Abstract
Historically, histomorphological and epidemiological data suggested that atypical ductal hyperplasia and ductal carcinoma in situ are the earliest recognizable neoplastic stages of breast cancer progression. Over the past several years, detailed high-throughput molecular genetic, gene expression and epigenetic analyses have enhanced our understanding of these early neoplastic lesions and have re-shaped our view of human breast cancer progression to include multiple distinct pathways of evolution.
Introduction
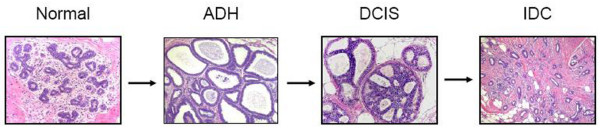
Atypical ductal hyperplasia (ADH) and ductal carcinoma in situ (DCIS), the earliest neoplastic stages of human breast cancer progression, are characterized by a proliferation of epithelial cells that is confined within the basement membrane of the mammary ductal network [1]. ADH and DCIS represent a diverse group of tumors that are detected in women undergoing screening mammography, and it is believed that the detection of breast cancer in these early neoplastic stages is largely responsible for the recent trend toward improvement in breast cancer mortality worldwide. Historically, the pathological distinction between ADH and DCIS is largely morphological and is considered by some to be based upon the size and extent of the epithelial proliferation [2]. The classic model of breast cancer progression (Figure 1) is seen as a linear multi-step process manifesting itself as a sequence of pathologically defined stages in which molecular alterations within normal breast epithelium give rise to ADH, the first premalignant stage of breast cancer progression, upon which progressive molecular alterations give rise to DCIS, the second premalignant stage of breast cancer [3,4]. Additional molecular alterations in DCIS are thought to give rise to the malignant stages of invasive and metastatic carcinoma.
Figure 1.
Classic linear multi-step model of human breast cancer progression based on histomorphological and epidemiological data. Molecular alterations occurring in breast epithelium of a normal terminal duct lobular unit result in atypical ductal hyperplasia (ADH), the earliest neoplastic stage of progression. Subsequent molecular alterations occur in ADH, resulting in ductal carcinoma in situ (DCIS), another early neoplastic stage, upon which additional events occur, resulting in invasive ductal carcinoma (IDC).
Until recently, a significant impediment to a better understanding of breast cancer progression has been our inability to accurately interrogate the early neoplastic or premalignant stages of this process. However, over the past several years the successful application of combining highly specific tissue microdissection technologies with advanced high-throughput genomic and gene expression technologies has re-shaped our view of breast cancer progression. Instead of viewing breast cancer as a simple single linear pathway, recent molecular genetic evidence supports a multiple linear pathway model of progression.
Genomic analysis of premalignant breast cancer
Loss of heterozygosity-based genomic studies of the premalignant stages of breast cancer progression supported the classic model of progression in which ADH and DCIS are genetically related to each other and give rise to invasive ductal carcinoma (IDC) [5-7]. However, the first significant evidence suggesting that the classic model of breast cancer progression required rethinking emerged from several comparative genomic hybridization (CGH) studies of DCIS and IDC [8]. Most notably, loss of 16q was seen almost exclusively in low and intermediate grade DCIS, while a higher frequency of 1q gain and 11q loss was observed in intermediate grade DCIS [8]. High grade DCIS, on the other hand, demonstrated complex genomic alterations that are characterized by loss of 8p, 11q, 13q and 14q, by gains of 1q, 5p, 8q and 17q, and high-level amplifications of 17q12 and 11q13. Analysis of synchronous IDC associated with DCIS revealed a near-identical genetic pattern supporting the direct precursor relationship between DCIS and IDC. Recently, Yao and colleagues [9], utilizing CGH in conjunction with serial analysis of gene expression (SAGE), have demonstrated an overall trend towards an increase in the number and amplitude of gains and losses during breast cancer progression, supporting the concept that the early neoplastic stage of DCIS is a direct precursor to IDC.
Together, these CGH-based studies continued to support the classic linear model of progression (that is, ADH gives rise to DCIS, which in turn gives rise to IDC), while providing evidence that human breast cancer progression may consist of several genetically distinct linear pathways that correlate with histological tumor grade.
Gene expression analysis of ADH and DCIS
Over the past several years considerable research interest has focused on understanding the gene expression changes that occur during breast cancer development and, in particular, during the early pre-invasive stage of progression [10,11]. One of the earliest and most extensive studies is by Ma and colleagues [10] in which patient-matched phenotypically normal breast epithelium from the terminal ductal lobular unit and epithelium constituting ADH, DCIS and IDC were microdissected and hybridized to a cDNA microarray containing 12,000 genes. Comparative gene expression profile analysis of patient-matched normal versus ADH, normal versus DCIS, and normal versus IDC revealed that the most pronounced transcriptome change occurred at the normal to ADH transition, and that such transcriptional alterations are maintained throughout the later stages (DCIS and IDC) of progression. On a global level, no consistent major transcriptional changes were identified between the pre-invasive and invasive stages, and this observation was consistent with SAGE data generated by Porter and colleagues [11]. These data support the idea that the different stages of breast cancer progression are evolutionary products of the same clonal origin, and suggest that genes expressed in the premalignant stages (ADH and DCIS) may reflect the progressive potential of the pathological lesion. The concept that gene expression of an early stage breast cancer can predict future clinical behavior is well documented in the literature as it relates to early stage invasive breast cancer and the risk of distant metastases [12-16].
Despite using different platforms (cDNA arrays versus oligonucleotide arrays versus SAGE), several studies have demonstrated that the transition from the premalignant stage of DCIS to the malignant stage of IDC is associated with quantitative, rather than qualitative, differences in gene expression [10,11]. Intriguingly, this quantitative relationship is most prominent in high grade (poorly differentiated/grade III) samples, revealing an intriguing link between tumor grade and tumor stage progression [10]. Similarly, the analysis of breast cancer development in transgenic mouse models also demonstrates that the transition from a pre-invasive to an invasive stage of progression is associated with quantitative, rather than qualitative, differences in gene expression [17]. All of these observations taken together suggest that breast cancer progression may be more complex than the current linear theory of activation and inactivation of oncogenes and tumor suppressor genes, respectively, and may be dependent upon such contingencies as quantitative levels and timing of gene expression.
Although distinct qualitative gene expression changes are not observed during breast cancer stage progression, several gene expression studies have demonstrated distinct expression alterations associated with different morphological phenotypes and, in particular, tumor grade [10,18,19]. Low grade (well differentiated/grade I) and high grade (poorly differentiated/grade III) breast cancers have been demonstrated to possess distinct reciprocal gene expression patterns and these data are consistent with CGH data suggesting that distinct gene expression pathways are associated with the low grade and high grade phenotypes [10]. Not unexpectedly, genes associated with the estrogen receptor phenotype were prominently expressed in the low grade DCIS, while genes associated with increased mitotic activity and cell cycle processes were most commonly expressed in cases of high grade DCIS [10]. Gene expression associated with intermediate grade (moderately differentiated/grade II) tumors exhibited a more complex pattern consisting of gene expression elements associated with low and high grade tumors. Using various class prediction algorithms, Ivshina and colleagues [20] and Sotiriou and colleagues [19] have identified gene expression indices that classify intermediate grade tumors into low grade-like and high grade-like tumors that display low and high risks of recurrence, respectively. These latter studies highlight the imperfect nature of the current histological grading system and, importantly, support the view that low and high grade tumors reflect distinct pathobiological entities rather than a direct continuum of cancer progression, and that intermediate grade tumors likely arise from these two distinct pathways [20].
An updated model of breast cancer progression
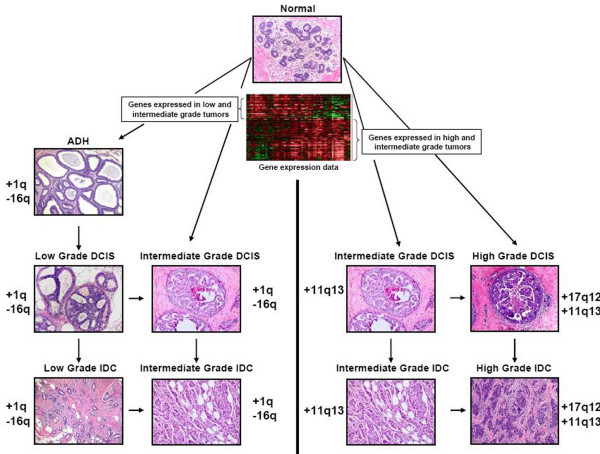
The development of breast cancer is a complex multi-step process consisting of genetic and gene expression changes that are frequently manifested in both the early (ADH and DCIS) and late stages of progression. Based upon histopathological observations as well as early genetic studies, a simple model of breast cancer progression was proposed in which the earliest neoplastic lesion, ADH, gives rise to low grade DCIS that progresses to high grade DCIS, which culminates in invasive carcinoma [4]. A number of recent molecular-based studies support modification of this general progression scheme. Firstly, and most importantly, CGH based studies provide evidence of two distinct pathways in the evolution of DCIS and IDC. One pathway is characterized by 16q loss and is observed predominantly in low grade tumors. The second pathway is characterized by 11q13 and 17q1 amplification. Interestingly, intermediate grade tumors share genetic alterations common to either pathway, suggesting that this population of tumors can arise from either pathway. Secondly, results from several gene expression-based studies suggest that breast cancers are stratified along two distinct gene expression pathways that correlate directly with tumor grade. These results together support a modified model of breast cancer progression (Figure 2). Notably, this tumor grade-based stratification scheme not only defines histopathological breast cancer progression, but also breast cancer clinical outcome.
Figure 2.
Contemporary model of breast cancer progression based on genetic and gene expression data. Distinct molecular events occur in normal breast epithelium giving rise to two divergent molecular pathways within which linear (denoted by vertical arrows) and horizontal (denoted by horizontal arrows) progression occurs. The first pathway is characterized by genetic alterations that include gain of 1q and loss of 16q, and this pattern of genetic alteration is seen predominantly in low grade ductal carcinoma in situ (DCIS) and invasive ductal carcinoma (IDC) and in a subset (low grade-like) of intermediate grade tumors. The second pathway is characterized by amplification of 11q13 and 17q12 in high grade tumors and 11q13 in a subset (high grade-like) of intermediate grade tumors. Additional support for the divergent two pathway model is provided by gene expression profiling data (depicted as a gene expression heatmap) generated from atypical ductal hyperplasia (ADH), DCIS and IDC. More specifically, low grade tumors express a unique set of genes that is rarely seen in high grade tumors and vice versa. Intermediate grade tumors express either 'low grade-like' gene expression signatures or 'high grade-like' gene expression signatures.
Abbreviations
ADH = atypical ductal hyperplasia; CGH = comparative genomic hybridization; DCIS = ductal carcinoma in situ; IDC = invasive ductal carcinoma; SAGE = serial analysis of gene expression.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
Acknowledgements
We thank M Erlander and X Ma for helpful discussions and M Lerwill for providing select photomicrographs. This work was supported in part by NIH RO1-1CA112021-01 (DCS), the Department of Defense grant W81XWH-04-1-0606 (DCS), Susan G Komen Breast Cancer Foundation grants BCTR0402932 (DCS) and the Avon Foundation (DCS).
References
- Pinder SE, Ellis IO. The diagnosis and management of pre-invasive breast disease: ductal carcinoma in situ (DCIS) and atypical ductal hyperplasia (ADH) – current definitions and classification. Breast Cancer Res. 2003;5:254–257. doi: 10.1186/bcr623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavassoli FA, Norris HJ. A comparison of the results of long-term follow-up for atypical intraductal hyperplasia and intraductal hyperplasia of the breast. Cancer. 1990;65:518–529. doi: 10.1002/1097-0142(19900201)65:3<518::AID-CNCR2820650324>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Dupont WD, Page DL. Risk factors for breast cancer in women with proliferative breast disease. N Engl J Med. 1985;312:146–151. doi: 10.1056/NEJM198501173120303. [DOI] [PubMed] [Google Scholar]
- Lakhani SR. The transition from hyperplasia to invasive carcinoma of the breast. J Pathol. 1999;187:272–278. doi: 10.1002/(SICI)1096-9896(199902)187:3<272::AID-PATH265>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Chuaqui RF, Zhuang Z, Emmert-Buck MR, Liotta LA, Merino MJ. Analysis of loss of heterozygosity on chromosome 11q13 in atypical ductal hyperplasia and in situ carcinoma of the breast. Am J Pathol. 1997;150:297–303. [PMC free article] [PubMed] [Google Scholar]
- O'Connell P, Pekkel V, Fuqua SA, Osborne CK, Clark GM, Allred DC. Analysis of loss of heterozygosity in 399 premalignant breast lesions at 15 genetic loci. J Natl Cancer Inst. 1998;90:697–703. doi: 10.1093/jnci/90.9.697. [DOI] [PubMed] [Google Scholar]
- Larson PS, de las MA, Cerda SR, Bennett SR, Cupples LA, Rosenberg CL. Quantitative analysis of allele imbalance supports atypical ductal hyperplasia lesions as direct breast cancer precursors. J Pathol. 2006;209:307–316. doi: 10.1002/path.1973. [DOI] [PubMed] [Google Scholar]
- Buerger H, Otterbach F, Simon R, Poremba C, Diallo R, Decker T, Riethdorf L, Brinkschmidt C, Dockhorn-Dworniczak B, Boecker W. Comparative genomic hybridization of ductal carcinoma in situ of the breast – evidence of multiple genetic pathways. J Pathol. 1999;187:396–402. doi: 10.1002/(SICI)1096-9896(199903)187:4<396::AID-PATH286>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Yao J, Weremowicz S, Feng B, Gentleman RC, Marks JR, Gelman R, Brennan C, Polyak K. Combined cDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Res. 2006;66:4065–4078. doi: 10.1158/0008-5472.CAN-05-4083. [DOI] [PubMed] [Google Scholar]
- Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, Payette T, Pistone M, Stecker K, Zhang BM, Zhou YX, Varnholt H, Smith B, Gadd M, Chatfield E, Kessler J, Baer TM, Erlander MG, Sgroi DC. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci USA. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter D, Lahti-Domenici J, Keshaviah A, Bae YK, Argani P, Marks J, Richardson A, Cooper A, Strausberg R, Riggins GJ, Schnitt S, Gabrielson E, Gelman R, Polyak K. Molecular markers in ductal carcinoma in situ of the breast. Mol Cancer Res. 2003;1:362–375. [PubMed] [Google Scholar]
- Goetz MP, Suman VJ, Ingle JN, Nibbe AM, Visscher DW, Reynolds CA, Lingle WL, Erlander M, Ma XJ, Sgroi DC, Perez EA, Couch FJ. A two-gene expression ratio of homeobox 13 and interleukin-17B receptor for prediction of recurrence and survival in women receiving adjuvant tamoxifen. Clin Cancer Res. 2006;12:2080–2087. doi: 10.1158/1078-0432.CCR-05-1263. [DOI] [PubMed] [Google Scholar]
- Jansen MP, Sieuwerts AM, Look MP, Ritstier K, Meijer-van Gelder ME, van Staveren IL, Klijn JG, Foekens JA, Berns EM. HOXB13-to-IL17BR expression ratio is related with tumor aggressiveness and response to tamoxifen of recurrent breast cancer: a retrospective study. J Clin Oncol. 2007;25:662–668. doi: 10.1200/JCO.2006.07.3676. [DOI] [PubMed] [Google Scholar]
- Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, Muir B, Mohapatra G, Salunga R, Tuggle JT, Tran Y, Tran D, Tassin A, Amon P, Wang W, Wang W, Enright E, Stecker K, Estepa-Sabal E, Smith B, Younger J, Balis U, Michaelson J, Bhan A, Habin K, Baer TM, Brugge J, Haber DA, Erlander MG, Sgroi DC. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de RM, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lønning PE, Børresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, Liu ET. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Trent S, Ionescu-Tiba V, Lan L, Shioda T, Sgroi D, Schmidt EV. Identification of cyclin D1- and estrogen-regulated genes contributing to breast carcinogenesis and progression. Cancer Res. 2006;66:11649–11658. doi: 10.1158/0008-5472.CAN-06-1645. [DOI] [PubMed] [Google Scholar]
- Desmedt C, Sotiriou C. Proliferation: the most prominent predictor of clinical outcome in breast cancer. Cell Cycle. 2006;5:2198–2202. doi: 10.4161/cc.5.19.3254. [DOI] [PubMed] [Google Scholar]
- Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, Desmedt C, Larsimont D, Cardoso F, Peterse H, Nuyten D, Buyse M, Van de Vijver MJ, Bergh J, Piccart M, Delorenzi M. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- Ivshina AV, George J, Senko O, Mow B, Putti TC, Smeds J, Lindahl T, Pawitan Y, Hall P, Nordgren H, Wong JEL, Liu ET, Bergh J, Kuznetsov VA, Miller LD. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]