Abstract
The Quiescin-sulfhydryl oxidase (QSOX) family of flavoenzymes catalyzes the direct and facile insertion of disulfide bonds into unfolded reduced proteins with concomitant reduction of oxygen to hydrogen peroxide. This review discusses the chemical mechanism of these enzymes and the involvement of thioredoxin and flavin-binding domains in catalysis. The variability of CxxC motifs in the QSOX family is highlighted and attention is drawn to the steric factors that may promote efficient thiol/disulfide exchange during oxidative protein folding. The varied cellular location of these multi-domain sulfhydryl oxidases is reviewed and potential intracellular and extracellular roles are summarized. Finally, this review identifies important unresolved questions concerning this ancient family of sulfhydryl oxidases.
Keywords: Sulfhydryl oxidase, QSOX, oxidative protein folding, disulfide exchange, protein disulfide isomerase
1. Introduction to sulfhydryl oxidases
The name “sulfhydryl oxidase” was first coined by Kiermeier and coworkers [1, 2] to describe an activity in fresh milk that counteracts the undesirable “burnt” flavors associated with ultra-high temperature pasteurization [3]. This reaction involves the generation of disulfide bonds between cysteine residues of denatured globular proteins [4]:
| (1) |
This first mammalian sulfhydryl oxidase has been extensively studied by Swaisgood and coworkers [3–7] and is reported to be an iron-dependent enzyme capable of oxidizing both glutathione and protein thiol groups. Subsequently, a number of metalloenzyme sulfhydryl oxidases have been shown to contain either iron [4, 8, 9] or copper [10–12]. Although they have been frequently suggested as participants in mammalian disulfide bond generation [4, 9, 12–14], these enzymes remain poorly understood without published sequences, descriptions of metal coordination environment, or crystal structures.
In contrast, the last decade has witnessed a growing interest in sulfhydryl oxidases that utilize flavin as an essential redox-active cofactor. Table 1 lists representatives of selected eukaryotic flavoprotein sulfhydryl oxidases in the approximate order that they were found to be flavin-linked. The first of them was isolated from mammalian seminal vesicles by Ostrowski and Kistler in 1979 [15]. Next came reports on glutathione oxidases from Pencillium species [16] and from Aspergillus niger [17]. These two enzymes are evolutionarily distinct from all of the other flavin-linked enzymes in Table 1 [18–20]: indeed their closest relatives appear to be thioredoxin reductase-like members of the pyridine nucleotide disulfide oxidoreductase family [21, 22]. The physiological roles of these fungal enzymes are unclear [20], and they will not be discussed further in this review.
Table 1.
Selected flavoprotein sulfhydryl oxidases
| Enzyme | Source | Flavin found | reference |
|---|---|---|---|
| Sulfhydryl oxidase | Seminal vesicles | 1979 | [107] |
| Glutathione oxidase | Penicillium spp. | 1982 | [16] |
| Aspergillus niger | 1987 | [17] | |
| Sulfhydryl oxidase | Avian egg white | 1996 | [23] |
| Erv1p | Yeast | 2000 | [108] |
| ALR | Rat & human | 2001 | [109] |
| Erv2p | Yeast | 2001 | [110, 111] |
| Ero1p | Yeast | 2000 | [31] |
| 2004 | [32] |
The avian egg white oxidase listed in Table 1 was found serendipitously in 1996 [23] and subsequently shown to be homologous with both the rat seminal vesicle flavoprotein [18, 24] and a series of human growth factors including Quiescin Q6 [25], bone-derived growth factor [26], cell growth inhibitory factor [27] and placental-derived prostate growth factor [28]. Enzymes of this family have come to be called Quiescin-sulfhydryl oxidases (here abbreviated QSOX). QSOX family members contain one or more thioredoxin domains and a highly helical flavin-binding domain (abbreviated here as Erv/ALR). In addition, this Erv/ALR fold [29, 30] is employed in a number of small stand-alone eukaryotic sulfhydryl oxidases (e.g. yeast Erv1p, its mammalian counterpart augmenter of liver regeneration (ALR), and yeast Erv2p; Table 1). At least one of these flavoproteins has been found in the genomes of almost every eukaryote so far examined, together with Ero1, an evolutionarily distant or unrelated flavoprotein sulfhydryl oxidase [31, 32]. These important flavin-linked oxidases are covered in separate reviews in this thematic issue, and so will only be addressed peripherally here.
2. Flavoprotein candidates for oxidative protein folding in higher eukaryotes
Two distinct flavoprotein oxidases have been proposed to play roles in the secretory systems of higher eukaryotes. The Ero1 proteins are believed to be major net generators of disulfides in the lumen of the ER [31, 33–38]. These ER-resident enzymes are proposed to regenerate PDI after it has served as the immediate oxidant of client proteins as shown in Fig. 1. This “PDI first” model of oxidative protein folding raises interesting issues of mechanism and regulation [39] that cannot be addressed here. In contrast to the Ero1 proteins, QSOXs apparently function in reverse. Their direct substrates are unfolded reduced proteins, and when disulfide bonds begin to accumulate, reduced PDI isomerizes those that are inserted incorrectly (see below). While Fig. 1 is a useful summary of current ideas of oxidative protein folding these models may require significant revision as they are further examined in vivo and in vitro. Finally, it should be acknowledged that there may be pathways for the net generation of disulfides that remain unrecognized or underappreciated [19].
Fig. 1.
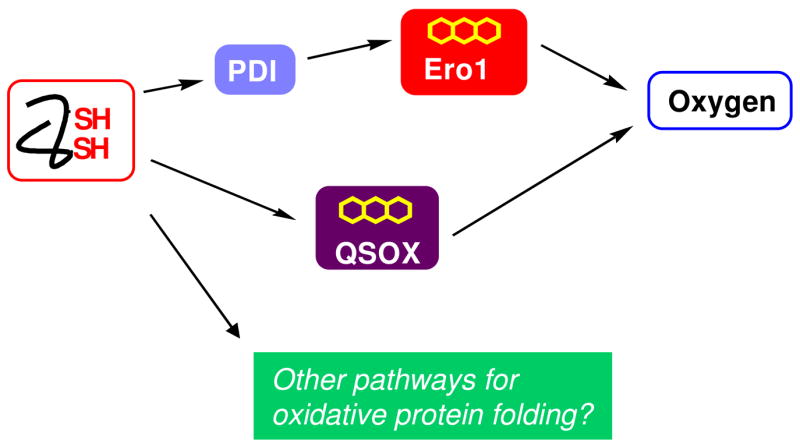
Pathways illustrating the flow of reducing equivalents during the net generation of structural disulfide bonds in metazoans. The Ero1 proteins are believed to accept reducing equivalents from PDI or PDI-like proteins as shown. In contrast QSOX may oxidize client reduced proteins directly. Additional pathways for net disulfide generation may exist. The arrows represent the flow of pairs of reducing equivalents formed during the generation of disulfides.
3. QSOX phylogenetics
It is interesting that the presence of QSOX and Erv2p in eukaryotes seems to be mutually exclusive [19, 20]. While a detailed phylogenetic analysis of the occurrence of flavin-dependent oxidases in sequenced genomes is beyond the scope of this review, a few general observations are pertinent. Erv2p, but not QSOX, is found in widely divergent fungi including Saccharomyces cerevisiae, Aspergillus nidulans, Neurospora crassa, and Encephalitozoon cuniculi [20]. In contrast QSOXs, but not Erv2p, are found in metazoans, plants and certain protists [19, 20, 39]. Gene duplication results in multiple paralogs of QSOX in metazoans (four in Drosophila melanogaster, three in Caenorhabditis elegans, and two in vertebrates and green plants [19, 20]).
The appearance of QSOX predates the emergence of multicellularity since the fusion of thioredoxin and Erv/ALR domains occurred before the divergence of yeast/fungi from other eukaryotes [19, 39]. A QSOX sequence is found in the smallest free-living unicellular eukaryote (a marine algae; Ostreococcus taurii) [40]. Additionally, a number of unicellular pathogenic protists (e.g. Plasmodium falciparum, Trypanosoma brucei, Leishmania major, and Cryptosporidium parvum) also contain a single QSOX gene. QSOXs from unicellular organisms might be involved in the elaboration of cell-surface proteins but nothing is known about their actual roles in vivo or their enzymological properties in vitro.
4. Models for sulfhydryl oxidase catalysis
Despite the apparent structural diversity of flavoprotein sulfhydryl oxidases, they show a remarkable convergence of mechanism [20, 29, 32, 39]. All utilize a flavin ring (contributed from FAD) in intimate redox communication with an active site disulfide bond (herein termed the proximal disulfide) as depicted in Fig. 2 (form 1). The communication between disulfide and flavin has been thoroughly explored for the pyridine nucleotide disulfide oxidoreductases [21, 22] and these insights continue to provide useful precedents for the mechanism of the sulfhydryl oxidases. A catalytic turnover for a prototypical sulfhydryl oxidase would commence by reduction of the proximal disulfide (either directly by the substrate (forms 2–5), or indirectly via a series of disulfide exchanges that bring reducing equivalents from a more distant locus).
Fig. 2.
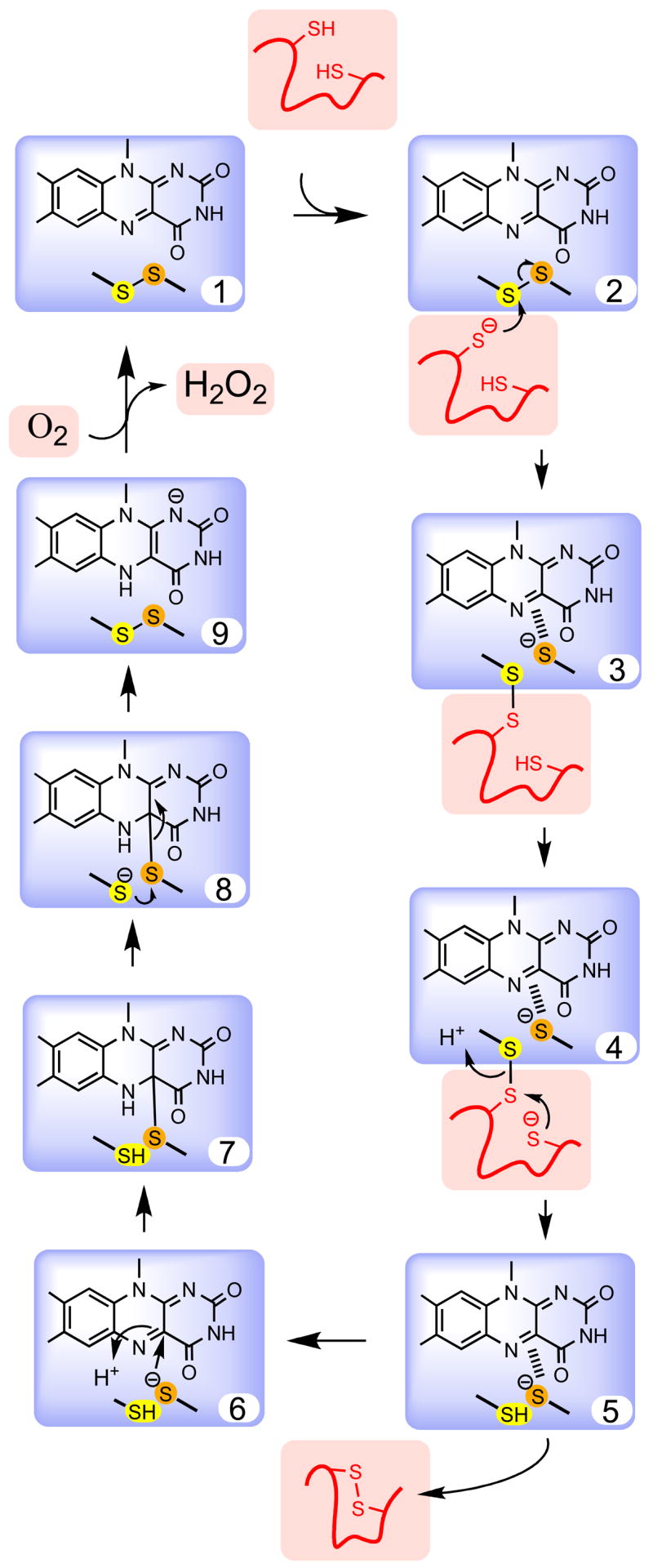
Steps in the mechanism of a hypothetical flavin-dependent sulfhydryl oxidase. The flavin and redox active disulfide common to all sulfhydryl oxidases is shown within the blue box. The reducing substrate (here a protein dithiol) and molecular oxygen are shown in salmon. Key steps in the mechanism are selected for emphasis and some of the deprotonation events are not shown explicitly.
The sulfur atoms of the proximal disulfide would be expected to have distinct functions. The outer “interchange” [21, 22] cysteine depicted in yellow in Fig. 2 forms mixed disulfides with incoming substrates (e.g. form 3). The inner “charge-transfer” sulfur (orange) interacts primarily with the flavin ring, frequently with the formation of a charge-transfer interaction [21–23, 41, 42] between the electron-rich thiolate and the electron-deficient flavin (depicted by the hatched lines in forms 3–5). Next, this thiolate attacks the flavin at the C4a position generating a transitory covalent adduct (form 7) [21, 43–46]. This adduct can then resolve, regenerating the proximal disulfide and forming the reduced flavin. Form 9 is thus the end of the reductive half-reaction. Finally, form 1 is regenerated in the oxidative half-reaction with the net 2-electron reduction of a molecule of oxygen yielding hydrogen peroxide. While this reoxidation step has been shown to proceed via the formation of a C4a-hydroperoxy adduct in certain flavoenzymes, it is not clear whether such a covalent species is formed in the sulfhydryl oxidases, or exactly how this formally spin-forbidden reaction occurs [47, 48]. Fass and coworkers have identified a potential site by which oxygen might approach the flavin in Erv2p [49].
Thus, flavins are well suited to serve as redox cofactors for disulfide generation because they can be readily reduced by dithiols and facilely reoxidized by oxygen. Enzymes exploiting this chemistry are true oxidases because they catalyze the oxidation of their substrates with the direct consumption of molecular oxygen [39]. However many reduced flavoproteins can utilize a range of alternate 1- and 2-electron acceptors in vitro suggesting that disulfide bond generation in eukaryotes might not always utilize oxygen as an immediate co-substrate. A particularly interesting example is raised by Herrmann and colleagues in this series. In that case, yeast Erv1 and its mammalian counterpart ALR, can generate disulfide bonds using cytochrome c as an effective terminal oxidant as suggested in vitro [50] and in vivo [51, 52].
Could the catalytic scheme in Fig. 2 be simplified, by dispensing with the proximal disulfide and using substrate thiol(ate)s to directly attack the flavin? In this mode the orange sulfur atom in form 6 would be replaced with the substrate thiolate in form 2. However such direct interactions would require that C4a flavin adducts be generated with a huge number of potential cysteinyl thiols. This may entail significant steric constraints when the substrates are large. In fact all sulfhydryl oxidases appear to channel reducing equivalent to the flavin by way of a common optimally-placed proximal disulfide bond (as shown in Fig. 2). Further, many of them employ an additional “distal” or “shuttle” disulfide on a mobile element of secondary structure that further separates the substrate from the flavin [29, 32, 49]. Distal and proximal disulfides are contained within distinct domains in QSOX proteins (see below).
One of the reasons for these apparently redundant multiple disulfide exchange steps may be to move the most sterically demanding interactions to flexible distal centers [29, 32, 49, 53, 54]. While not widely discussed, disulfide exchange steps, reactions that are central to both the oxidative and isomerization phases of oxidative protein folding, are subject to significant steric constraints [55, 56]. As a simple illustration Fig. 3 shows a mixed disulfide intermediate A–B formed after the attack of a dithiol-containing protein B on a disulfide-containing protein A. The subsequent resolution of this disulfide, yielding reduced A and oxidized B, is favored when the thiolate nucleophile in B attacks along the S-S axis of the A–B disulfide [55, 56]. Placement of this “resolving” thiolate in an unfavorable non-linear geometry (e.g. as depicted in the lower panel of Fig. 3) would then require sufficient protein flexibility in B (and/or in A) to allow the reacting sulfur atoms to attain approximate co-linearity. If this reaction cannot be accomplished efficiently, the mixed disulfide may simply reverse in favor of the reactants (oxidized A and reduced B).
Fig. 3.
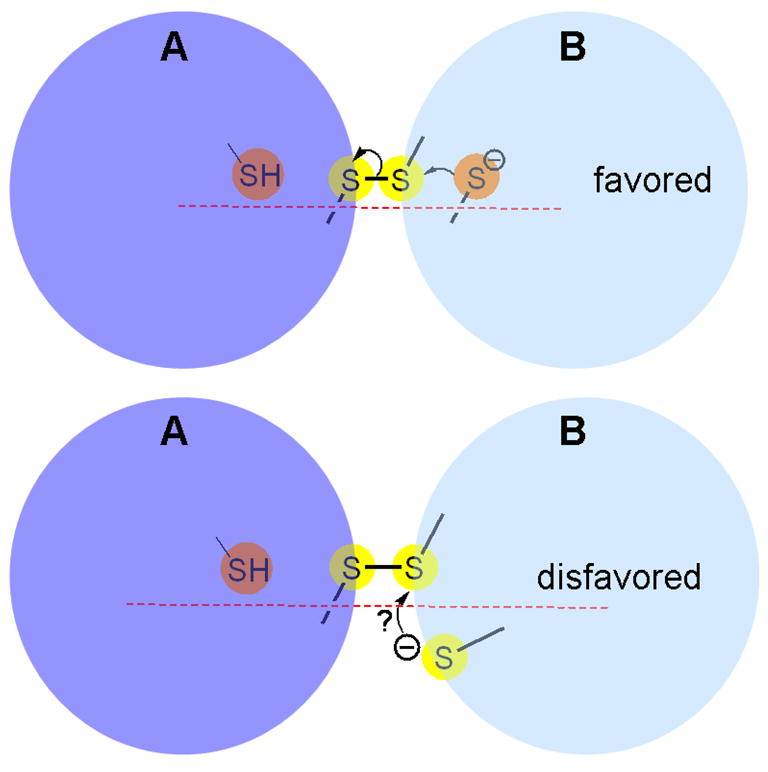
A representation of favored and disfavored orientation for disulfide exchange. Top and bottom panels depict reactions involving a mixed disulfide between proteins A and B. Surface accessible and buried sulfur atoms are shown as yellow and orange spheres, respectively.
5. Domain structure and catalysis in QSOX enzymes
Fig. 4 shows the amino acid sequence of the long form of human QSOX1 with key features highlighted in color. Two thioredoxin domains (Trx1 and 2) follow an N-terminal signal sequence. Trx1 contains a CxxC motif (hereafter CxxC-trx) with the sequence CGHC that is frequently associated with eukaryotic protein disulfide isomerases (PDI; see below [57–60]). Partial proteolysis studies of avian QSOX showed that this domain is crucial for the rapid oxidation of unfolded protein substrates [61]. These experiments also suggested that it is this CxxC-trx motif that is first reduced by protein substrates and subsequently passes reducing equivalents to the Erv/ALR domain [61]. Homology models of both these catalytic domains, together with an indication of the flow of reducing equivalents between them, are shown in Fig. 5.
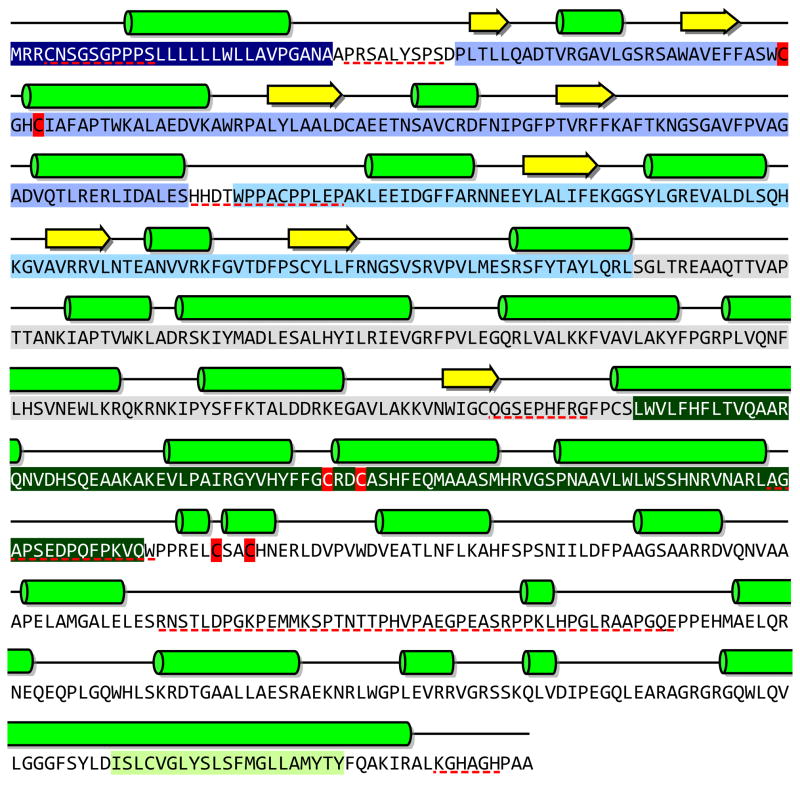
Fig. 4.
Amino acid sequence, secondary structure and domain organization of the long form of human QSOX1 (NP_002817). The three CxxC motifs are shown in red (CxxC-trx, CxxC-erv, and CxxC-trm). The predicted signal sequence (navy blue), and Trx1 (blue), Trx2 (light blue), spacer (grey), and Erv/ALR (dark green) domains, and the transmembrane span (light green) are highlighted. Helices and strands are shown by bright green cylinders and yellow arrows respectively. Predicted disordered regions are underlined in dashed red [112–114].
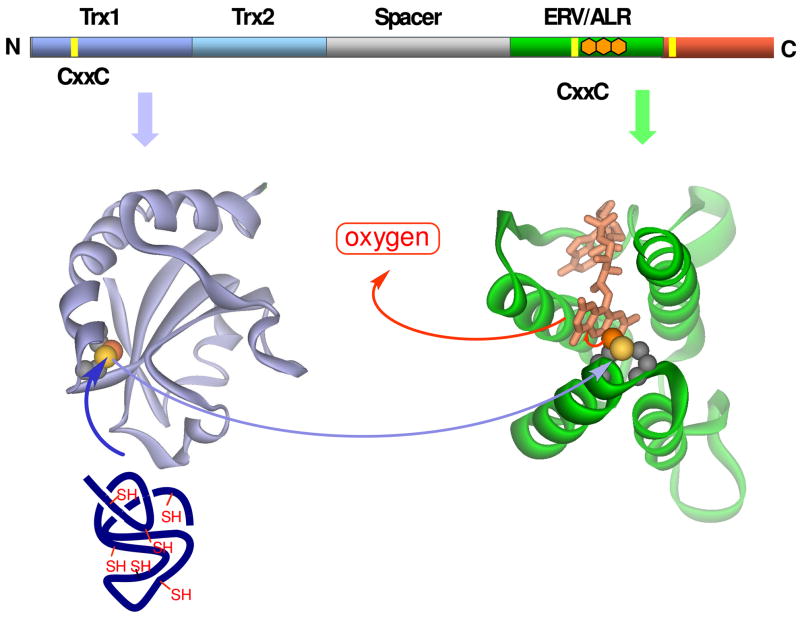
Fig. 5.
The flow of reducing equivalents in a monomeric QSOX. Homology models of the Trx1 and Erv/ALR domains were constructed using the crystal structures of yeast PDI1 a domain [59] and yeast Erv2p [29] respectively. Surface accessible and buried sulfur atoms are shown as yellow and orange spheres, respectively.
The second thioredoxin module in metazoan QSOXs is not identified by standard sequence analysis programs: it contains the same alternation of secondary structural elements found for Trx1 but omits the first short beta-strand (Fig. 4). This Trx2 domain lacks a CxxC motif and is fused to a third “spacer” domain of about 100 amino acids. Database searches with this spacer sequence do not reveal compelling non-QSOX matches, but secondary structure prediction programs suggest that it is substantially helical (Fig. 4). The final domain in QSOXs is the Erv/ALR module whose novel flavin-binding fold, first established by Fass and coworkers for yeast Erv2p [29], serves as the model for the corresponding domain in QSOX (Fig. 5). The isoalloxazine ring is inserted into the mouth of a four-helix bundle with the proximal disulfide situated at a turn between helices 2 and 3. The interchange sulfur is solvent exposed whereas the charge-transfer sulfur is buried and almost touching the C4a position of the flavin. Overall, the placement of both interchange and charge transfer sulfur atoms seem perfectly adapted for the reactions depicted in Fig. 2: for the accommodation of a mixed disulfide intermediate with external protein reductants, and for the subsequent formation and resolution of the C4a flavin adduct.
Domain motions are likely to be an important feature of catalysis in QSOX: the CxxC-trx motif must alternately form covalent intermediates between potentially large protein substrates and the Erv/ALR domain (Fig. 5). The observation that a relatively slow (3/sec at 4 °C) protein isomerization event limits oxidation of thiol substrates by the avian enzyme [41] is certainly consistent with these expectations. Such motions might be accommodated via the disordered regions that are predicted to flank both the N- and C-termini of Trx1 and Erv1/ALR domains of QSOX (Fig. 4).
The catalytic model in Fig. 5 envisages critical roles for two CxxC motifs, but it omits a third CxxC feature which is conserved in every metazoan, plant and protist QSOX sequence examined to date (see below). This feature is located at a comparable position to a CxC motif believed to act as the mobile shuttle disulfide in Erv2p [29, 49]. This analogy, together with the finding that avian QSOX can accept a total of 8-electrons under forcing conditions (three disulfides and one flavin), prompted our earlier proposal that this C-terminal CxxC motif plays a comparable shuttle role in QSOX [61]. Surprisingly, recent site-directed mutagenesis experiments with human QSOX1, demonstrate that this motif is not essential for the oxidation of the reduced forms of glutathione, dithiothreitol, or RNase (Heckler, E., Alon, A., Fass, D., and Thorpe, C., unpublished observations). The roles of this conserved CxxC motif are currently unknown.
While metazoan QSOXs contain two thioredoxin domains [20, 61], plants and protists lack a complete Trx2 domain but retain the spacer and Erv/ALR domains [20]. All QSOX sequences so far examined share a rather divergent and partly unstructured C-terminal sequence of amino acids which often ends with a single transmembrane span [20]. Differential splicing leads to two forms of human QSOX. The longer (747 residues) form retains the membrane span, and is likely to be anchored at a membrane surface, or subsequently freed from its attachment by proteolysis. The shorter form contains 604 residues and lacks the transmembrane helix. A priori one would not expect long and short forms of QSOX to differ in their intrinsic catalytic specificity because both retain the same core catalytic domains. Conventional C-terminal ER retention sequences are absent in QSOX.
6. The variability of CxxC sequences in QSOX
There has been considerable interest in the influence of the intervening residues on the redox potential and reactivity of CxxC motifs in DsbA, PDI and thioredoxin, [62–66]. As mentioned above, the CxxC-trx motif occurs at the junction between a loop and a helix. Interestingly, the CxxC-erv motif is also placed at a loop-helix junction, but occurs within the context of a 4 helix-bundle (Fig. 5). The third CxxC (CxxC-trm) is located immediately C-terminal to the Erv/ALR domain (Fig. 5, top) in what may be a flexible region of the protein. Although the role CxxC-trm is now uncertain, we have included this conserved feature in the following sequence analysis.
Fig. 6 is a compilation based on the translated sequences from 72 unique QSOX genes. By far the most conserved are those for CxxC-trx: those of CxxC-erv and CxxC-trm are much more variable. Thus the most prevalent CxxC-trx sequence is CGHC: a motif widely found in eukaryotic PDIs [57, 58, 60]. Since the CxxC-trx and CxxC-erv motifs are links in a chain of redox centers between reduced protein client and molecular oxygen (Fig. 5), one might expect a significant degree of mutual sequence conservation in these motifs. Contrary to this prediction, CxxC-erv is highly variable, with 29 different combinations of the intervening dipeptide among the sequences examined (Fig. 6, middle panel). The intervening xx dipeptide is generally polar: 80% of these sequences contain at least one charged amino acid and about 50% of the animal sequences have two oppositely charged residues (KE, RD, RE or EH). All plant CxxC-erv sequences examined comprise negatively charged residues (DD, EE, DE, ED). It will be interesting to learn what factors direct the relative conservation of the CxxC-trx motif compared to the diversity of CxxC-erv sequences. It seems paradoxical that the former likely interacts with a multiplicity of client peptide and proteins [20, 67, 68], whereas the more variable CxxC-erv motif may have just two partners: the reduced thioredoxin domain and the oxidized flavin prosthetic group (Fig. 5).
Fig. 6.
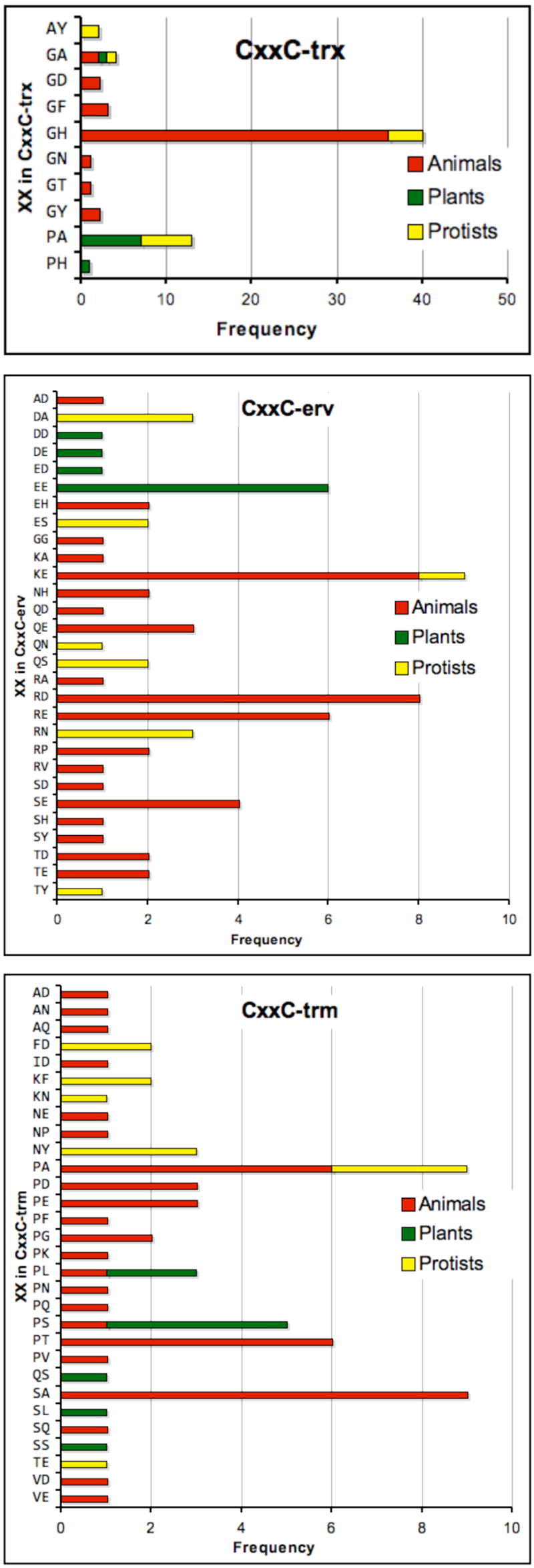
Sequence variability within the three CxxC motifs of QSOXs of animals, plants and protists. A total of 72 unique QSOX gene products (omitting differentially spliced products) are color coded by animals (red), plants and algae (green), and protists (yellow). Currently animals are overrepresented in the available sequences (comprising about 50% of the total).
Finally, CxxC-trm sequences are also highly variable with 30 different intervening dipeptides. There appears no clear correlation between sequence and organism type. Two modest generalities emerge from the CxxC-trm sequence comparisons in Fig. 6: greater than half of X1 residues are proline and about 2/3 of X1 and X2 residues are neutral.
7. Catalytic specificity of vertebrate QSOX enzymes
QSOX enzymes are the only flavoprotein sulfhydryl oxidases described to date that are capable of a direct and facile formation of disulfide bonds in unfolded reduced proteins [20, 39, 67]. They remain the only flavin-linked sulfhydryl oxidases for which catalytic efficiencies (kcat/KM values) have been measured for their presumed cognate substrates [23, 41, 67].
Steady state kinetic parameters for the avian, bovine (Jaje, J., and Thorpe, C., unpublished data) and human QSOX1 (Heckler, E., Alon, A., Fass, D, and Thorpe, C., unpublished observations) enzymes are comparable: averaging about 1000 disulfide bonds inserted into unfolded reduced proteins per minute with KM values of about 150 μM per thiol residue. Hence the insights gained from mechanistic studies of the avian enzyme are likely to be generally applicable to other vertebrate QSOXs.
Typically, “unfolded substrates” for QSOX are obtained by exhaustive reduction of secreted proteins (such as RNase or lysozyme) under denaturing conditions followed by the removal of excess reductant [41, 67]. These reduced proteins, freed from the restraints of their native disulfide pairings, assume a somewhat expanded molten-globule-like state that allows rapid access of their cysteine residues to small oxidants like 5,5′ dithiobis(2-nitrobenzoate) and, evidently, to the catalytic apparatus of the QSOX enzymes (see above). However, avian QSOX cannot reach buried thiols in folded proteins structures: these cysteines remain unreactive to QSOX unless they are previously exposed by denaturation [67]. It is not yet clear whether QSOX can efficiently oxidize vicinal cysteine residues when they are located at the surface of a well-ordered native folded structure, or whether it can effectively catalyze intersubunit or interprotein disulfide bonds. In these cases the steric constraints on disulfide exchange reactions that were mentioned earlier may assume great significance.
The avian, bovine and human QSOX1 enzymes are all rather weakly reactive towards reduced GSH [23, 41, 67]. Here kcat/KM values are some 100-fold lower than those shown by a typical unfolded substrate. By this measure, unfolded proteins are clearly preferred substrates over glutathione. However the effective concentrations of these potentially competing substrates are unknown in the several intra- and extra-cellular locations in which QSOX has been found (see below).
8. QSOX cellular locations and potential intracellular substrates
Immunohistochemical localization shows that QSOXs have been found in a variety of cellular locations including the ER [19, 69], Golgi [19, 68–70], secretory granules [69, 70], and at the nuclear and plasma membrane surfaces [71]. A V5 tagged construct of the long form, HsQSOX1a, is found to concentrate in the Golgi of Chinese hamster ovary cells [68]. Short forms of QSOX, either the products of differential splicing or possibly of proteolysis, transit the secretory apparatus as they exit the cell (there are no obvious retention sequences in this form). Further, glycosylation of the QSOX short form, HsQSOX1b, is not required for enzymatic activity (Heckler, E., Alon, A., Fass, D., and Thorpe, C., unpublished observations). An important unresolved issue [68] is whether these shorter forms contribute to disulfide generation in the ER. Can QSOX serve multiple functions: first contributing to net disulfide bond generation within the ER, and then playing additional roles after secretion? Before this suggestion can be dismissed, one needs to ensure that the localization methods for QSOX are of sufficient sensitivity to detect what may be very low steady-state concentrations of QSOX in the ER. High concentrations of QSOX might not be needed here: nanomolar levels of QSOX rapidly insert disulfides into micromolar levels of reduced proteins [67].
It should be stressed that there is currently no direct evidence that the short forms of QSOX are important players in the net generation of disulfide bonds in the ER. Nevertheless, if active QSOX is present, what potential thiol substrates would it encounter? First, the general permeability of the ER membrane to GSH [72, 73] suggests that this tripeptide will be an abundant thiol species within the ER lumen. However, the relatively high KM for GSH (20 mM for the avian enzyme), and its 100-fold lower kcat/KM value compared to the cysteine residues of unfolded reduced proteins, suggest that GSH is not the preferred substrate of QSOX within the ER [39, 67]. A second category of potential substrates are the PDIs (and PDI-like proteins). In aggregate the PDIs probably contribute mM levels of redox-active thiols to the mammalian ER; presumably maintained largely in their reduced states by rapid equilibration with the prevailing GSH/GSSG levels [74, 75]:
| (2) |
However, reduced PDI (up to 1 mM CxxC thiols) is not a significant substrate of QSOX (Rancy, P., Winther, J.R., and Thorpe, C., unpublished observations). This notable unreactivity may partly reflect steric arguments (see above) and the fact that reduced mammalian PDI is a relatively weak thermodynamic reductant compared to an average pair of juxtaposed thiols in an unfolded protein [76].
QSOX is likely to be highly active towards a third class of potential substrates: the nascent protein chains extruded into the ER [67]. In vitro experiments demonstrate a facile cooperation between QSOX and PDI in introducing and isomerizing disulfides in a range of protein clients. In the initial work [67], catalytic levels of PDI were used with QSOX to effect the rapid oxidative refolding of reduced pancreatic RNase. Here 1 mM GSH was included to maintain PDI in its reduced state. However glutathione is unnecessary when concentrations of reduced PDI are used that approach the levels believed to be present in the mammalian ER [36, 58, 77]. With this simple system (1–100 nM QSOX, 30–200 μM reduced PDI and low micromolar concentrations of reduced client protein), QSOX is able to selectively oxidize a range of unfolded proteins while reduced PDI efficiently isomerizes incorrect pairings as they arise (Rancy, P., Winther, J.R., and Thorpe, C., unpublished observations). These in vitro studies suggest that if QSOX were active in the ER it could selectively introduce disulfide bonds in unfolded proteins without significantly changing the PDI or glutathione redox poise [39].
9. QSOX tissue distribution
The most extensive work on the physiological locations and roles of the QSOXs has focused on the rat. Immunohistochemistry of rat QSOX showed that expression is seen in the immune, reproductive, respiratory, and digestive systems along with the retina and skin [78]. Specifically, expression is highest in various secretory endocrine glands (e.g. hypothalamus, pituitary, pineal, and adrenal) and in the pancreatic islets of Langerhans; secretory and epithelial cells of the trachea, stomach, small intestine, salivary glands, esophagus and lung; and in the reproductive organs (testis, seminal vesicles, vagina, and ovary) [78]. The expression pattern of rat QSOX is consistent with data from guinea pig and human QSOX [19, 20, 39, 79].
Extensive studies on the distribution and ontogenesis of QSOX in rodent brain have shown specific expression in many locales, especially those dealing with motor and sensory aspects and with hormone secretion [69, 70, 78, 80–83]. These investigations suggest roles for QSOX in the secretion, maintenance, and maturation of a wide range of disulfide containing peptides and proteins as well as in neuronal maturation and synaptic strengthening [70, 82, 83]. QSOX may also contribute to redox cell signaling in the brain via the generation of hydrogen peroxide [70], which might play a role in a variety of signaling cascades both within [84] and outside the brain [85].
Musard and coworkers report that guinea pig QSOX is involved in negative cell cycle control and is down-regulated by estrogen in the endometrium [79]. A similar effect of estrogen on QSOX expression is seen in breast cancer cells [86]. Tury et al. observed changes in QSOX levels during the rat estrus cycle, but determined that estrogen up-regulated QSOX expression in the pituitary of ovary-less rats [69]. These divergent results may be due to tissue or species differences, or reflect the complexity of estrogen action [69]. High levels of expression of QSOX in the rat hypothalamic-pituitary neuroendocrine axis were described by Mairet-Coello et al. and suggest a role for QSOX in the redox regulation of hormone action [70]. QSOX and disulfide-containing hormones, especially luteinizing and follicle stimulating hormones, co-localize to the same neuronal clusters in rat brain [69, 70]. Interestingly, QSOX is also up-regulated in pituitary tumors from mice overexpressing luteinizing hormone [87].
10. QSOX2
Database profiles for the abundance of QSOX1 and QSOX2 in a range of tissues show that the former appears to be usually several-fold more abundant than the latter [20]. HsQSOX2 shows 37% identify with its more abundant paralog and shares all of the key features of the sequence depicted in Fig. 4. QSOX2 has yet to be characterized enzymatically and is therefore not given extensive coverage here. There are, however, two interesting aspects of mammalian QSOX2 that deserve comment.
First, Schwab and coworkers identified QSOX2 in a screen for genes that maintain sensitivity to proapoptotic stimuli in neuroblastoma cells [71]. They suggested that QSOX2 is a positive mediator of programmed cell death [71]. Second, examination of the expression of QSOX2 during mouse development shows that by far the highest abundance of transcripts occurs in the oocyte [88]. This level drops precipitously (about 100-fold) seemingly following fertilization. Conceivably a sulfhydryl oxidase may be involved in the deployment of protein disulfide bonds that accompanies the hardening of the oocyte extracellular matrix during the slow block to polyspermy [89, 90].
11. QSOX1 and growth regulation
Human QSOX1 was first identified as a gene product (Quiescin Q6) that was strongly up-regulated when fibroblasts approach confluence [25, 91]. Several other growth-regulating proteins with names suggestive of significant cellular roles appear to be QSOX1 [19, 20]: bone-derived growth factor [26], cell growth inhibiting factor [27], placental derived prostrate growth factor [92], and erythroid cell stimulating factor [93]. A molecular explanation of the effects of QSOX in any of these processes has yet to appear. Some of these effects may involve the proposed role of QSOX in the fabrication/remodelling of the extracellular matrix [19]. Attention has already been drawn to the correlated expression between QSOX1 and a number of proteins of the extracellular matrix (including collagen IV α1 and lysyl oxidase) in human cancer cell lines [20, 39].
12. QSOX secreted from cells
QSOX1 was first found in rat seminal vesicles [15, 24] and then as a secreted product of the avian oviduct [23]. A particularly interesting observation is the presence of appreciable levels of QSOX in mammalian sera [94, 95]. Recently QSOX1 has been isolated from bovine milk (Jaje, J., and Thorpe, C., unpublished data). QSOX1 is also released from a range of cell types in tissue culture including human embryo lung fibroblasts particularly as they approach confluence [25, 91], from Chinese hamster ovary cells [24], from prostrate cancer cells [92, 96], breast cancer cell lines [97], and osteosarcoma cells [92].
A variety of hypotheses have been proposed to address why QSOX enzymes are released from cells. These include: to generate hydrogen peroxide as part of an antimicrobial system [15] perhaps driven by the export of reduced glutathione [20]; for hydrogen peroxide signaling in the brain [70]; in the fabrication of disulfide bridged structures that are too bulky to be assembled intracellularly [20], to continue protein folding outside the cell [78]; to counteract the effects of extracellular reductants [68], and in the preservation of sperm membrane integrity [15, 24]. Several of these proposed functions would be expected to modulate extracellular thiol/disulfide redox poise and therefore may affect a range of important redox-linked processes. These include: platelet activation [98, 99], cellular adhesion [100], viral fusion [99, 101], cellular proliferation [102, 103], tumor immune evasion [104] and metastatic potential [105, 106]. In these contexts the reported presence of long-form QSOX enzymes at the cell surface is intriguing.
13. Some outstanding questions
Despite a number of important recent contributions to the QSOX literature, many fundamental questions remain to be addressed definitively. For example, do the physiological functions of QSOX reflect their impressive abilities to insert disulfide bonds in unfolded proteins? Can QSOX isoforms generate disulfides in the ER, or would their activities be deleterious at that location [68]? What roles do secreted QSOX proteins assume in the extracellular matrix and why have they been reported to be mammalian growth factors? Do mammalian QSOX1 and QSOX2 have fundamentally different cellular roles? Finally, what are the outcomes of QSOX knockouts in the mouse? Hopefully the next few years will reveal a clearer understanding of the mechanisms and physiological roles of these enigmatic flavin-dependent sulfhydryl oxidases.
Acknowledgments
The authors’ laboratory is supported by NIH grant GM26643. P.C.R. was supported, in part, by NIH Training Grant T32GM08550.
Abbreviations
- ALR
augmenter of liver regeneration
- ER
endoplasmic reticulum
- Ero1
endoplasmic reticulum oxidoreductin 1
- Erv
a protein essential for respiration and vegetative growth
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- PDI
protein disulfide isomerase
- QSOX
quiescin-sulfhydryl oxidase
- Trx
thioredoxin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kiermeier F, Ranfft K. About Some Characteristics of Sulfhydryloxidase in Milk. Z Lebensm-Unters-Forsch. 1970;143:11–15. [Google Scholar]
- 2.Kiermeier F, Petz E. A Sulfhydryl group-oxidizing Enzyme in milk: II Influence of heating on milk and whey. Z Lebensm-Unters-Forsch. 1967;134:97–102. [Google Scholar]
- 3.Swaisgood H, Janolino V. Mammalian sulfhydryl oxidase. Food Science and Technology. 2003;122:539–546. [Google Scholar]
- 4.Janolino VG, Swaisgood HE. Isolation and characterization of sulfhydryl oxidase from bovine milk. J Biol Chem. 1975;250:2532–2538. [PubMed] [Google Scholar]
- 5.Janolino VG, Swaisgood HE. Sulfhydryl oxidase-catalyzed formation of disulfide bonds in reduced ribonuclease. Arch Biochem Biophys. 1987;258:265–271. doi: 10.1016/0003-9861(87)90344-4. [DOI] [PubMed] [Google Scholar]
- 6.Janolino VG, Swaisgood HE. A comparison of sulfhydryl oxidase from bovine milk and Aspergillus niger. Milchwissenschaft. 1992;47:143–146. [Google Scholar]
- 7.Swaisgood HE, Horton HR. Sulfhydryl oxidase: oxidation of sulphydryl groups and the formation of three-dimensional structure in proteins. Ciba Found Symp. 1980;72:205–222. doi: 10.1002/9780470720554.ch13. [DOI] [PubMed] [Google Scholar]
- 8.Clare DA, Horton HR, Stabel TJ, Swaisgood HE, Lecce JG. Tissue Distribution of Mammalian Sulfhydryl Oxidase. Arch Biochem Biophys. 1984;230:138–145. doi: 10.1016/0003-9861(84)90094-8. [DOI] [PubMed] [Google Scholar]
- 9.Clare DA, Pinnix IB, Lecce JG, Horton HR. Purification and properties of sulfhydryl oxidase from bovine pancreas. Arch Biochem Biophys. 1988;265:351–361. doi: 10.1016/0003-9861(88)90138-5. [DOI] [PubMed] [Google Scholar]
- 10.Yamada H. Localization in skin, activation and reaction mechanisms of skin sulfhydryl oxidase. Nippon Hifuka Gakkai Zasshi. 1989;99:861–869. [PubMed] [Google Scholar]
- 11.Lash LH, Jones DP. Characterization of the membrane-associated thiol oxidase activity of rat small-intestinal epithelium. Arch Biochem Biophys. 1983;225:344–352. doi: 10.1016/0003-9861(83)90039-5. [DOI] [PubMed] [Google Scholar]
- 12.Roth RA, Koshland ME. Identification of a lymphocyte enzyme that catalyzes pentamer immunoglobulin M assembly. J Biol Chem. 1981;256:4633–4639. [PubMed] [Google Scholar]
- 13.Lash LH, Jones DP. Purification and properties of the membranal thiol oxidase from porcine kidney. Arch Biochem Biophys. 1986;247:120–130. doi: 10.1016/0003-9861(86)90540-0. [DOI] [PubMed] [Google Scholar]
- 14.Janolino VG, Sliwkowski MX, Swaisgood HE, Horton HR. Catalytic effect of sulfhydryl oxidase on the formation of three-dimensional structure in chymotrypsinogen A. Arch Biochem Biophys. 1978;191:269–277. doi: 10.1016/0003-9861(78)90089-9. [DOI] [PubMed] [Google Scholar]
- 15.Ostrowski MC, Kistler WS. Properties of a flavoprotein sulfhydryl oxidase from rat seminal vesicle secretion. Biochemistry. 1980;19:2639–2645. doi: 10.1021/bi00553a016. [DOI] [PubMed] [Google Scholar]
- 16.Kusakabe H, Kuninaka A, Yoshino H. Purification and properties of a new enzyme, glutathione oxidase from Penicillium sp. K-6-5. Agric Biol Chem. 1982;46:2057–2067. [Google Scholar]
- 17.de la Motte RS, Wagner FW. Aspergillus niger sulfhydryl oxidase. Biochemistry. 1987;26:7363–7371. doi: 10.1021/bi00397a025. [DOI] [PubMed] [Google Scholar]
- 18.Hoober KL, Glynn NM, Burnside J, Coppock DL, Thorpe C. Homology between egg white sulfhydryl oxidase and quiescin Q6 defines a new class of flavin-linked sulfhydryl oxidases. J Biol Chem. 1999;274:31759–31762. doi: 10.1074/jbc.274.45.31759. [DOI] [PubMed] [Google Scholar]
- 19.Thorpe C, Hoober K, Raje S, Glynn N, Burnside J, Turi G, Coppock D. Sulfhydryl oxidases: emerging catalysts of protein disulfide bond formation in eukaryotes. Arch Biochem Biophys. 2002;405:1–12. doi: 10.1016/s0003-9861(02)00337-5. [DOI] [PubMed] [Google Scholar]
- 20.Coppock DL, Thorpe C. Multidomain flavin-dependent sulfhydryl oxidases. Antioxid Redox Signal. 2006;8:300–311. doi: 10.1089/ars.2006.8.300. [DOI] [PubMed] [Google Scholar]
- 21.Williams CH., Jr . Lipoamide dehydrogenase, glutathione reductase, thioredoxin reductase, and mercuric ion reductase-A family of flavoenzyme transhydrogenases. In: Müller F, editor. Chemistry and Biochemistry of Flavoenzymes. CRC Press; Chemistry and Biochemistry of Flavoenzymes: 1992. pp. 121–211. [Google Scholar]
- 22.Argyrou A, Blanchard JS. Flavoprotein disulfide reductases: advances in chemistry and function. Prog Nucleic Acid Res Mol Biol. 2004;78:89–142. doi: 10.1016/S0079-6603(04)78003-4. [DOI] [PubMed] [Google Scholar]
- 23.Hoober KL, Joneja B, White HB, III, Thorpe C. A Sulfhydryl Oxidase from Chicken Egg White. J Biol Chem. 1996;271:30510–30516. doi: 10.1074/jbc.271.48.30510. [DOI] [PubMed] [Google Scholar]
- 24.Benayoun B, Esnard-Fève A, Castella S, Courty Y, Esnard F. Rat seminal vesicle FAD-dependent sulfhydryl oxidase:biochemical characterization and molecular cloning of a member of the new sulfhydryl oxidase/quiescin Q6 gene family. J Biol Chem. 2001;276:13830–13837. doi: 10.1074/jbc.M010933200. [DOI] [PubMed] [Google Scholar]
- 25.Coppock DL, Kopman C, Scandalis S, Gillerman S. Preferential gene expression in quiescent human lung fibroblasts. Cell Growth Differ. 1993;4:483–493. [PubMed] [Google Scholar]
- 26.Gao C, Zhau HE, Chen B-Q, Chung LWK. GenBank accession: AAA89173. 1996. [Google Scholar]
- 27.Sasada R, Igarashi K, Takeyama M. Takeda Chem. Ind. Ltd; Japan: 1998. [Google Scholar]
- 28.Farrell CL, Martin FH, Yabkowitz R. Amgen Inc; USA: 2000. pp. 1–28. [Google Scholar]
- 29.Gross E, Sevier CS, Vala A, Kaiser CA, Fass D. A new FAD-binding fold and intersubunit disulfide shuttle in the thiol oxidase Erv2p. Nat Struct Biol. 2002;9:61–67. doi: 10.1038/nsb740. [DOI] [PubMed] [Google Scholar]
- 30.Wu CK, Dailey TA, Dailey HA, Wang BC, Rose JP. The crystal structure of augmenter of liver regeneration: A mammalian FAD-dependent sulfhydryl oxidase. Protein Sci. 2003;12:1109–1118. doi: 10.1110/ps.0238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tu BP, Ho-Schleyer SC, Travers KJ, Weissman JS. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science. 2000;290:1571–1574. doi: 10.1126/science.290.5496.1571. [DOI] [PubMed] [Google Scholar]
- 32.Gross E, Kastner DB, Kaiser CA, Fass D. Structure of Ero1p, source of disulfide bonds for oxidative protein folding in the cell. Cell. 2004;117:601–610. doi: 10.1016/s0092-8674(04)00418-0. [DOI] [PubMed] [Google Scholar]
- 33.Frand AR, Kaiser CA. Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol Cell. 1999;4:469–477. doi: 10.1016/s1097-2765(00)80198-7. [DOI] [PubMed] [Google Scholar]
- 34.Cabibbo A, Pagani M, Fabbri M, Rocchi M, Farmery MR, Bulleid NJ, Sitia R. ERO1-L, a human protein that favors disulfide bond formation in the endoplasmic reticulum. J Biol Chem. 2000;275:4827–4833. doi: 10.1074/jbc.275.7.4827. [DOI] [PubMed] [Google Scholar]
- 35.Sevier CS, Kaiser CA. Conservation and diversity of the cellular disulfide bond formation pathways. Antioxid Redox Signal. 2006;8:797–811. doi: 10.1089/ars.2006.8.797. [DOI] [PubMed] [Google Scholar]
- 36.Mezghrani A, Fassio A, Benham A, Simmen T, Braakman I, Sitia R. Manipulation of oxidative protein folding and PDI redox state in mammalian cells. EMBO J. 2001;20:6288–6296. doi: 10.1093/emboj/20.22.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molteni SN, Fassio A, Ciriolo MR, Filomeni G, Pasqualetto E, Fagioli C, Sitia R. Glutathione limits Ero1-dependent oxidation in the endoplasmic reticulum. J Biol Chem. 2004;279:32667–32673. doi: 10.1074/jbc.M404992200. [DOI] [PubMed] [Google Scholar]
- 38.van Anken E, Braakman I. Versatility of the endoplasmic reticulum protein folding factory. Crit Rev Biochem Mol Biol. 2005;40:191–228. doi: 10.1080/10409230591008161. [DOI] [PubMed] [Google Scholar]
- 39.Thorpe C, Coppock DL. Generating disulfides in multicellular organisms: Emerging roles for a new flavoprotein family. J Biol Chem. 2007;282:13929–13933. doi: 10.1074/jbc.R600037200. [DOI] [PubMed] [Google Scholar]
- 40.Derelle E, Ferraz C, Rombauts S, Rouze P, Worden AZ, Robbens S, Partensky F, Degroeve S, Echeynie S, Cooke R, Saeys Y, Wuyts J, Jabbari K, Bowler C, Panaud O, Piegu B, Ball SG, Ral JP, Bouget FY, Piganeau G, De Baets B, Picard A, Delseny M, Demaille J, Van de Peer Y, Moreau H. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci U S A. 2006;103:11647–11652. doi: 10.1073/pnas.0604795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoober KL, Thorpe C. Egg white sulfhydryl oxidase: Kinetic mechanism of the catalysis of disulfide bond formation. Biochemistry. 1999;38:3211–3217. doi: 10.1021/bi9820816. [DOI] [PubMed] [Google Scholar]
- 42.Hofhaus G, Lee JE, Tews I, Rosenberg B, Lisowsky T. The N-terminal cysteine pair of yeast sulfhydryl oxidase Erv1p is essential for in vivo activity and interacts with the primary redox centre. Eur J Biochem. 2003;270:1528–1535. doi: 10.1046/j.1432-1033.2003.03519.x. [DOI] [PubMed] [Google Scholar]
- 43.O’Donnell ME, Johnson FA, Williams CH., Jr Proton nuclear magnetic resonance investigation of the mechanism of flavin C-4a adduct formation induced by oxidized nicotinamide adenine dinucleotide binding to monoalkylated pig heart lipoamide dehydrogenase. Biochemistry. 1983;22:3792–3796. doi: 10.1021/bi00285a012. [DOI] [PubMed] [Google Scholar]
- 44.O’Donnell ME, Williams CH., Jr Reconstitution of Escherichia coli thioredoxin reductase with 1-deazaFAD. Evidence for 1-deazaFAD C-4a adduct formation linked to the ionization of an active site base. J Biol Chem. 1984;259:2243–2251. [PubMed] [Google Scholar]
- 45.Thorpe C, Williams CH. Spectral evidence for a flavin adduct in a monoalkylated derivative of pig heart lipoamide dehydrogenase. J Biol Chem. 1976;251:7726–7728. [PubMed] [Google Scholar]
- 46.Miller SM, Massey V, Ballou D, Williams CH, Jr, Distefano MD, Moore MJ, Walsh CT. Use of a site-directed triple mutant to trap intermediates: demonstration that the flavin C(4a)-thiol adduct and reduced flavin are kinetically competent intermediates in mercuric ion reductase. Biochemistry. 1990;29:2831–2841. doi: 10.1021/bi00463a028. [DOI] [PubMed] [Google Scholar]
- 47.Massey V. Activation of molecular oxygen by flavins and flavoproteins. J Biol Chem. 1994;269:22459–22462. [PubMed] [Google Scholar]
- 48.Mattevi A. To be or not to be an oxidase: challenging the oxygen reactivity of flavoenzymes. Trends Biochem Sci. 2006;31:276–283. doi: 10.1016/j.tibs.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 49.Vitu E, Bentzur M, Lisowsky T, Kaiser CA, Fass D. Gain of function in an ERV/ALR sulfhydryl oxidase by molecular engineering of the shuttle disulfide. J Mol Biol. 2006;362:89–101. doi: 10.1016/j.jmb.2006.06.070. [DOI] [PubMed] [Google Scholar]
- 50.Farrell SR, Thorpe C. Augmenter of liver regeneration: a flavin dependent sulfhydryl oxidase with cytochrome C reductase activity. Biochemistry. 2005;44:1532–1541. doi: 10.1021/bi0479555. [DOI] [PubMed] [Google Scholar]
- 51.Allen S, Balabanidou V, Sideris DP, Lisowsky T, Tokatlidis K. Erv1 Mediates the Mia40-dependent Protein Import Pathway and Provides a Functional Link to the Respiratory Chain by Shuttling Electrons to Cytochrome c. J Mol Biol. 2005 doi: 10.1016/j.jmb.2005.08.049. [DOI] [PubMed] [Google Scholar]
- 52.Herrmann JM, Kohl R. Catch me if you can! Oxidative protein trapping in the intermembrane space of mitochondria. J Cell Biol. 2007;176:559–563. doi: 10.1083/jcb.200611060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sevier CS, Kaiser CA. Disulfide transfer between two conserved cysteine pairs imparts selectivity to protein oxidation by Ero1. Mol Biol Cell. 2006;17:2256–2266. doi: 10.1091/mbc.E05-05-0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vala A, Sevier CS, Kaiser CA. Structural determinants of substrate access to the disulfide oxidase Erv2p. J Mol Biol. 2005;354:952–966. doi: 10.1016/j.jmb.2005.09.076. [DOI] [PubMed] [Google Scholar]
- 55.Fernandes PA, Ramos MJ. Theoretical insights into the mechanism for thiol/disulfide exchange. Chemistry. 2004;10:257–266. doi: 10.1002/chem.200305343. [DOI] [PubMed] [Google Scholar]
- 56.Rosenfield RE, Parthasarathy R, Dunitz JD. Directional Preferences of Nonbonded Atomic Contacts with Divalent Sulfur.1. Electrophiles and Nucleophiles. J Amer Chem Soc. 1977;99:4860–4862. [Google Scholar]
- 57.Ellgaard L, Ruddock LW. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2004;1699:35–44. doi: 10.1016/j.bbapap.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 59.Tian G, Xiang S, Noiva R, Lennarz WJ, Schindelin H. The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell. 2006;124:61–73. doi: 10.1016/j.cell.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 60.Gruber CW, Cemazar M, Heras B, Martin JL, Craik DJ. Protein disulfide isomerase: the structure of oxidative folding. Trends Biochem Sci. 2006;31:455–464. doi: 10.1016/j.tibs.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 61.Raje S, Thorpe C. Inter-domain redox communication in flavoenzymes of the quiescin/sulfhydryl oxidase family: role of a thioredoxin domain in disulfide bond formation. Biochemistry. 2003;42:4560–4568. doi: 10.1021/bi030003z. [DOI] [PubMed] [Google Scholar]
- 62.Huber-Wunderlich M, Glockshuber R. A single dipeptide sequence modulates the redox properties of a whole enzyme family. Fold Des. 1998;3:161–171. doi: 10.1016/S1359-0278(98)00024-8. [DOI] [PubMed] [Google Scholar]
- 63.Chivers PT, Prehoda KE, Raines RT. The CXXC motif: a rheostat in the active site. Biochemistry. 1997;36:4061–4066. doi: 10.1021/bi9628580. [DOI] [PubMed] [Google Scholar]
- 64.Chivers PT, Laboissiere MC, Raines RT. The CXXC motif: imperatives for the formation of native disulfide bonds in the cell. EMBO J. 1996;15:2659–2667. [PMC free article] [PubMed] [Google Scholar]
- 65.Krause G, Lundstrom J, Barea JL, Pueyo de la Cuesta C, Holmgren A. Mimicking the active site of protein disulfide-isomerase by substitution of proline 34 in Escherichia coli thioredoxin. J Biol Chem. 1991;266:9494–9500. [PubMed] [Google Scholar]
- 66.Quan S, Schneider I, Pan J, Hacht AV, Bardwell JC. The CXXC motif is more than a redox rheostat. J Biol Chem. 2007 doi: 10.1074/jbc.M705291200. [DOI] [PubMed] [Google Scholar]
- 67.Hoober KL, Sheasley SS, Gilbert HF, Thorpe C. Sulfhydryl oxidase from egg white: a facile catalyst for disulfide bond formation in proteins and peptides. J Biol Chem. 1999;274:22147–22150. doi: 10.1074/jbc.274.32.22147. [DOI] [PubMed] [Google Scholar]
- 68.Chakravarthi S, Jessop CE, Willer M, Stirling CJ, Bulleid NJ. Intracellular catalysis of disulphide bond formation by the human sulphydryl oxidase, QSOX1. Biochem J. 2007;404:403–411. doi: 10.1042/BJ20061510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tury A, Mairet-Coello G, Poncet F, Jacquemard C, Risold PY, Fellmann D, Griffond B. QSOX sulfhydryl oxidase in rat adenohypophysis: localization and regulation by estrogens. J Endocrinol. 2004;183:353–363. doi: 10.1677/joe.1.05842. [DOI] [PubMed] [Google Scholar]
- 70.Mairet-Coello G, Tury A, Esnard-Feve A, Fellmann D, Risold PY, Griffond B. FAD-linked sulfhydryl oxidase QSOX: topographic, cellular, and subcellular immunolocalization in adult rat central nervous system. J Comp Neurol. 2004;473:334–363. doi: 10.1002/cne.20126. [DOI] [PubMed] [Google Scholar]
- 71.Wittke I, Wiedemeyer R, Pillmann A, Savelyeva L, Westermann F, Schwab M. Neuroblastoma-derived sulfhydryl oxidase, a new member of the sulfhydryl oxidase/Quiescin6 family, regulates sensitization to interferon gamma-induced cell death in human neuroblastoma cells. Cancer Res. 2003;63:7742–7752. [PubMed] [Google Scholar]
- 72.Le Gall S, Neuhof A, Rapoport T. The endoplasmic reticulum membrane is permeable to small molecules. Mol Biol Cell. 2004;15:447–455. doi: 10.1091/mbc.E03-05-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chakravarthi S, Jessop CE, Bulleid NJ. The role of glutathione in disulphide bond formation and endoplasmic-reticulum-generated oxidative stress. EMBO Rep. 2006;7:271–275. doi: 10.1038/sj.embor.7400645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Darby NJ, Creighton TE. Characterization of the active site cysteine residues of the thioredoxin-like domains of protein disulfide isomerase. Biochemistry. 1995;34:16770–16780. doi: 10.1021/bi00051a027. [DOI] [PubMed] [Google Scholar]
- 75.Gilbert HF. Catalysis of thiol/disulfide exchange: single-turnover reduction of protein disulfide-isomerase by glutathione and catalysis of peptide disulfide reduction. Biochemistry. 1989;28:7298–7305. doi: 10.1021/bi00444a023. [DOI] [PubMed] [Google Scholar]
- 76.Gilbert HF. Redox control of enzyme activities by thiol/disulfide exchange. Methods Enzymol. 1984;107:330–351. doi: 10.1016/0076-6879(84)07022-1. [DOI] [PubMed] [Google Scholar]
- 77.Jessop CE, Chakravarthi S, Watkins RH, Bulleid NJ. Oxidative protein folding in the mammalian endoplasmic reticulum. Biochem Soc Trans. 2004;32:655–658. doi: 10.1042/BST0320655. [DOI] [PubMed] [Google Scholar]
- 78.Tury A, Mairet-Coello G, Esnard-Feve A, Benayoun B, Risold PY, Griffond B, Fellmann D. Cell-specific localization of the sulphydryl oxidase QSOX in rat peripheral tissues. Cell Tissue Res. 2006;323:91–103. doi: 10.1007/s00441-005-0043-x. [DOI] [PubMed] [Google Scholar]
- 79.Musard JF, Sallot M, Dulieu P, Fraichard A, Ordener C, Remy-Martin JP, Jouvenot M, Adami P. Identification and expression of a new sulfhydryl oxidase SOx-3 during the cell cycle and the estrus cycle in uterine cells. Biochem Biophys Res Commun. 2001;287:83–91. doi: 10.1006/bbrc.2001.5440. [DOI] [PubMed] [Google Scholar]
- 80.Mairet-Coello G, Tury A, Fellmann D, Jouvenot M, Griffond B. Expression of SOx-2, a member of the FAD-dependent sulfhydryl oxidase/quiescin Q6 gene family, in rat brain. Neuroreport. 2002;13:2049–2051. doi: 10.1097/00001756-200211150-00012. [DOI] [PubMed] [Google Scholar]
- 81.Radom J, Colin D, Thiebault F, Dognin-Bergeret M, Mairet-Coello G, Esnard-Feve A, Fellmann D, Jouvenot M. Identification and expression of a new splicing variant of FAD-sulfhydryl oxidase in adult rat brain. Biochim Biophys Acta. 2006;1759:225–233. doi: 10.1016/j.bbaexp.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 82.Mairet-Coello G, Tury A, Fellmann D, Risold PY, Griffond B. Ontogenesis of the sulfhydryl oxidase QSOX expression in rat brain. J Comp Neurol. 2005;484:403–417. doi: 10.1002/cne.20411. [DOI] [PubMed] [Google Scholar]
- 83.Amiot C, Musard JF, Hadjiyiassemis M, Jouvenot M, Fellmann D, Risold PY, Adami P. Expression of the secreted FAD-dependent sulfydryl oxidase (QSOX) in the guinea pig central nervous system. Mol Brain Res. 2004;125:13–21. doi: 10.1016/j.molbrainres.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 84.Avshalumov MV, Bao L, Patel JC, Rice ME. H2O2 signaling in the nigrostriatal dopamine pathway via ATP-sensitive potassium channels: issues and answers. Antioxid Redox Signal. 2007;9:219–231. doi: 10.1089/ars.2007.9.219. [DOI] [PubMed] [Google Scholar]
- 85.Veal EA, Day AM, Morgan BA. Hydrogen peroxide sensing and signaling. Mol Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 86.Moggs JG, Murphy TC, Lim FL, Moore DJ, Stuckey R, Antrobus K, Kimber I, Orphanides G. Anti-proliferative effect of estrogen in breast cancer cells that re-express ERalpha is mediated by aberrant regulation of cell cycle genes. J Mol Endocrinol. 2005;34:535–551. doi: 10.1677/jme.1.01677. [DOI] [PubMed] [Google Scholar]
- 87.Mohammad HP, Seachrist DD, Quirk CC, Nilson JH. Reexpression of p8 contributes to tumorigenic properties of pituitary cells and appears in a subset of prolactinomas in transgenic mice that hypersecrete luteinizing hormone. Mol Endocrinol. 2004;18:2583–2593. doi: 10.1210/me.2004-0163. [DOI] [PubMed] [Google Scholar]
- 88.Boon K, Osorio EC, Greenhut SF, Schaefer CF, Shoemaker J, Polyak K, Morin PJ, Buetow KH, Strausberg RL, De Souza SJ, Riggins GJ. An anatomy of normal and malignant gene expression. Proc Natl Acad Sci U S A. 2002;99:11287–11292. doi: 10.1073/pnas.152324199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang X, Rutledge J, Armstrong DT. Studies on zona hardening in rat oocytes that are matured in vitro in a serum-free medium. Mol Reprod Dev. 1991;28:292–296. doi: 10.1002/mrd.1080280312. [DOI] [PubMed] [Google Scholar]
- 90.Iwamoto K, Ikeda K, Yonezawa N, Noguchi S, Kudo K, Hamano S, Kuwayama M, Nakano M. Disulfide formation in bovine zona pellucida glycoproteins during fertilization: evidence for the involvement of cystine cross-linkages in hardening of the zona pellucida. J Reprod Fertil. 1999;117:395–402. doi: 10.1530/jrf.0.1170395. [DOI] [PubMed] [Google Scholar]
- 91.Coppock DL, Cina-Poppe D, Gilleran S. The Quiescin Q6 gene (QSCN6) is a fusion of two ancient gene families: thioredoxin and ERV1. Genomics. 1998;54:460–468. doi: 10.1006/geno.1998.5605. [DOI] [PubMed] [Google Scholar]
- 92.Farrell CL, Martin FH, Yabkowitz R. Amgen Inc; USA: 2001. [Google Scholar]
- 93.Udupa KB, Bose C. ASH Annual Meeting; American Society of Hematology. 2005. p. 4270. [Google Scholar]
- 94.Zanata SM, Luvizon AC, Batista DF, Ikegami CM, Pedrosa FO, Souza EM, Chaves DF, Caron LF, Pelizzari JV, Laurindo FR, Nakao LS. High levels of active quiescin Q6 sulfhydryl oxidase (QSOX) are selectively present in fetal serum. Redox Rep. 2005;10:319–323. doi: 10.1179/135100005X83699. [DOI] [PubMed] [Google Scholar]
- 95.States DJ, Omenn GS, Blackwell TW, Fermin D, Eng J, Speicher DW, Hanash SM. Challenges in deriving high-confidence protein identifications from data gathered by a HUPO plasma proteome collaborative study. Nat Biotechnol. 2006;24:333–338. doi: 10.1038/nbt1183. [DOI] [PubMed] [Google Scholar]
- 96.Martin DB, Gifford DR, Wright ME, Keller A, Yi E, Goodlett DR, Aebersold R, Nelson PS. Quantitative proteomic analysis of proteins released by neoplastic prostate epithelium. Cancer Res. 2004;64:347–355. doi: 10.1158/0008-5472.can-03-2062. [DOI] [PubMed] [Google Scholar]
- 97.Kulasingam V, Diamandis EP. Proteomic analysis of conditioned media from three breast cancer cell lines: A mine for biomarkers and therapeutic targets. Mol Cell Proteomics. 2007 doi: 10.1074/mcp.M600465-MCP200. [DOI] [PubMed] [Google Scholar]
- 98.Essex DW, Li M. Redox modification of platelet glycoproteins. Curr Drug Targets. 2006;7:1233–1241. doi: 10.2174/138945006778559193. [DOI] [PubMed] [Google Scholar]
- 99.Jordan PA, Gibbins JM. Extracellular disulfide exchange and the regulation of cellular function. Antiox Redox Signal. 2006;8:312–324. doi: 10.1089/ars.2006.8.312. [DOI] [PubMed] [Google Scholar]
- 100.Go YM, Jones DP. Intracellular proatherogenic events and cell adhesion modulated by extracellular thiol/disulfide redox state. Circulation. 2005;111:2973–2980. doi: 10.1161/CIRCULATIONAHA.104.515155. [DOI] [PubMed] [Google Scholar]
- 101.Matthias LJ, Hogg PJ. Redox Control on the Cell Surface: Implications for HIV-1 Entry. Antioxid Redox Signal. 2003;5:133–138. doi: 10.1089/152308603321223621. [DOI] [PubMed] [Google Scholar]
- 102.Jonas CR, Ziegler TR, Gu LH, Jones DP. Extracellular thiol/disulfide redox state affects proliferation rate in a human colon carcinoma (Caco2) cell line. Free Radic Biol Med. 2002;33:1499–1506. doi: 10.1016/s0891-5849(02)01081-x. [DOI] [PubMed] [Google Scholar]
- 103.Ramirez A, Ramadan B, Ritzenthaler JD, Rivera HN, Jones DP, Roman J. Extracellular cysteine/cystine redox potential controls lung fibroblast proliferation and matrix expression through upregulation of transforming growth factor-{beta} Am J Physiol Lung Cell Mol Physiol. 2007 doi: 10.1152/ajplung.00010.2007. [DOI] [PubMed] [Google Scholar]
- 104.Kaiser BK, Yim D, Chow IT, Gonzalez S, Dai Z, Mann HH, Strong RK, Groh V, Spies T. Disulphide-isomerase-enabled shedding of tumour-associated NKG2D ligands. Nature. 2007;447:482–486. doi: 10.1038/nature05768. [DOI] [PubMed] [Google Scholar]
- 105.Goplen D, Wang J, Enger PO, Tysnes BB, Terzis AJ, Laerum OD, Bjerkvig R. Protein disulfide isomerase expression is related to the invasive properties of malignant glioma. Cancer Res. 2006;66:9895–9902. doi: 10.1158/0008-5472.CAN-05-4589. [DOI] [PubMed] [Google Scholar]
- 106.Gumireddy K, Sun F, Klein-Szanto AJ, Gibbins JM, Gimotty PA, Saunders AJ, Schultz PG, Huang Q. In vivo selection for metastasis promoting genes in the mouse. Proc Natl Acad Sci. 2007;104:6696–6701. doi: 10.1073/pnas.0701145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ostrowski MC, Kistler MK, Kistler WS. Purification and cell-free synthesis of a major protein from rat seminal vesicle secretion. A potential marker for androgen action. J Biol Chem. 1979;254:383–390. [PubMed] [Google Scholar]
- 108.Lee J, Hofhaus G, Lisowsky T. Erv1p from Saccharomyces cerevisiae is a FAD-linked sulfhydryl oxidase. FEBS Lett. 2000;477(1–2):62–66. doi: 10.1016/s0014-5793(00)01767-1. [DOI] [PubMed] [Google Scholar]
- 109.Lisowsky T, Lee JE, Polimeno L, Francavilla A, Hofhaus G. Mammalian augmenter of liver regeneration protein is a sulfhydryl oxidase. Dig Liver Dis. 2001;33:173–180. doi: 10.1016/s1590-8658(01)80074-8. [DOI] [PubMed] [Google Scholar]
- 110.Gerber J, Muhlenhoff U, Hofhaus G, Lill R, Lisowsky T. Yeast ERV2p is the first microsomal FAD-linked sulfhydryl oxidase of the Erv1p/Alrp protein family. J Biol Chem. 2001;276:23486–23491. doi: 10.1074/jbc.M100134200. [DOI] [PubMed] [Google Scholar]
- 111.Sevier CS, Cuozzo JW, Vala A, Aslund F, Kaiser CA. A flavoprotein oxidase defines a new endoplasmic reticulum pathway for biosynthetic disulphide bond formation. Nat Cell Biol. 2001;3:874–882. doi: 10.1038/ncb1001-874. [DOI] [PubMed] [Google Scholar]
- 112.Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 114.Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]