, measured by Bradford assay, Bio-Rad, UK) from the four samples were reduced (5 mM tris-(2-carboxyethyl)phosphine, 60 °C, 1 hr), alkylated (10 mM methyl methanethiosulphate, room temperature, 10 min) and digested with trypsin (1:10 w/w, 37 °C) overnight. Peptides were labelled with iTRAQ reagents as described in the manufacturer’s instructions (Applied Biosystems, UK). In brief peptides derived from samples (a) - (d) were labelled with iTRAQ reagents 114 - 117 respectively. To check the efficiency of digestion and labelling, an aliquot of each sample was purified by a strong cation exchange (SCX) spin column (Vivascience, UK) and then analysed by liquid chromatography-tandem mass spectrometry (LC-MS/MS), see below. The four labelled samples were then mixed.
Two Dimensional Liquid Chromatography (2D-LC)
Two different protocols were applied. Initially an on-line 2D-LC-MS/MS approach was taken. Using a Dionex UltiMate 3000 Dual LC system (Camberley, Surrey, UK), 10 μg of peptides from the total digest was injected on a SCX column (150 × 0.3 (i.d.) mm, 5 μm PolySULFOETHYL A, PolyLC, Columbia, MD, USA). Using an NH4Cl linear gradient (0 - 500 mM, in 2% acetonitrile, 5 μL/min) twelve 2 hr fractions were collected on-line on two C18 trap columns arranged in parallel to one another (5 × 0.3 (i.d.) mm, 5 μm, Dionex). Initially trap column 1 was positioned in-line with the SCX column and fraction 1 collected on it. After 2 hr trap column 2 was switched in-line with the SCX column in place of trap column 1, and fraction 2 collected on it. Trap column 1 was then washed off-line for 10 min with 2% acetonitrile at 20 μL/min, and then positioned in-line with a reversed phase (RP) nano-column (150 × 0.075 (i.d) mm, 3 μm C18, Dionex) interfaced to a Q-TOF Global mass spectrometer (Waters, Manchester, UK). Peptides were eluted from the RP column using a binary gradient over 120 min (including washing time). Mobile phase A was 2% acetonitrile containing 0.1% formic acid, and mobile phase B was 90% acetonitrile containing 0.1% formic acid. The gradient employed after switching the trap column in-line with the RP column was: 4% B to 40% B over 80 min, then 40% B to 90% B over 1 min, maintaining 90% B for 20 min, followed by re-equilibration at 4% B for 18 min. The flow-rate through the RP column was 0.3 μL/min. After 2 hr trap column 1 was repositioned in-line with the SCX column to collect fraction 3, while trap column 2 was washed and positioned in-line with the RP column. This procedure was repeated until twelve 2 hr fractions were collected from SCX column and eluted to the mass spectrometer.
Despite providing the advantage of total automation, on-line 2D-LC is limited by the loading and peak capacity of the RP nano-column. This can be overcome by performing multiple repeat injections and employing “gas-phase fractionation”, where small m/z ranges are sequentially interrogated30,31. In a preliminary investigation it was established that the number of proteins identified by performing multiple repeat injections and analysis over five m/z ranges rather than just one injection and analysis over a single m/z range was increased by a factor of five. These results encouraged us to perform off-line 2D-LC, where we perform just one SCX separation (100 μg of total digest injected on column) using a 150 × 1 (i.d.) mm (5 μm PolySULFOETHYL A, PolyLC) column rather than making multiple injections on the smaller column. The gradient employed was 0 - 500 mM NH4Cl in 2% acetonitrile and 12 - 28 fractions were manually collected. Each fraction was then desalted on a C18 spin column (Pierce, UK), re-constituted in 25 μL of 0.1% formic acid, and 5 μL aliquots injected repeatedly onto the RP-nano column. For off-line 2D-LC the subsequent RP separation was performed using a Micromass CapLC system (Waters, UK) with sample loading onto a trap column (5 mm × 0.3 mm, 5 μm, PepMap C18 guard column, Dionex) at a flow-rate of 15 μl/min, delivered isocratically with solvent C (0.1% formic acid) by auxiliary pump C. Sample was washed on the trap column for 3 min with solvent C before being switched in-line with the RP nano-column (150 × 0.075 (i.d) mm, 3 μm C18, Dionex), which was equilibrated with 95% solvent A (0.1% formic acid in 5% acetonitrile), 5% solvent B (0.1% formic acid in 95% acetonitrile) at a flow-rate of approximately 200 nl/min. Five min after sample loading the proportion of solvent B was increased linearly to 28% over 75 min, then to 80% over 20 min; maintained at 80% solvent B for 15 min (wash phase) then re-equilibrated at 95% solvent A, 5% solvent B for 10 min. The column effluent was continuously directed into a Micromass Q-TOF Global mass spectrometer. Five repeat injections were made per SCX fraction and analysis performed over successive m/z ranges (see below).
Mass Spectrometry
The Q-TOF Global mass spectrometer was operated in the positive-ion electrospray (ES) mode using data dependent analysis (DDA) for the identification and quantification of peptides. DDA employs an initial “survey scan” which identifies the four most abundant multiply charged ions (tryptic peptides usually appear as 2+ and/or 3+ ions), which are then fragmented by MS/MS which provides amino acid sequence information, before another “survey scan” is performed and the cycle is repeated throughout the chromatographic run. DDA analysis was performed using a 0.5 s MS survey scan (m/z range 420-1000 or e.g. 420-600, 600-700, 700-800, 800-900, or 900-1000) followed by 1 s MS/MS scans (0.1 s interscan time) on up to four different precursor ions (intensity threshold 10 counts per second). In the DDA mode, MS/MS spectrum acquisition (in the m/z range 50 - 1800) was allowed for up to a total of 2.2 s on each precursor ion, or stopped when the signal intensity fell below three counts per second, and a new MS to MS/MS cycle was started. Precursors were excluded from any further MS/MS fragmentation for 45 s (retention time) to minimize repeated identification of the same peptide; singly charged ions were also excluded as precursors for MS/MS.
Protein identification and quantification
Each raw data file was first processed by MassLynx 4.1 The parameters for creating pkl files were as follows: Spectrum selection criteria, Combine sequential scans with same precursor, Process all combined scans; Mass measure, Smooth window 3 channels, Number of smooths 2, Smooth mode Savitzky-Golay; Centroid, Minimum peak width at half height 4, Centroid mode Centroid top 80%. Peptide and protein identifications were performed using the Mascot search engine (version 2.2, MatrixScience, London, UK) located on a local server. Database searching was restricted to tryptic peptides of rat proteins, using the IPI rat database (April 7th, 2007, 41506 entries). The IPI database provides a minimally redundant, yet maximally complete set of proteins for the featured species (one sequence per transcript). Methionine oxidation and N-terminal acetylation were selected as variable modifications, cysteine blocked with methyl methanethiosulphate and iTRAQ 114 - 117 were selected as fixed modifications at peptide N-terminus and side-chain of lysine; one missed cleavage was allowed. Precursor and MS/MS tolerances were < 0.3 Da, monoisotopic mass.
The level of confidence for peptide identifications was based on the Mascot assignment of “identity” (p<0.05). When peptide scores were only just above “identity” (e.g. score of 33 when identity is 32) their MS/MS spectra were checked manually and only accepted when criteria outlined by the MCP document on protein identification were met (e.g. see Knapp at http://www.mcponline.org/misc/PariReport_PP.shtml). Protein identifications were only made when two or more peptides from that protein were identified with Mascot scores above the identity threshold. (To generate Table 1 presented in supplementary data, the reporting criteria were relaxed so that in cases where a protein had been identified by two peptides in one of the biological replicates, only a single peptide with score above the identity threshold was required to identify it in each of the other two replicates). Furthermore, where the identified protein was a member of a multi-protein family with similar sequences, the protein identified was the one with the highest number of matched peptides and Mascot score. To asses the false positive peptide identification rate, the data was searched as above but against a randomised version of the IPI rat database.
Quantification was performed using Mascot 2.2 (MatrixScience). Using the Mascot quantification method, protein quantification was only performed on proteins identified by two or more peptides with scores above the “identity” threshold. Protein ratios were “weighted” according to signal intensity, where the individual iTRAQ reporter-ion intensity values of the assigned peptides are summed and the protein ratio calculated from the summed values. iTRAQ ratios were “normalisation”, where a correction factor is applied such that for each reported ratio the “median” for that ratio for all peptide matches in the data set is unity.
Immunoblot analysis
Twenty μg of protein from each cell lysate was loaded onto a 4-12% Novex Bis-Tris gel (Invitrogen, UK). After electrophoresis, the proteins were transferred onto a nitrocellulose membrane. Anti-squalene synthase (sc-49758) and anti-goat IgG HRP conjugate (sc-2020) were from Santa Cruz Biotechnology (CA, USA). Apolipoprotein E precursor antibody (ab20874) was purchased from Abcam (UK) and anti-rabbit IgG HRP conjugate was from GE Healthcare (UK) Immunoreactivities were detected using the ECL Western blotting substrate (Pierce, UK). β-Tubulin was used as a loading control.
Results
Derivatisation with iTRAQ reagent
The derivatisation reactions at the peptide N-terminal amino and lysine ε-amino groups were complete (>99%), as assessed by modification of peptides derived from porcine trypsin. Derivatisation with a single iTRAQ tag, increases the peptide mass by 145 Da (290 Da if a double derivative), made up by a reporter group of 114, 115, 116 or 117 Da and a balance group of 31, 30, 29 or 28 Da respectively26. Peptides of the same amino acid sequence but different reporter/balance groups were found to chromatograph identically and were not resolved in terms of mass in mass spectra. MS/MS spectra of such peptides gave identical patterns of b and y ions but different reporter ions (m/z 114 - 117) from which their relative abundance was determined.
For a protein whose expression pattern did not alter upon 24S-hydroxycholesterol administration to the cell culture medium, the ratio of reporter ion (114/115/116/117) abundance in the MS/MS spectra of its tryptic peptides will theoretically be 1:1:1:1. This was in fact the case for most proteins (e.g. fatty acid synthase EC 2.3.1.85 and acetyl-CoA carboxylase EC 6.4.1.2, Table 1).
Table 1.
Differential expression1 of proteins upon the addition of 24S-hydroxycholesterol in HPBCD to cortical neurons as compared to the addition of HPBCD vehicle, determined by iTRAQ labelling
| IPI No | EC Number | Protein Name | Number of Peptides Identified (Measurement Made)2 in each Biological Replicate | Protein Ratio(117.1/116.1)/(SD geo)3 | ||||
|---|---|---|---|---|---|---|---|---|
| Biological Replicate4 | ||||||||
| 1 | 2 | 3 | 1 | 2 | 3 | |||
| IPI00214665 | EC 2.3.3.8 | ATP-citrate synthase | 10(12) | 14(23) | 26(50) | 0.75/(1.151) | 0.772/(1.173) | 0.751/(1.205) |
| IPI00480766 | EC 2.3.1.9 | Cytosolic acetoacetyl-CoA thiolase | 2(2) | 5(5) | 5(15) | 0.873/(1.184) | 0.82/(1.068) | 0.804/(1.222) |
| IPI00188158 | EC 2.3.3.10 | 3-hydroxy-3-methylglutaryl-CoA synthase 1 cytoplasmic | 5(12) | 10(38) | 10(28) | 0.468/(1.134) | 0.556/(1.048) | 0.475/(1.182) |
| IPI00210317 | EC 4.1.1.33 | diphosphomevalonate decarboxylase/ mevalonate-5-pyrophosphate decarboxylase | 2(2) | 1(1) | 3(5) | 0.779/(1.082) | --- | 0.749/(1.064) |
| IPI00212309 | EC 5.3.3.2 | isopentenyl-diphosphate Δ-isomerase/ isopentenylpyrophosphate isomerase | 2(2) | 2(3) | 7(13) | 0.705/(1.002) | 0.603/(1.093) | 0.651/(1.119) |
| IPI00325147 | EC 2.5.1.10 | geranyltranstransferase/ farnesyl pyrophosphate synthetase/ farnesyl-diphosphate synthase | 3(7) | 5(14) | 6(24) | 0.76/(1.066) | 0.628/(1.357) | 0.663/(1.284) |
| IPI00210233 | EC 2.5.1.21 | squalene synthase/ farnesyl-diphosphate farnesyltransferase 1 | 2(4) | 1(3) | 2(2) | 0.644/(1.154) | 0.542/(1.184) | 0.621/(1.101) |
| IPI00210465 | EC 1.14.13.72 | methylsterol monooxygenase | 0(0) | 2(2) | 2(2) | --- | 0.446/(1.155) | 0.7/(1.112) |
| IPI00190701 | Apolipoprotein E precursor (Apo-E) | 1(1) | 5(17) | 5(8) | --- | 1.307/(1.029) | 1.364/(1.042) | |
| IPI00194102 | EC 6.4.1.2 | acetyl-Co A carboxylase5 | 3(4) | 1(1) | 12(16) | 0.992/(1.07) | --- | 0.937/(1.102) |
| IPI00200661 | EC 2.3.1.85 | fatty acid synthase5 | 29(40) | 35(109) | 51(144) | 0.935/(1.219) | 0.959/(1.021) | 0.93/(1.164) |
A protein is regarded to be differentially expressed when the ratio of iTRAQ reporter ions 117.1/116.1 (i.e. addition of 24S-hydroxycholesterol in HPBCD vehicle / addition of HPBCD vehicle alone) is above 1.30 or below 0.83 (see Figure 3) i.e. proteins which show a significant (p<0.05) change in protein abundance. Table 1 includes all such proteins which were shown to be differentially expressed in at least two of the three biological replicates.
For each protein and biological replicate the number of peptides identified is given. Listed in parenthesis is the number of times these peptides were identified by the Mascot search procedure.
The geometric standard deviation is given in parenthesis.
In biological replicate 1, 2 and 3, 12, 22 and 28 fractions were collected from the cation-exchange column.
Examples of proteins not differentially expressed.
Protein identification and classification
In our initial study using on-line 2D-LC and a single injection equivalent to 10 μg of protein we were able to identify only 128 proteins. By performing multiple repeat injections on the 2D-LC system and performing DDA analysis over five different m/z ranges this number was increased to 686. As we are not sample limited, we decided to change to an off-line 2D-LC system where it was possible to inject the equivalent of 100 - 150 μg of protein onto the SCX column, manually collect 12 (exp 1), 22 (exp 2) or 28 (exp 3) fractions according to chromatographic peak elution, and inject aliquots of each of these fractions, after desalting, successively five times onto the RP column and perform DDA mass spectrometric analysis. In this manner 935 (exp 1), 1227 (exp 2) and 2071 (exp 3) proteins were identified and quantified. This lead to 693 proteins in common in the three replicates, with protein identification based on two peptides with scores above the identity threshold of 32 in each experiment. The peptide false positive identification were rate 1.63 (exp 1), 0.7 (exp 2) and 0.8% (exp 3). To further compare the results of the three experiments, and generate a final protein list, the reporting criteria were relaxed so that in cases where a protein had been identified by two peptides in one of the biological replicates, only a single peptide with score above the identity threshold was required to identify it in each of the other two replicates. This gave a final list of 1040 proteins reproducibly identified in all three replicates (see Table 1 in supplementary data).
Three biological replicates were carried out with analysis performed at least once by off-line 2D-LC-MS/MS for each replicate. Identified proteins were classified according to cellular location using Protein Centre software (Proxeon, Denmark) and are shown in Figure 1 of supplementary data.
Up- and down-regulated proteins
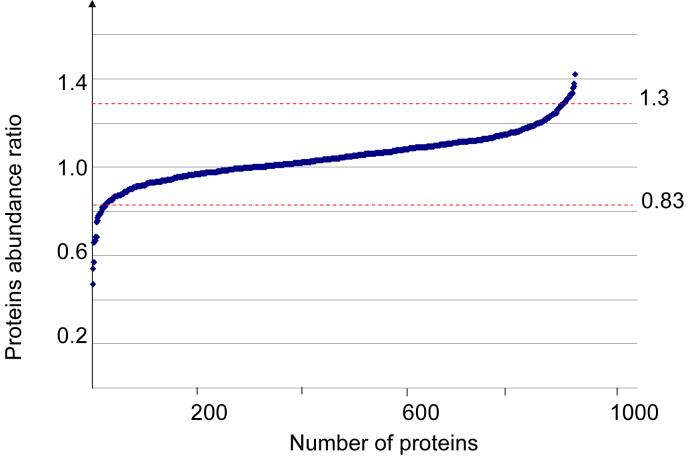
In this section only protein differentially expressed upon treatment with 24S-hydroxycholesterol in HPBCD as compared to treatment with HPBCD vehicle alone will be discussed. This is on account of the unsatisfactory addition of the sterol to cells in the absence of HPBCD (see Experimental). The expression of most proteins did not change upon the treatment of cells with 24S-hydroxycholesterol in HPBCD as compared to treatment with HPBCD vehicle alone (Figure 2). However, a small group of proteins did show a significant (p<0.05) change in protein abundance.
Figure 2.
iTRAQ determined change in protein abundance upon treatment of cortical neurons with 10 μM 24S-hydroxycholesterol in HPBCD vehicle as opposed to treatment of neurons with HPBCD vehicle alone. Protein abundance ratios were determined using Mascot quantification software as described in the text. The dashed lines represent thresholds for up- and down-regulated proteins. Individual protein ratios were converted to a log ratio and then for all proteins an overall mean ratio and standard deviation calculated. Ratios which fall outside two standard deviations from the mean are regarded as significant changes.
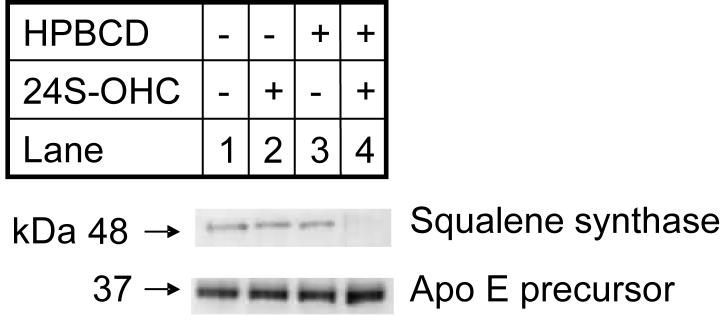
In each of our iTRAQ analysis HMG-CoA synthase 1 (EC 2.3.3.10) was found to be down-regulated by a factor of about two upon addition of 24S-hydroxycholesterol in HPBCD as compared to the effect of HPBCD vehicle alone (Table 1, Figure 3a). As HMG CoA synthase 1 is an enzyme of the cholesterol synthesis pathway (Figure 1), this alerted our attention to other enzymes involved in this pathway. The expression of enzymes in this pathway are regulated by SREBPs, which also regulate enzymes involved in the synthesis pathways of fatty acids and triglycerides1,2 [see Figure 2 in reference 2]. Table 1 shows all the proteins found to be differentially regulated in rat cortical neurons upon the addition of 24S-hydroxycholesterol in HPBCD as compared to the addition of HPBCD vehicle alone (p<0.05). The results from the triplicate analysis age given in the Table, where there is a requirement for differential expression in at least two of the three biological replicates. Listed in Table 1 of supplementary data are all proteins identified, including those whose expression did not change upon incubation with 24S-hydroxycholesterol. In contrast to enzymes of the cholesterol synthesis pathway, the apo E precursor protein was found to be up-regulated in cells treated with 24S-hydroxycholesterol (Figure 3b), the expression of apo E is regulated by the LXR transcription factor18. iTRAQ data for the expression of apo E precursor (a protein whose expression was up-regulated), and squalene synthase (a protein whose expression was down regulated), a representative member of the cholesterol synthesis pathway, were confirmed by immunoblot analysis (Figure 4).
Figure 4.
Immunoblot analysis of squalene synthase and apo E precursor in cortical neurons. Neurons were incubated for 24 hr in medium containing: 0.014% ethanol (v/v) (lane 1); 10 μM 24S-hydroxycholesterol (24S-OHC) in 0.014% ethanol (v/v) (lane 2); HPBCD 0.13% (w/v) (lane 3); and 10 μM 24S-hydroxycholesterol in HPBCD 0.13% (w/v) (lane 4).
Discussion
The results presented in Table 1 demonstrate that 24S-hydroxycholesterol down-regulates the expression of enzymes involved in the cholesterol synthesis pathway (Figure 1), while not effecting those in the fatty acid and triglyceride synthetic pathway (see Figure 2 in reference 2, and Table 2 in supplementary data). 24S-Hydroxycholesterol also up-regulates the expression of apo E, the cholesterol trafficking lipoprotein.
The results in primary cortical neurons are particularly relevant as cholesterol 24-hydroxylase (CYP46A1) is normally only expressed in neurons. Therefore, 24S-hydroxycholesterol is exclusively synthesised in these cells and its level is likely to reflect that of cholesterol itself. Furthermore, the pool of cholesterol in neurons has a high turnover rate which is in contrast to the bulk of brain cholesterol found in cells devoid of CYP46A120,25.
24S-Hydroxycholesterol is a ligand for the LXR32, the β form of which is expressed in brain24. Apo E is under transcriptional regulation of the LXR4, as is SREBP-1c, but not SERBP-1a or SREBP-219. Thus, the up-regulation of apo E in neurons observed when treated with 24S-hydroxycholesterol can be explained as a LXR regulated event (See Table 2 in supplementary data for other LXR target gene products). Further evidence for this was provided by iTRAQ experiments using the synthetic LXR ligand T1317 rather than 24S-hydroxycholesterol where the expression of apo E precursor was found to increase (data not shown). What then accounts for the down-regulation of enzymes in the cholesterol synthesis pathway? The expression of these enzymes is regulated by SREBPs 1a and 2. SREBPs are synthesised as inactive precursors bound to the ER and must be escorted to the golgi by SCAP to be proteolytically cleaved to nuclear SREBP (nSREBP), which translocates to the nucleus, where it activates transcription by binding to nonpalindromic sterol response elements (SREs) in the promoter regions of target genes2. When cholesterol builds up in ER membranes, the sterol binds to SCAP and triggers a conformational change that causes SCAP to bind to INSIGs, so SCAP can no longer escort SREBPs to the golgi for processing to their active form. Adams et al have demonstrated in Chinese hamster ovary (CHO) cells that 25-hydroxycholesterol also inhibits cholesterol synthesis via SREBPs15. While cholesterol has been shown in cross-linking experiments to bind directly to SCAP and trigger a conformational change, 25-hydroxycholesterol does not bind directly to SCAP. Instead, 25-hydroxycholesterol binds to INSIGs, causing INSIG binding to SCAP and anchoring of SREBPs in the ER6,7. We suggest that in neurons 24S-hydroxycholesterol behaves in a similar manner to 25-hydroxycholesterol in CHO cells, interacting with INSIG causing it to bind to SCAP and preventing SREBP-1a and 2 from reaching the golgi for processing to nSREBPs. Our data also suggests that 24S-hydroxycholesterol in neurons has a greater effect on SREBP-2 and -1a, than on -1c. This can be reconciled with the fact that SREBP-1c regulates enzymes involved in fatty acid synthesis, and that one way of removing excess free sterol is by ester formation with fatty acids mediated by the cholesterol trafficking protein apo E.
In conclusion, 24S-hydroxycholesterol behaves as a biologically active form of cholesterol in neurons, which when present in elevated levels leads to decreased cholesterol synthesis and increased cholesterol trafficking.
Supplementary Material
Acknowledgements
This work was supported by the UK Biotechnology and Biological Sciences Research Council (BBSRC grant no. BB/C515771/1, BB/C511356/1 and BB/C507237/1).
Footnotes
Supporting information available. This information is available free of charge via the Internet at http://pubs.acs.org.
Reference List
- (1).Goldstein JL, Bose-Boyd RA, Brown MS. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- (2).Horton JD, Goldstein JL, Brown MS. J. Clin. Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Lund EG, Xie C, Kotti T, Turley SD, Dietschy JM, Russell DW. J. Biol. Chem. 2003;278:22980–22988. doi: 10.1074/jbc.M303415200. [DOI] [PubMed] [Google Scholar]
- (4).Abildayeva K, Jansen PJ, Hirsch-Reinshagen V, Bloks VW, Bakker AH, Ramaekers FC, de VJ, Groen AK, Wellington CL, Kuipers F, Mulder M. J. Biol. Chem. 2006;281:12799–12808. doi: 10.1074/jbc.M601019200. [DOI] [PubMed] [Google Scholar]
- (5).Kotti TJ, Ramirez DM, Pfeiffer BE, Huber KM, Russell DW. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3869–3874. doi: 10.1073/pnas.0600316103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Radhakrishnan A, Ikeda Y, Kwon HJ, Brown MS, Goldstein JL. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6511–6518. doi: 10.1073/pnas.0700899104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Sun LP, Seemann J, Goldstein JL, Brown MS. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6519–6526. doi: 10.1073/pnas.0700907104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Ong SE, Mann M. Nat. Chem. Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- (9).Zhang Y, Zhang Y, Adachi J, Olsen JV, Shi R, de SG, Pasini E, Foster LJ, Macek B, Zougman A, Kumar C, Wisniewski JR, Jun W, Mann M. Nucleic Acids Res. 2007;35:D771–D779. doi: 10.1093/nar/gkl784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Ying W, Jiang Y, Guo L, Hao Y, Zhang Y, Wu S, Zhong F, Wang J, Shi R, Li D, Wan P, Li X, Wei H, Li J, Wang Z, Xue X, Cai Y, Zhu Y, Qian X, He F. Mol. Cell Proteomics. 2006;5:1703–1707. doi: 10.1074/mcp.M500344-MCP200. [DOI] [PubMed] [Google Scholar]
- (11).Forner F, Foster LJ, Campanaro S, Valle G, Mann M. Mol. Cell Proteomics. 2006;5:608–619. doi: 10.1074/mcp.M500298-MCP200. [DOI] [PubMed] [Google Scholar]
- (12).Lane CS, Wang Y, Betts R, Griffiths WJ, Patterson LH. Mol. Cell Proteomics. 2007;6:953–962. doi: 10.1074/mcp.M600296-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Yu LR, Conrads TP, Uo T, Kinoshita Y, Morrison RS, Lucas DA, Chan KC, Blonder J, Issaq HJ, Veenstra TD. Mol. Cell Proteomics. 2004;3:896–907. doi: 10.1074/mcp.M400034-MCP200. [DOI] [PubMed] [Google Scholar]
- (14).Yang JW, Rodrigo R, Felipo V, Lubec G. J. Proteome. Res. 2005;4:768–788. doi: 10.1021/pr049774v. [DOI] [PubMed] [Google Scholar]
- (15).Adams CM, Reitz J, De Brabander JK, Feramisco JD, Li L, Brown MS, Goldstein JL. J. Biol. Chem. 2004;279:52772–52780. doi: 10.1074/jbc.M410302200. [DOI] [PubMed] [Google Scholar]
- (16).Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- (17).Lehmann JM, Kliewer SA, Moore LB, Smith-Oliver TA, Oliver BB, Su JL, Sundseth SS, Winegar DA, Blanchard DE, Spencer TA, Willson TM. J. Biol. Chem. 1997;272:3137–3140. doi: 10.1074/jbc.272.6.3137. [DOI] [PubMed] [Google Scholar]
- (18).Liang Y, Lin S, Beyer TP, Zhang Y, Wu X, Bales KR, DeMattos RB, May PC, Li SD, Jiang XC, Eacho PI, Cao G, Paul SM. J. Neurochem. 2004;88:623–634. doi: 10.1111/j.1471-4159.2004.02183.x. [DOI] [PubMed] [Google Scholar]
- (19).Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Dietschy JM, Turley SD. J. Lipid Res. 2004;45:1375–1397. doi: 10.1194/jlr.R400004-JLR200. [DOI] [PubMed] [Google Scholar]
- (21).Lutjohann D. Acta Neurol. Scand. Suppl. 2006;185:33–42. doi: 10.1111/j.1600-0404.2006.00683.x. [DOI] [PubMed] [Google Scholar]
- (22).Karu K, Hornshaw M, Woffendin G, Bodin K, Hamberg M, Alvelius G, Sjovall J, Turton J, Wang Y, Griffiths WJ. J. Lipid Res. 2007;48:976–987. doi: 10.1194/jlr.M600497-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Lund EG, Guileyardo JM, Russell DW. Proc. Natl. Acad. Sci. U. S. A. 1999;96:7238–7243. doi: 10.1073/pnas.96.13.7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Teboul M, Enmark E, Li Q, Wikstrom AC, Pelto-Huikko M, Gustafsson JA. Proc. Natl. Acad. Sci. U. S. A. 1995;92:2096–2100. doi: 10.1073/pnas.92.6.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Bjorkhem I, Lutjohann D, Diczfalusy U, Stahle L, Ahlborg G, Wahren J. J. Lipid Res. 1998;39:1594–1600. [PubMed] [Google Scholar]
- (26).Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Mol. Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- (27).Jovanovic JN, Thomas P, Kittler JT, Smart TG, Moss SJ. J. Neurosci. 2004;24:522–530. doi: 10.1523/JNEUROSCI.3606-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Zhang J, Akwa Y, el-Etr M, Baulieu EE, Sjovall J. Biochem. J. 1997;322:175–84. doi: 10.1042/bj3220175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Zhang J, Larsson O, Sjovall J. Biochim. Biophys. Acta. 1995;1256:353–9. doi: 10.1016/0005-2760(95)00045-e. [DOI] [PubMed] [Google Scholar]
- (30).Yi EC, Marelli M, Lee H, Purvine SO, Aebersold R, Aitchison JD, Goodlett DR. Electrophoresis. 2002;23:3205–3216. doi: 10.1002/1522-2683(200209)23:18<3205::AID-ELPS3205>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- (31).Dumont D, Noben JP, Verhaert P, Stinissen P, Robben J. Proteomics. 2006;6:4967–4977. doi: 10.1002/pmic.200600080. [DOI] [PubMed] [Google Scholar]
- (32).Janowski BA, Grogan MJ, Jones SA, Wisely GB, Kliewer SA, Corey EJ, Mangelsdorf DJ. Proc. Natl. Acad. Sci. U. S. A. 1999;96:266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.