Short abstract
TG dinucleotides functioning as alternative 3' splice sites were identified and experimentally verified in 36 human genes.
Abstract
Background
Despite some degeneracy of sequence signals that govern splicing of eukaryotic pre-mRNAs, it is an accepted rule that U2-dependent introns exhibit the 3' terminal dinucleotide AG. Intrigued by anecdotal evidence for functional non-AG 3' splice sites, we carried out a human genome-wide screen.
Results
We identified TG dinucleotides functioning as alternative 3' splice sites in 36 human genes. The TG-derived splice variants were experimentally validated with a success rate of 92%. Interestingly, ratios of alternative splice variants are tissue-specific for several introns. TG splice sites and their flanking intron sequences are substantially conserved between orthologous vertebrate genes, even between human and frog, indicating functional relevance. Remarkably, TG splice sites are exclusively found as alternative 3' splice sites, never as the sole 3' splice site for an intron, and we observed a distance constraint for TG-AG splice site tandems.
Conclusion
Since TGs splice sites are exclusively found as alternative 3' splice sites, the U2 spliceosome apparently accomplishes perfect specificity for 3' AGs at an early splicing step, but may choose 3' TGs during later steps. Given the tiny fraction of TG 3' splice sites compared to the vast amount of non-viable TGs, cis-acting sequence signals must significantly contribute to splice site definition. Thus, we consider TG-AG 3' splice site tandems as promising subjects for studies on the mechanisms of 3' splice site selection.
Background
Intervening sequences (introns), primary transcript regions that are removed during mRNA maturation, are an outstanding feature of eukaryotic gene structure. Introns are excised through two transesterification reactions involving the collaboration of five different small nuclear ribonucleoprotein particles and additional proteins that associate to form the spliceosome. Rearrangements of the spliceosome and, consequently, splicing catalysis is driven by the sequential action of ATP-dependent helicases [1,2]. The assembly of the early spliceosomal complex relies on sequence-specific contacts between the intron terminal regions and the spliceosome subunits U1, U2, and U2AF [1,3,4]. From accumulating intron sequence data, it was noted that invariant dinucleotides must represent important signals for the definition of intron termini, the so-called GT-AG rule (for simplicity, we use the nucleotide symbol T to denote thymidine in DNA as well as uridine in RNA sequences). With respect to the role of intron termini in the transesterification reactions, the 5' GT site and the 3' AG site were named donor and acceptor splice sites, respectively.
Early work on unusual splice signals revealed introns with the terminal dinucleotides AT-AC [5], and these were later shown to be processed by an independent splicing pathway, the U12 spliceosome. The U12 spliceosome recognizes highly specific donor site and branchpoint motifs [6] while recognition of 3' splice sites is rather unspecific. As a result, there are several variants of intron termini besides the prominent combinations GT-AG and AT-AC [7,8].
Among U2-dependent introns, the most frequent exception to the GT-AG rule are GC-AG intron termini, which comprise 0.7-0.9% of vertebrate introns [5,8,9]. Other rare exceptions are GA-AG intron termini in the FGFR gene family [10] and AT-AC termini, mostly found in introns of the SCN gene family [8,11,12]. While the latter cases are the only reported non-AG 3' splice sites, results from in silico studies have repeatedly suggested that other unusual 3' splice sites occur in U2-type introns [13,14]. An in-depth, systematic screening effort could not reveal significant evidence for additional unusual intron 3' termini above the noise level brought by annotation errors [9]. However, it was noted that a few exceptional U2-spliceosomal introns exist that involve unusual 3' splice sites in scenarios of alternative splice site choice. For example, intron 3 of the human guanine nucleotide binding protein gene GNAS is spliced at either TG or AG in the 3' intron sequence CTGCAG [15,16]. Remarkably, the homologous Drosophila gene shows the same unusual splicing pattern for another intron [17]. Moreover, unusual TG splice acceptors appear to be involved in alternative splicing of the human gene for presynaptic density protein 95, DLG4 [18], and the human dopamine D2 receptor gene, DRD2 [19].
We have previously reported a widespread type of alternative splicing mediated by the tandem splice acceptor motif NAGNAG [20]. From the analysis of single-nucleotide polymorphisms (SNPs) we concluded that a NAGNAG motif is necessary and sufficient to explain three-nucleotide variant splicing at intron-exon boundaries [21]. In contrast, alternative splicing of an intron 3' terminus in the GNAS gene appears to occur independently of a NAGNAG motif. Furthermore, it has been suggested that unusual splice sites could be selectively involved in alternative splicing [5,9], although this was never examined in detail. Here, we report a systematic screening of the human transcriptome that identified 36 introns with bona fide TG 3' splice sites. These TG splice sites are exclusively found as alternative 3' splice sites, each associated with a canonical AG 3' splice site. The evolutionary conservation of these introns and their alternative splicing patterns indicate physiological relevance and point to the requirement for cis-regulatory sequence elements to promote usage of TG 3' splice sites.
Results
Prior considerations
We used an in silico approach based on expressed sequence tags (ESTs) to identify unusual 3' splice sites that are found in pairs of 3' splice variants. ESTs, as first-pass results from high-throughput cDNA sequencing projects, are clearly prone to errors. Therefore, we assumed that single ESTs are insufficient to indicate genuine subtle splice variants since technical artifacts contribute to false positives. We considered a variant as sufficiently evident if it is supported by at least two independent ESTs, and expect the EST variant ratio to serve as an approximation for the natural ratio of splice variants. An additional threshold was applied to the relative abundance of splice variants, since our experimental approach, that is, sequencing of 100 individual RT-PCR clones, had a detection limit. Using a random binomial distribution to model the occurrence of splice variants in the RT-PCR clones, we calculated a diagnostic power of 95% (β error 5%) if a splice variant occurs with at least 3% frequency. It is important to note that we have not inferred anything for the cases that failed the threshold criteria. It is possible that they actually represent natural splice variants; however, the evidence for such cases is weak and the experimental approach did not provide sufficient sensitivity for validation.
TG dinucleotides function as non-canonical alternative 3' splice sites
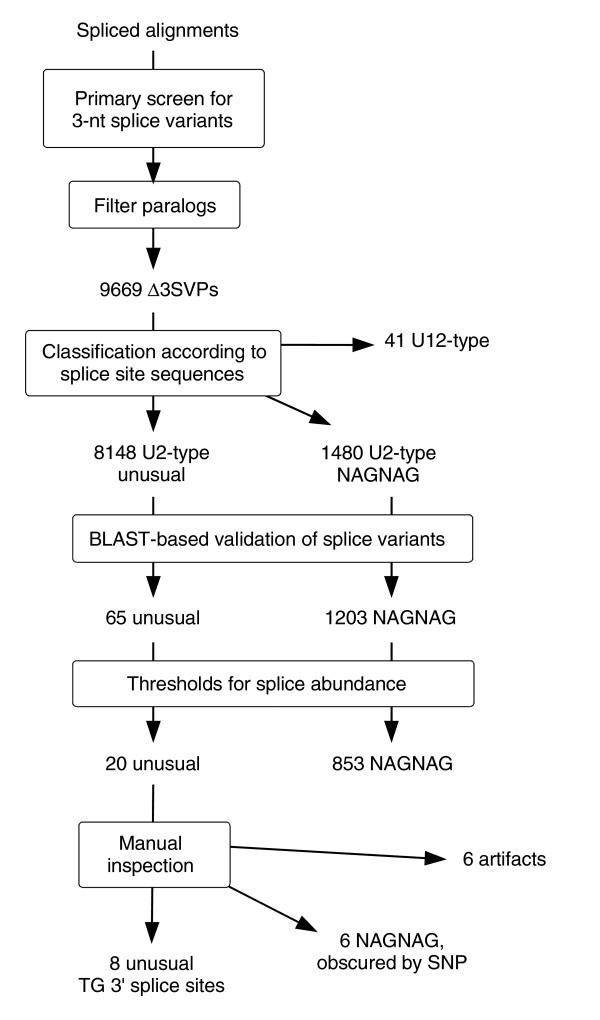
We initially aimed to identify unusual 3' splice sites that are found in pairs of 3' splice variants that differ by 3 nucleotides (nt), such as in the GNAS intron 3 splice site tandem [15,16]. Identification of 3 nt splice variant pairs (Δ3SVPs) was based on 3' splice sites as indicated by spliced alignments of human ESTs [22]. After a reduction of false positives performed by a series of filtering steps (Figure 1), we identified 65 'unusual' Δ3SVPs that were supported by high-quality local EST alignments. Of these, 20 meet the requirements that the minor splice variant is supported by at least two ESTs and 3% of the matching ESTs (see considerations below). However, after close inspection and re-sequencing, we identified 6 of the 20 unusual Δ3SVPs as false positives (Additional data file 1), explained by: 3 nt deletion variants due to sequencing errors; mouse ESTs erroneously attributed to human; or alignment artifacts. Another six Δ3SVPs can be explained by SNPs, where the SNP allele corresponding to a NAGNAG splice site motif is not displayed by the human reference genome sequence [21]. Strikingly, all the remaining eight Δ3SVPs suggest that TG dinucleotides function as alternative 3' splice sites (Table 1).
Figure 1.
Screening procedure for unusual 3' splice sites found in pairs of 3' splice variants that differ by 3 nt (Δ3SVPs). Processing of AG-AG tandem cases ('NAGNAG', parallel branch on the right) was performed as a comparison to unusual 3' splice site tandems.
Table 1.
Unusual TG splice acceptors identified in the human transcriptome
| Intron | 3' Splice site pair | ESTs for unusual 3' splice sites | ||||
| Gene | No. | Length | Distance | Motif | Fraction | No. |
| GNAS | 3 | 7843 | 3 | CTG,CAG| | 0.15-0.62*† | 282 |
| PCGF2 | 1 | 224 | 3 | AAG|ATG, | 0.50 | 4 |
| CNBP | 3 | 168 | 3 | TTGTTG,AAG| | 0.25 | 257 |
| 3 | 168 | 6 | TTG,TTGAAG| | 0.01 | 10 | |
| FBXO17‡ | 3 | 1999 | 3 | CAG|ATG, | 0.14 | 4 |
| C21orf63 | 3 | 9975 | 3 | CAG|ATG, | 0.09 | 2 |
| BRUNOL4 | 6 | 1147 | 3 | CAG|CTG, | 0.07 | 2 |
| PCID2 | 2 | 2162 | 3 | CAG|ATG, | 0.04 | 7 |
| TNNT2 | 1 | 4354 | 3 | TTG,GAG| | 0.04 | 2 |
| CACNA1A | 9 | 2532 | 3 | TTGTTG,GAG| | ?† | - |
| 9 | 2532 | 6 | TTG,TTGGAG| | 0.17† | - | |
| GPBP1 | 9 | 1377 | 4 | CAG|GATG, | 0.03 | 2 |
| KIAA0494 | 1 | 1459 | 5 | TTG,AGCAG| | 0.09 | 2 |
| OSBPL8 | 2 | 36530 | 6 | CTG,TTGTAG| | 0.11 | 2 |
| SAP30 | 1 | 1892 | 6 | CTG,TTTCAG| | 0.04 | 2 |
| DRD2 | 6 | 1485 | 6 | CTG,GTGCAG| | 0.02† | 5† |
| SUV420H2 | 5 | 134 | 7 | CTG,GCTCCAG| | 0.20 | 3 |
| SSRP1 | 1 | 489 | 7 | TTG,AATTCAG| | 0.20 | 16 |
| FREQ | 2 | 16849 | 7 | CTG,CCTCCAG| | 0.04 | 2 |
| IL21 | 3 | 2753 | 8 | TTG,ATTTCTAG| | 0.13 | 2 |
| RYK | 7 | 3107 | 9 | TTG,GCTCCTTAG| | 0.77 | 27 |
| DLG4 | 5 | 131 | 9 | CTG,GAGTTGCAG| | 0.62 | 8 |
| SMARCA4 | 29 | 6174 | 9 | TTG,ACCCTGAAG| | 0.41 | 34 |
| FBXL10 | 15 | 177 | 9 | TTG,GCCTACAAG| | 0.21 | 3 |
| HNRPR | 7 | 2839 | 9 | TTG,GTTTAACAG| | 0.13 | 15 |
| RRAD | 1 | 214 | 9 | CTG,ATCCCCTAG| | 0.06 | 2 |
| TGM1 | 6 | 454 | 10 | CTG,TCCTGGGCAG| | 0.13 | 2 |
| ALAS1 | 11 | 1599 | 11 | CTG,TTTCTCCTCAG| | 0.04 | 5 |
| ARS2 | 18 | 182 | 12 | TTG,TACTCCCCCCAG| | 0.74 | 75 |
| PCBP2 | 7 | 1337 | 12 | CTG,ACTCTCTCCCAG| | 0.43 | 169 |
| PTPN11 | 10 | 4269 | 12 | TTG,GCTCTACTCCAG| | 0.33 | 3 |
| MSH5 | 6 | 164 | 12 | CTG,ATCCCCTCCCAG| | 0.25 | 5 |
| SYTL2 | 9 | 1259 | 13 | TTG,CCCTCCTGAGTAG| | 0.09 | 3 |
| TOMM40 | 1 | 95 | 16 | CTG,ACCTCTCCCCTAGCAG| | 0.07 | 2 |
| MARK3 | 3 | 20478 | 17 | TTG,TTTGTTTTTTTTTTTAG| | 0.07 | 3 |
| BAT3 | 6 | 832 | 18 | CTG,ACTCTCCCCTACCTTCAG| | 0.01 | 1 |
| SH3D19 | 6 | 838 | 21 | TTG,GTTTTGTTTTGGTCTCGTCAG| | 0.07 | 1 |
| LOC346653 | 1 | 3097 | 27 | CTG,ACCCATGTACCTGAGGCTGATTTCCAG| | 0.60 | 3 |
| ACAD9 | 10 | 253 | 28 | TTG,TTTCTTGTGTTTTTTCTGAACACTCCAG| | 0.09 | 4 |
Entries in bold have RefSeq transcripts supporting the unusual TG acceptor site. Each TG splice variant is supported by at least two ESTs and at least 3% of all covering ESTs, except for some RefSeq-supported cases, CACNA1A [24,35], DRD2 [19] and BAT3. In the 'Motif' column, a vertical line (|) indicates a canonical splice site, and a comma (,) marks the TG splice site. Splice ratios are given as absolute EST counts (No.) as well as the fraction of TG splice variants. A question mark indicates that an explicit fraction is not given in the referenced article, although the authors performed quantitative experiments. *EST ratio depends on the exon junction; the upstream exon 3 may be skipped. †Splice variants were previously quantified by others: GNAS [16,26], CACNA1A (splice ratio cited from [24,35]), DRD2 (splice ratio cited from [19]). ‡Alternative splicing at FBXO17 intron 3 was not experimentally reproducible in this study.
Since all the unusual alternative Δ3-nt splice acceptors identified display TG dinucleotides, we investigated their occurrence in a wider scope. Analogously to the screen for Δ3SVPs (Figure 1), we performed a search for alternative TG splice acceptors at larger distances, up to 36 nt from from the canonical splice site. The same filter procedures were applied, and close inspection did not reveal obvious artifacts or explanatory SNP alleles. We identified 26 additional EST-supported splice variant pairs that suggest alternative TG splice acceptors functioning in U2-dependent introns (Table 1).
We sought to screen for unusual splice acceptors using an independent approach in order to cross-validate our initial findings and to make a link to previous studies that were primarily based on curated transcript data [5,9]. An analysis of RefSeq-to-genome alignments identified 122 putative introns with terminal TG dinucleotides (120 unique genomic sites) out of 228,925 total introns (171,605 unique genomic sites). Of these, 39 introns have a canonical GT donor dinucleotide (Additional data file 1). A previous study, performing a similar screening approach for unusual splice sites using curated transcript data, showed high enrichment of annotation artifacts [9]. Therefore, we checked the identified TG acceptor cases thoroughly. In fact, cases failed this quality check for several reasons: known SNPs masking existing canonical AG splice sites; RefSeqs lacking transcript evidence; and misleading RefSeq-to-genome alignments. Since the overall false positive rate seemed very high, we additionally required independent transcript entries (mRNA or EST) to support the unusual splice site. In summary, 9 of the 39 RefSeqs showed robust support for unusual TG acceptor sites (Table 1, entries in bold), and 6 of these overlap with cases obtained from the EST screening approach while the others are exclusively identified by the RefSeq-based approach (SH3D19, BAT3, CACNA1A). Intriguingly, these three EST-independent RefSeq-supported cases all comprise 'alternative' splice sites, although this was not a screening criterion. Taking into account that about 1% of all introns have alternative 3' termini [23], this strongly indicates that TG splice acceptors are functionally linked to nearby AG splice sites and cannot function in a constitutive manner (P = 0.000001, binomial test). Altogether, the two screens identified 37 introns with 39 alternative TG splice acceptors (Table 1).
Negation of genome sequence errors and polymorphisms
Since six putatively unusual 3' splice sites can be explained by SNP-affected NAGNAG acceptors (which were filtered; Figure 1), we asked whether undiscovered SNPs, or even inaccuracies in the available human genome sequence, may explain some of the remaining candidates. The genomic sequence of the splice site regions of GNAS and CACNA1A had been experimentally verified by others [16,24]. For 10 other genes (listed in Table 2), we analyzed PCR products obtained from genomic DNA, pooled from 100 individuals, for sequence variations. The re-sequenced genomic regions were in perfect agreement with the available genome sequence, negating the possibility that unusual splice sites are trivial sequencing errors (data not shown). Moreover, we identified no SNP alleles that confer explanatory AG splice sites on any of the observed unusual splice variants, demonstrating that the TG splice sites are real and genetically invariant.
Table 2.
Validation and quantification of alternative splice variants
| Splice junction | Tissue | Fraction of TG splice | Method | |
| GNAS | Intron 3, exon junction 3-4 | Leukocytes | 0.14 | n = 115 |
| PCGF2 | Intron 1 | Placenta | 0.99 | n = 89 |
| CNBP | Intron 3, indel AAG | Leukocytes | 0.52 | n = 69 |
| Brain | 0.38 | n = 58 | ||
| Placenta | 0.33 | n = 70 | ||
| Intron 3, indel TTGAAG | Leukocytes | 0.01 | n = 69 | |
| FBXO17 | Intron 3 | Leukocytes | - | n = 96 |
| Liver | - | n = 151 | ||
| C21orf63 | Intron 3 | Leukocytes | 0.09 | n = 110 |
| Brain | - | Direct sequencing | ||
| BRUNOL4 | Intron 6 | Lung | - | n = 90 |
| Brain | 0.19 | n = 92 | ||
| PCID2 | Intron 2 | Leukocytes | 0.02 | n = 142 |
| TNNT2 | Intron 1 | Heart | 0.03 | n = 91 |
| CACNA1A | Intron 9, indel GAG | Brain | 0.85 | n = 90 |
| Intron 9, indel TTGGAG | Brain | 0.03 | n = 90 | |
| ARS2 | Intron 18 | Leukocytes | 0.55 | n = 37 |
| PCBP2 | Intron 7 | Leukocytes | 0.54 | n = 47 |
| LOC346653 | Intron 1 | Testis | 0.50 | Direct sequencing |
In the 'Methods' column, n represents the number of subclones sequenced.
Validation and quantification of splice variants
To verify the existence of TG-derived splice variants, we performed RT-PCR experiments designed to yield amplicons that cover the exon-exon junctions under consideration. Cloning of the PCR products and sequencing allowed us to detect splice variants. Subclassification and counting of clones gave measures of splice variant ratios. This way, the alternative splicing pattern was reproduced and quantified for 11 out of 12 analyzed cases (Table 2). Generally, the splice ratios obtained from clone counting agree well with the EST data. The observed deviations can be explained by significant fluctuations depending on the analyzed tissue (C21orf63, BRUNOL4, and CNBP in Table 2). The splice variant validation failed for FBXO17, a gene for which 4 out of 29 ESTs had suggested a TG-derived splice variant. All supporting ESTs originated from the same EST library, NIH_MGC_100, derived from a hepatocellular carcinoma. A peculiarity of the source material, either the NIH_MGC_100 cell line or the single-individual liver sample used for our RT-PCR experiments, may be the reason for the inconsistent results concerning this putative splice variant. This example illustrates that at least two ESTs from independent sources are required to indicate a natural splice variant with high reliability. Overall, the success rate of the validation experiments was high (92%), and extrapolated to the 25 non-tested cases, about 2 false positives are expected.
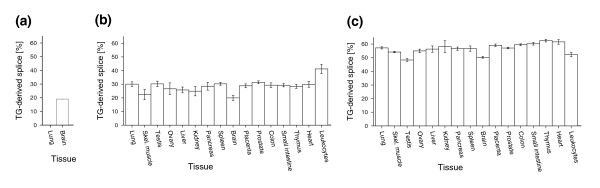
Tissue-specificity of splice ratios
According to the results of the PCR cloning approach, BRUNOL4 displayed remarkable tissue-specific splice ratios. The TG-derived splice variant was not detected in lung cDNA whereas the same variant constituted 20% of brain BRUNOL4 transcripts (Table 2, Figure 2a). So we asked if splice ratios of TG-derived and AG-derived variants generally show tissue-specific differences. We analyzed the splice variant ratios more extensively in other genes using pyrosequencing, a method that allows accurate and cost-effective quantification of mixtures of polymorphic DNA populations [25,26]. ARS2 and CNBP, both having a ubiquitous expression profile, show tissue-dependent fluctuations in splice ratios (55-65% and 20-40%, respectively; Figure 2). While these differences are numerically significant in each of these genes (α = 0.01, ANOVA), their biological relevance is debatable. We conclude that splice ratios of TG-AG tandems are tissue-specific for particular introns but are rather stable for others.
Figure 2.
Tissue-specific fractions of TG-derived splice variants. (a) BRUNOL4 (values are as shown in Table 2); (b) CNBP; and (c) ARS2. Pyrosequencing assays (for (b,c)) were performed multiple times for each sample (two to four times). Error bars depict the standard deviation of individual measurements.
Evolutionary conservation of introns with TG 3' splice sites
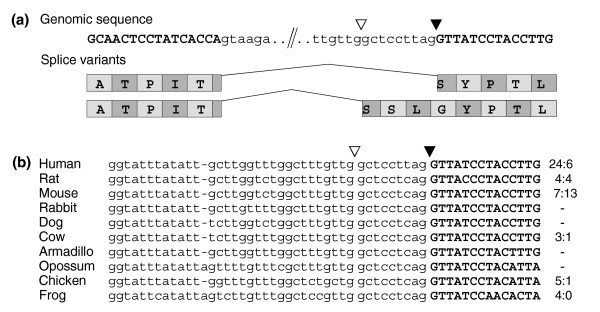
Since splicing at TG sites occurs in a very small number of introns, one might argue that these represent 'accidental' events attributable to spliceosome dysfunction. To address this question, we first analyzed the conservation of splice sites in homologous introns as an indication of alternative splice variants being under purifying selection. Out of 36 introns with 3' TG splice sites (37 minus the false-positive FBXO17 intron), 26 (72%) are conserved between the human and mouse genomes. In 14 of these cases (39% of the total), mouse ESTs indicate homologous TG-derived splice variants. For comparison, this rate is three- to four-fold higher than that of alternative exons found in both human and mouse [27-29]. In some cases, EST evidence for orthologous TG-derived splice variants even exists for distantly related species, such as chicken (CNBP, BRUNOL4, RYK), and frog or fish (BRUNOL4, RYK, FBXL10). An outstanding example of conserved intron sequence and homologous splice variants is intron7 of the RYK gene (Figure 3). The ratio of 3' splice variants is remarkably similar between human and chicken, as can be inferred from the available EST data (EST ratios of 24:6 and 5:1 for human and chicken, respectively). In general, it should be noted that homologous splice variants may remain undetected due to the limited depth of EST coverage [23,27].
Figure 3.
Conservation of the TG splice site found in intron7 of the RYK gene from human to chicken. (a) Human genomic sequence and derived splice variants. Canonical (filled triangle) and TG 3' splice site (open triangle) are marked. (b) Alignment of orthologous exon-intron boundary regions from several vertebrate genomes, splice sites highlighted as in (a). Numbers on the right display the ratios of species-specific ESTs for the TG and AG splice sites, respectively.
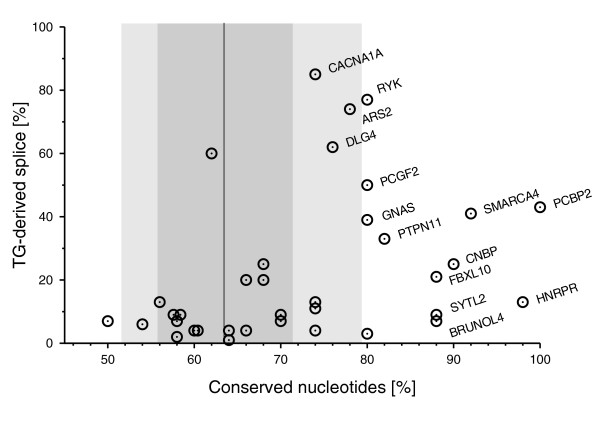
Independently, we analyzed intron sequence conservation as an indication of the functional relevance of alternative splicing [27-29]. A data set of human-mouse orthologous intron-exon boundaries was used to determine the degree of conservation within a 50 nt intron sequence upstream of the splice acceptor, or acceptor tandem. Intronic flanks of TG splice sites show an average sequence similarity of 74%, whereas flanks of AG splice sites within canonical (AG-only) introns are 65% similar on average (P << 0.00001, permutation test). A plot of flanking sequence conservation against the abundance of the TG-derived splice variant (Figure 4) shows that these two measures are positively correlated. Introns that give rise to less than 10% TG-derived splice variants have an average human-mouse intron sequence identity of 64%, indistinguishable from canonical introns. In contrast, introns with a TG-derived splice variant making up more than 10% of the transcripts show an average sequence identity as high as 80%. This parallels a previous finding that the abundance of splice variants correlates with sequence conservation of alternative exons [27]. Consistently, high intron sequence conservation is strongly correlated with conservation of the TG splice site (13 of the 14 cases with gene labels in Figure 4). Our results indicate that splicing at TG acceptors may arise from neutral evolution, presumably showing low splicing efficiency. However, efficiently spliced TG 3' splice sites seem to evolve and to be maintained by evolutionary selection.
Figure 4.
Intron flank conservation of TG-AG splice acceptor tandems. Orthologous human/mouse intron-exon boundaries involving TG splice sites are displayed in a two-dimensional plot according to two properties: horizontal axis = sequence identity of 50 nt sequence upstream of both splice sites; vertical axis = relative abundance of the TG-derived splice variant, as reflected by the fraction of TG-spliced ESTs (except for CACN1A1, where the data are taken from Table 2). Data points are labeled with the gene symbol if the conservation score and/or the fraction of TG-derived splice variant are significantly high. Conservation properties of canonical introns are indicated by shaded intervals: black line = median; dark gray = 66% percentile; light gray = 90% percentile.
Structural and sequence characteristics of TG 3' splice sites
We analyzed the context properties of TG 3' splice sites in order to find an explanation for these rare exceptions to the GT-AG rule. With regard to the gene structure context, TG-splicing introns are indistinguishable from canonical introns in several respects: length of the affected as well as the downstream introns, and length of the upstream and downstream exons (data not shown). TG 3' splice sites are significantly often found in the first intron of the gene; 8 of 36 (22%) TG-AG introns compared to 11% of other introns (P = 0.02, Fisher's Exact). This bias is also found for AG-AG splice site tandems and is certainly due to neutral evolution of introns located in the 5' untranslated transcript region.
TG 3' splice sites were exclusively found within a context of alternative splice site choice (Table 1). This is clearly significant for results from the RefSeq-based screening procedure, which are unbiased with respect to constitutive or alternative transcripts. Taking into account that about 1% of all introns have alternative 3' termini [23], it strongly indicates that TG 3' splice sites are functionally linked to nearby AG splice sites and cannot function in a constitutive manner. This conclusion is further supported by studies of human AG>TG 3' splice site mutants [30-32], which always resulted in activation of neighboring AG splice sites, but not splicing at TG. Furthermore, we observed that TG-AG tandems display a splice site distance restriction with a limit of 28 nt, which is not seen for AG-AG tandems (Table 1, Additional data file 1). Thus, splicing of a 3' TG does not depend only on an additional AG splice site, this dependency also seems to pose a constraint on TG-AG splice site distance. The observed distance limit corresponds well with the distance between the branch site and the 3' splice site, which is typically 20-40 nt [33].
Splice site strength was scored using the maximum entropy method 'maxent' of Yeo and Burge [34]. The 5' splice site scores are indistinguishable between TG-AG introns and canonical introns. On the other hand, the AG 3' splice sites in TG-AG tandems score significantly lower than canonical introns (6.1 ± 3.5 versus 8.5 ± 2.8, respectively; P = 0.00001, Student's t-test). The 3' splice site score remains significantly small if low-scoring outliers are excluded from the analysis (6.7). Interestingly, the sequence context of TG 3' splice sites is very similar to canonical AG splice sites in that it shows a preference for pyrimidines at position -3 and preference for purines at position +1 (Additional data file 1). However, TG 3' splice sites changed to AG yield an average score of 6.6, again significantly lower than that of canonical AG splice sites (P < 0.00001, Student's t-test). This disfavors the simple explanation that TG and AG splice sites compete for the same recognizing factors and the neighboring nucleotide composition (that is, the feature scored by maxent) alone acts to direct splice site choice towards TGs. Finally, it remains questionable if maxent, trained on canonical AG 3' splice sites, has any predictive power for the functionality of TG splice sites.
The fraction of TGs functioning as splice acceptors is extremely small, about 0.01% of candidate motifs. Thus, cis-regulatory elements must play a crucial role in the definition of TG splice acceptors. From the ratio of functional/non-functional TG-AG tandems, in comparison with AG-AG tandems, we estimate that at least 6 nt of cis-regulatory sequence information is required to promote splice site usage of 3' TGs (Additional data file 1). This is in agreement with 5-20 unchanged nucleotides in excess over the average intron mutation rate, found in about half of the TG-splicing introns, that is, those considered to be subject to purifying selection for splicing of 3' TGs. However, we failed to identify specific regulatory motifs (data not shown). This may be due to the dispersed arrangement of cis-regulatory elements, or the contextual cooperation of diverse elements. Due to the relatively small sample size for TG 3' splice sites, available methods for motif discovery have limited detection power.
Discussion
Previous studies provided incidental evidence for unusual 3' terminal dinucleotides in U2-dependent introns, particularly TG dinucleotides that are used as alternative 3' splice sites. Few directed efforts have been made so far to verify such instances and to elucidate underlying mechanisms and consequences [16,35]. Here, we report 36 human U2-spliceosomal introns with TG dinucleotides functioning as 3' splice sites, identified by thoroughly filtered EST-to-genome alignments. The high accuracy of the EST-based screening approach was validated by RT-PCR with a success rate of 92%. Though it might seem paradoxical, the analysis of EST data gave superior results compared to an analysis of curated data, that is, RefSeq transcripts. We found that the abundance of EST data allows the application of statistical methods for obtaining valid results whereas curated data sets, which are typically devoid of redundancy, may contain errors that are rarely captured by filtering criteria, consistent with the findings of others [36]. In practice, we found that two independent ESTs are strong evidence for a natural splice variant. Given this rather permissive threshold [9,37], we expect that the established screening protocol achieves high sensitivity.
Since our screening procedure is EST-based, certainly more unusual 3' splice sites remain undiscovered in transcript regions that lack sufficient EST coverage. Moreover, there are indications that even other unusual dinucleotides, apart from TG, may function as alternative 3' splice sites. For example, others reported an AT 3' splice site in the mammalian DGCR2 gene [8], a CG 3' splice site in the Drosophila per gene [38], and we found that a TG splice acceptor in human CNBP intron 3 is replaced by a viable GG in the chicken ortholog (results not shown). The occurrence of a TG splice acceptor in the Drosophila gnas gene suggests that they occur throughout metazoan organisms.
Other studies have questioned the extent to which alternative splicing is functionally relevant [27-29]. Since TG splice acceptors are extremely rare compared to AG acceptors, one might think that these cases reflect a fuzziness of the splicing reaction. However, multiple findings support the idea that TG splice sites are activated by directed mechanisms and that the resulting splice variants fulfill functional roles: first, several TG splice acceptors are used with a high frequency or can even be the preferred splice site, which excludes splicing errors as a plausible explanation (Table 1, Figure 4); second, TG splice acceptors and their adjacent intron sequence are remarkably conserved between orthologous mammalian genes (Figure 4); third, tissue-specific splice patterns are observed for GNAS [16,26] as well as BRUNOL4 (this study; Figure 2), suggestive of specific regulatory processes; and fourth, the TG splice site-mediated protein isoform of the mammalian calcium channel subunit α1A (CACNA1A) has been shown to result in significant differences in neuronal excitability [35].
Thinking of splice site evolution as a process of functional engineering, we might ask about the functional options that distinguish TG-AG splice acceptor tandems from AG-AG tandems. During analysis of orthologs of human TG splice acceptors, we did not identify any case of orthologous AG splice sites, suggesting that TG and AG splice site dinucleotides are functionally non-equivalent. The inserted/deleted nucleotide sequence differs only if TG is positioned downstream of the tandem splice site. Apart from the possible impact on the protein sequence, an NAGATG tandem acceptor allows insertion of a start codon. For example, this seems to be realized in intron 1 of human PCGF2, where the observed splice variants differ by the presence of an upstream open reading frame. Preliminary results indicate that this ATG insertion has an effect on the translation efficiency of the mRNA (results not shown). It is also worth noting that the Drosophila gnas gene has a TG splice acceptor, like the human gene, but it is located in a non-homologous intron [17]. Given the overall low frequency of TG 3' splice sites (0.02%), this example of convergent evolution indicates a functional benefit of the unusual splice site, independent of its impact on protein sequence. It is tempting to speculate that splicing of TG splice acceptors, rather than providing a pathway for alternative transcripts or protein isoforms, may play a role as a regulatory bottleneck for maturation of the transcript, as was suggested for U12-type introns [39].
Considering functional classes, a significant fraction of TG-spliced genes represent regulators of chromatin structure (PCGF2, GPBP1, SAP30, SUV420H2, SSRP1, SMARCA4) as well as splicing factors and translational modulators (CNBP, BRUNOL4, HNRPR, PCBP2). Interestingly, two of the affected RNA-binding proteins are reported to bind DNA as well [40,41]. Together, these enrichments suggest a regulatory cross-talk between transcription on the one hand, and splicing, mRNA maintenance, and translation on the other. Together with another subgroup associated with receptor-mediated signal transduction (GNAS, DRD2, FREQ, IL21, RYK, DLG4, RRAD, PTPN11, SYTL2, MARK3, SH3D19), most of the genes' functions may be circumscribed with 'information processing', a term that was introduced to describe the functional characteristics of U12-dependent introns [6]. However, as a statistical analysis of Gene Ontology functional classification terms does not reveal any significant over- or under-representation (results not shown), further work is required to determine the relevance of these findings.
TG-AG splice acceptor tandems illustrate the flexibility as well as the specificity of splice site selection by the U2-type spliceosome. The spliceosome is flexible enough to choose TG dinucleotides as splice acceptors. Despite this flexibility, a TG splice site depends on a neighboring AG splice acceptor, since constitutive TG splice acceptors are not found, and TG-AG acceptor tandems show a distance constraint. We assume that an AG splice acceptor, within the typical context of a branch-point motif and polypyrimidine tract, is essentially required for intron definition to promote splicing stepI in vivo. Consistent with this, a recent report showed that the essential splicing factor U2AF35 in cooperation with other factors mediates the spliceosome's specificity for AG 3' intron termini during splicing stepI [42]. Assuming that splicing stepI does not ultimately define the 3' splice site, we hypothesize that definite splice site choice takes place during reaction stepII, allowing TG dinucleotides to function as 3' splice sites. Since U2AF dissociates from the spliceosomal complex after stepI [43,44], other factors may influence splice site choice at a later step. Two different modes of 3' splice site selection after splicing stepI have been suggested for AG-AG splice site tandems. First, a second 3' AG may be chosen as the site of exon ligation during splicing stepII if it is located a few nucleotides downstream of the first-step AG, defined by U2AF binding [45]. This rather unspecific mechanism is the likely explanation for the high propensity of small-distance AG-AG tandems to result in alternative splicing, and may also be relevant for TG-AG acceptor tandems, which are found overrepresented at a 3-nt distance compared to larger distances (Figure S1 in Additional data file 1). Another mechanism is exemplified by intron 2 of the Drosophila sxl gene [46] as well as intron 1 of the β-globin mutant β110 [47,48]. Here, the downstream AG is essential for splicing while the dispensable upstream AG may be chosen in splicing stepII, even as the preferred splice site. The splicing factor SPF45 was shown to bind to the upstream AG dinucleotide during splicing stepII, promoting splice site choice [46]. It remains to be tested if SPF45 or other factors contribute to TG splice site choice.
Given the extremely low ratio of viable versus non-viable TG-AG tandems at intron-exon boundaries, contextual sequence signals must contribute to TG splice site definition and influence splice site choice. In agreement, half of the TG splice acceptors are associated with outstandingly high intron sequence conservation. Notably, the alternative TG splice acceptor of GNAS intron 3 has been shown to be flanked by three putative exonic splice enhancer motifs (specific for SF2/ASF, SC35, and SRp40), and TG splice site choice has been experimentally shown to be modulated by the ratios of SF2/ASF and hnRNPA1 [16]. We could not identify specific sequence motifs associated with TG splice sites (results not shown). Due to the relatively small sample size for TG 3' splice sites, available methods for motif discovery have limited detection power, especially if cis-regulatory elements are highly dispersed, or if diverse elements cooperate in a contextual manner. Presumably, each individual TG-AG tandem recruits a characteristic ensemble of splice regulators to facilitate unusual splice site choice. Thus, the compilation of TG splice sites could serve as a rich source of splicing-relevant contextual sequence signals to be examined in future experimental studies.
Materials and methods
Screening for non-canonical 3' splice sites
From the UCSC Genome Browser site [22] we obtained spliced alignments of human ESTs (file all_est.txt, released 2005-07-14) and of human RefSeq transcripts (refGene.txt, 2005-07-23) [49], as well as a compilation of all human EST sequences (est.fa, 2005-11-26). First, we sampled EST-supported 3' splice sites to identify 3-nt splice variant pairs (Δ3SVPs). In parallel, we identified ESTs that were mapped to multiple genome locations, indicative of paralogous gene loci including pseudogenes. We discarded those Δ3SVPs whose EST support for the minor splice variant did not exceed the number of these ambiguously mapped ESTs. Furthermore, we retained only those Δ3SVPs that have at least one splice site corresponding to a RefSeq transcript, according to the RefSeq-to-genome alignment. Then, we separated cases that involve the dinucleotide AG at both 3' splice sites, that is, NAGNAG tandem splice acceptors, as well as U12-dependent introns, identified by their characteristic donor site and branch-point motifs [6]. The remaining Δ3SVPs were considered 'unusual' since they comprise at least one non-AG splice acceptor in a U2-spliceosomal intron. The splice variants of these Δ3SVPs were validated and quantified by a WU-BLASTN search of 60-nt sequence windows around the resulting exon-exon junctions against all human ESTs, using parameters W = 13, N = -8, nogap S = 180, hspmax = 1. BLAST matches were considered valid if perfect sequence identity was found in a 12-nt window around the exon-exon junctions [37]. Finally, Δ3SVPs were considered highly reliable if the minor 3' splice site was found in at least two ESTs and was used in at least 3% of the covering ESTs. A screen for splice variant pairs for distances of 4-36 nt was performed analogously, restricting the search to tandems of TG-AG splice sites.
PCR and RT-PCR
For validation of splice variants, nested PCR was performed using 1 ng cDNA templates from the Human Multiple Tissue cDNA PanelsI and II (Clontech, Mountain View, CA, USA). For a given gene, suitable tissues were determined from expression data obtained from the Stanford SOURCE database [50]. However, pooled leukocyte cDNA from 200 individuals was preferably chosen in order to obtain comparable results. Verification of the genomic sequence and an analysis of potential polymorphisms were done by nested PCR using 200 ng of pooled genomic DNA from 100 Caucasian individuals (Roche, Mannheim, Germany) as template. Primers were obtained from Metabion (Martinsried, Germany) (Additional data file 1). Reactions were set up with PuReTaq Ready-To-Go PCR beads (GE Healthcare, Munich, Germany) and 10 pmol primer in 25 μl total volume, according to the manufacturer's instructions. A typical thermocycle protocol was 3 minutes initial denaturation at 94°C, followed by 25 cycles of 1 minute denaturation at 94°C, 1 minute annealing at 53-55°C, 1 minute extension at 72°C, and a final 10 minute extension step at 72°C. In the second round of nested PCR, 1 μl of the first-round product was amplified for 30 cycles. For cloning, PCR products were separated on agarose, DNA was extracted applying the Millipore (Billerica, MA, USA) Montage Gel Extraction kit, followed by ethanol precipitation. Isolated fragments were cloned in pCR2.1-TOPO (Invitrogen, Karlsruhe, Germany), and cloned DNA was Sanger sequenced using M13 standard reverse primer (17-mer).
Splice variant quantification by pyrosequencing
Templates for pyrosequencing were generated using universal biotinylated primers [51]. RT-PCR amplicons of the exon-exon junctions were ligated into pCR2.1-TOPO (Invitrogen) according to the supplier's recommendations and subsequently re-amplified with all four possible combinations of 5'-biotinylated M13 standard primers (17-mers) and unlabeled insert-specific primers (Additional data file 1). The latter also served to prime the pyrosequencing reaction. Biotin-labeling of DNA, single strand preparation and sequencing were performed as described [51].
Orthologous intron-exon boundaries
A data set of orthologous intron-exon boundaries was constructed automatically to obtain sufficient data (especially reference data) to test for evolutionary constraints on intron flanking sequence. Sets of human (data as described for the splice site screen) and mouse transcript annotations (UCSC genome assembly mm7, RefSeq-to-genome alignment 2006-05-21) were processed as described earlier (supplementary methods in [20]). For 97,107 unambiguous orthologous pairs (57% of unique human intron-exon boundaries, including 23 of 36 TG-AG splice site tandems), 100 nt flanking intron sequences were aligned using CLUSTALW [52], using the optimized parameter -gapopen = 2.2. The degree of conservation was determined for 50 nt of the human intron sequence upstream of the splice site (tandem), giving a score of 1 for an identical aligned nucleotide in mouse, a score of 0 for a mismatch, and a penalty of -1 for inserted mouse sequence. Since a histogram of sequence conservation in canonical introns showed a non-normal distribution, statistical testing was performed using a permutation test. Intron samples of given size were simulated by random drawings from the intron data set, and the average sequence identity was calculated, repeating the sampling procedure 10,000 times.
Where automated processing failed (13 of 36 TG-AG splice site tandems), orthologous intron-exon boundaries were retrieved using the UCSC genome browser. These cases were not used for the statistical analysis since these represent a likely biased subset with regard to sequence conservation, and an appropriate large data set for comparison is not available.
Additional data files
The following additional data are available with the online version of this paper. Additional data file 1 is a Word file containing a description of the estimation of the amount of cis-regulatory sequence context, three supplementary tables and two supplementary figures. Supplementary Table 1 lists the putative unusual splice sites evident from EST-to-genome alignments that failed the quality checks. Supplementary Table 2 provides data about the comprehensive analysis of putative 3' TG splice sites suggested by spliced alignments of RefSeq transcripts. Supplementary Table 3 contains all primer sequences. Supplementary Figure S1 shows the distance-dependent occurrence of TG-AG and AG-AG splice acceptor tandems. Supplementary Figure S2 shows a LOGO representation of the TG 3' splice site sequence context.
Supplementary Material
Supplementary Table 1 lists the putative unusual splice sites evident from EST-to-genome alignments that failed the quality checks. Supplementary Table 2 provides data about the comprehensive analysis of putative 3' TG splice sites suggested by spliced alignments of RefSeq transcripts. Supplementary Table 3 contains all primer sequences. Supplementary Figure S1 shows the distance-dependent occurrence of TG-AG and AG-AG splice acceptor tandems. Supplementary Figure S2 shows a LOGO representation of the TG 3' splice site sequence context.
Acknowledgments
Acknowledgements
We thank M-L Schmidt and I Görlich for expert technical assistance, F Liu and Z-G Han for providing clone material, members of the RefSeq Division staff of the National Center for Biotechnology Information for helpful discussions, many members of the FLI and two anonymous referees for critical reading of the manuscript and helpful suggestions. This work was supported by grants from the German Ministry of Education and Research to SS (01GS0426) and MP (01GR0504, 0313652D) as well as from the Deutsche Forschungsgemeinschaft (SFB604-02) to MP.
Contributor Information
Karol Szafranski, Email: szafrans@fli-leibniz.de.
Stefanie Schindler, Email: sschindl@fli-leibniz.de.
Stefan Taudien, Email: stau@fli-leibniz.de.
Michael Hiller, Email: hiller@informatik.uni-freiburg.de.
Klaus Huse, Email: khuse@fli-leibniz.de.
Niels Jahn, Email: nielsj@fli-leibniz.de.
Stefan Schreiber, Email: s.schreiber@mucosa.de.
Rolf Backofen, Email: backofen@informatik.uni-freiburg.de.
Matthias Platzer, Email: mplatzer@fli-leibniz.de.
References
- Burge CB, Tuschl TH, Sharp PA. Splicing precursors to mRNAs by the spliceosomes. In: Gesteland RF, Cech T, Atkins JF, editor. The RNA World. 2. Plainview, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- Konarska MM, Query CC. Insights into the mechanisms of splicing: more lessons from the ribosome. Genes Dev. 2005;19:2255–2260. doi: 10.1101/gad.1363105. [DOI] [PubMed] [Google Scholar]
- Reed R. Mechanisms of fidelity in pre-mRNA splicing. Curr Opin Cell Biol. 2000;12:340–345. doi: 10.1016/S0955-0674(00)00097-1. [DOI] [PubMed] [Google Scholar]
- Moore MJ. Intron recognition comes of AGe. Nat Struct Biol. 2000;7:14–16. doi: 10.1038/71207. [DOI] [PubMed] [Google Scholar]
- Jackson IJ. A reappraisal of non-consensus mRNA splice sites. Nucleic Acids Res. 1991;19:3795–3798. doi: 10.1093/nar/19.14.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge CB, Padgett RA, Sharp PA. Evolutionary fates and origins of U12-type introns. Mol Cell. 1998;2:773–785. doi: 10.1016/S1097-2765(00)80292-0. [DOI] [PubMed] [Google Scholar]
- Levine A, Durbin R. A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res. 2001;29:4006–4013. doi: 10.1093/nar/29.1.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth N, Roca X, Hastings ML, Roeder T, Krainer AR, Sachidanandam R. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 2006;34:3955–3967. doi: 10.1093/nar/gkl556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burset M, Seledtsov IA, Solovyev VV. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 2000;28:4364–4375. doi: 10.1093/nar/28.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackenridge S, Wilkie AOM, Screaton GR. Efficient use of a 'dead-end' GA 5' splice site in the human fibroblast growth factor receptor genes. EMBO J. 2003;22:1620–1631. doi: 10.1093/emboj/cdg163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Krainer AR. Splicing of a divergent subclass of AT-AC introns requires the major spliceosomal snRNAs. RNA. 1997;3:586–601. [PMC free article] [PubMed] [Google Scholar]
- Dietrich RC, Incorvaia R, Padgett RA. Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol Cell. 1997;1:151–160. doi: 10.1016/S1097-2765(00)80016-7. [DOI] [PubMed] [Google Scholar]
- Chong A, Zhang G, Bajic VB. Information for the Coordinates of Exons (ICE): a human splice sites database. Genomics. 2004;84:762–766. doi: 10.1016/j.ygeno.2004.05.007. [DOI] [PubMed] [Google Scholar]
- van Nimwegen E, Paul N, Sheridan R, Zavolan M. SPA: a probabilistic algorithm for spliced alignment. PLoS Genet. 2006;2:e24. doi: 10.1371/journal.pgen.0020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozasa T, Itoh H, Tsukamoto T, Kaziro Y. Isolation and characterization of the human Gs alpha gene. Proc Natl Acad Sci USA. 1988;85:2081–2085. doi: 10.1073/pnas.85.7.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard AJ, Krainer AR, Robson SC, Europe-Finner GN. Alternative splicing of the adenylyl cyclase stimulatory G-protein G alpha(s) is regulated by SF2/ASF and heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) and involves the use of an unusual TG 3'-splice site. J Biol Chem. 2002;277:15241–15251. doi: 10.1074/jbc.M109046200. [DOI] [PubMed] [Google Scholar]
- Quan F, Forte MA. Two forms of Drosophila melanogaster Gs alpha are produced by alternate splicing involving an unusual splice site. Mol Cell Biol. 1990;10:910–917. doi: 10.1128/mcb.10.3.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathakis DG, Udar N, Sandgren O, Andreasson S, Bryant PJ, Small K, Forsman-Semb K. Genomic organization of human DLG4, the gene encoding postsynaptic density 95. J Neurochem. 1999;73:2250–2265. doi: 10.1046/j.1471-4159.1999.0732250.x. [DOI] [PubMed] [Google Scholar]
- Seeman P, Nam D, Ulpian C, Liu IS, Tallerico T. New dopamine receptor, D2(Longer), with unique TG splice site, in human brain. Brain Res Mol Brain Res. 2000;76:132–141. doi: 10.1016/S0169-328X(99)00343-5. [DOI] [PubMed] [Google Scholar]
- Hiller M, Huse K, Szafranski K, Jahn N, Hampe J, Schreiber S, Backofen R, Platzer M. Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nat Genet. 2004;36:1255–1257. doi: 10.1038/ng1469. [DOI] [PubMed] [Google Scholar]
- Hiller M, Huse K, Szafranski K, Jahn N, Hampe J, Schreiber S, Backofen R, Platzer M. Single-nucleotide polymorphisms in NAGNAG acceptors are highly predictive for variations of alternative splicing. Am J Hum Genet. 2006;78:291–302. doi: 10.1086/500151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugnet CW, Kent WJ, Ares MJ, Haussler D. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac Symp Biocomput. 2004:66–77. doi: 10.1142/9789812704856_0007. [DOI] [PubMed] [Google Scholar]
- Soong TW, DeMaria CD, Alvania RS, Zweifel LS, Liang MC, Mittman S, Agnew WS, Yue DT. Systematic identification of splice variants in human P/Q-type channel alpha1(2.1) subunits: implications for current density and Ca2+dependent inactivation. J Neurosci. 2002;22:10142–10152. doi: 10.1523/JNEUROSCI.22-23-10142.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve B, Froguel P, Corset L, Vaillant E, Vatin V, Boutin P. Rapid SNP allele frequency determination in genomic DNA pools by pyrosequencing. Biotechniques. 2002;32:1138–1142. doi: 10.2144/02325dd03. [DOI] [PubMed] [Google Scholar]
- Frey UH, Nuckel H, Dobrev D, Manthey I, Sandalcioglu IE, Eisenhardt A, Worm K, Hauner H, Siffert W. Quantification of G protein Gaalphas subunit splice variants in different human tissues and cells using pyrosequencing. Gene Expr. 2005;12:69–81. doi: 10.3727/000000005783992124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Z, States D, Gish W. Selecting for functional alternative splices in ESTs. Genome Res. 2002;12:1837–1845. doi: 10.1101/gr.764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Van Nostrand E, Holste D, Poggio T, Burge CB. Identification and analysis of alternative splicing events conserved in human and mouse. Proc Natl Acad Sci USA. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Dror G, Shamir R. Assessing the number of ancestral alternatively spliced exons in the human genome. BMC Genomics. 2006;7:273. doi: 10.1186/1471-2164-7-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno H, Fujii H, Wei DC, Chan LC, Hirono A, Tsukimoto I, Miwa S. Frame shift mutation, exon skipping, and a two-codon deletion caused by splice site mutations account for pyruvate kinase deficiency. Blood. 1997;89:4213–4218. [PubMed] [Google Scholar]
- Thongnoppakhun A, Rungroj N, Wilairat P, Vareesangthip K, Sirinavin C, Yenchitsomanus PT. A novel splice-acceptor site mutation (IVS13-2A>T) of polycystic kidney disease 1 (PKD1) gene resulting in an RNA processing defect with a 74-nucleotide deletion in exon 14 of the mRNA transcript. Hum Mutat. 2000;15:115. doi: 10.1002/(SICI)1098-1004(200001)15:1<115::AID-HUMU22>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Yamada H, Shinmura K, Tsuneyoshi T, Sugimura H. Effect of splice-site polymorphisms of the TMPRSS4, NPHP4 and ORCTL4 genes on their mRNA expression. J Genet. 2005;84:131–136. doi: 10.1007/BF02715838. [DOI] [PubMed] [Google Scholar]
- Kol G, Lev-Maor G, Ast G. Human-mouse comparative analysis reveals that branch-site plasticity contributes to splicing regulation. Hum Mol Genet. 2005;14:1559–1568. doi: 10.1093/hmg/ddi164. [DOI] [PubMed] [Google Scholar]
- Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Bourinet E, Soong TW, Sutton K, Slaymaker S, Mathews E, Monteil A, Zamponi GW, Nargeot J, Snutch TP. Splicing of alpha 1A subunit gene generates phenotypic variants of P- and Q-type calcium channels. Nat Neurosci. 1999;2:407–415. doi: 10.1038/8070. [DOI] [PubMed] [Google Scholar]
- Furey TS, Diekhans M, Lu Y, Graves TA, Oddy L, Randall-Maher J, Hillier LW, Wilson RK, Haussler D. Analysis of human mRNAs with the reference genome sequence reveals potential errors, polymorphisms, and RNA editing. Genome Res. 2004;14:2034–2040. doi: 10.1101/gr.2467904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanaraj TA. A clean data set of EST-confirmed splice sites from Homo sapiens and standards for clean-up procedures. Nucleic Acids Res. 1999;27:2627–2637. doi: 10.1093/nar/27.13.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri Y, Colot HV, Jacquier AC, Yu Q, Hall JC, Baltimore D, Rosbash M. A family of unusually spliced biologically active transcripts encoded by a Drosophila clock gene. Nature. 1987;326:42–47. doi: 10.1038/326042a0. [DOI] [PubMed] [Google Scholar]
- Patel AA, McCarthy M, Steitz JA. The splicing of U12-type introns can be a rate-limiting step in gene expression. EMBO J. 2002;21:3804–3815. doi: 10.1093/emboj/cdf297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SS, Pandey KK, Choi HS, Kim SY, Law PY, Wei LN, Loh HH. Poly(C) binding protein family is a transcription factor in mu-opioid receptor gene expression. Mol Pharmacol. 2005;68:729–736. doi: 10.1124/mol.105.012245. [DOI] [PubMed] [Google Scholar]
- Michelotti EF, Tomonaga T, Krutzsch H, Levens D. Cellular nucleic acid binding protein regulates the CT element of the human c-myc protooncogene. J Biol Chem. 1995;270:9494–9499. doi: 10.1074/jbc.270.16.9494. [DOI] [PubMed] [Google Scholar]
- Soares LMM, Zanier K, Mackereth C, Sattler M, Valcarcel J. Intron removal requires proofreading of U2AF/3' splice site recognition by DEK. Science. 2006;312:1961–1965. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- Bennett M, Michaud S, Kingston J, Reed R. Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev. 1992;6:1986–2000. doi: 10.1101/gad.6.10.1986. [DOI] [PubMed] [Google Scholar]
- Chiara MD, Palandjian L, Feld Kramer R, Reed R. Evidence that U5 snRNP recognizes the 3' splice site for catalytic step II in mammals. EMBO J. 1997;16:4746–4759. doi: 10.1093/emboj/16.15.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K, Reed R. An upstream AG determines whether a downstream AG is selected during catalytic step II of splicing. Mol Cell Biol. 2001;21:1509–1514. doi: 10.1128/MCB.21.5.1509-1514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallena MJ, Chalmers KJ, Llamazares S, Lamond AI, Valcarcel J. Splicing regulation at the second catalytic step by Sex-lethal involves 3' splice site recognition by SPF45. Cell. 2002;109:285–296. doi: 10.1016/S0092-8674(02)00730-4. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Reed R, Maniatis T. Mechanisms of human-globin-pre-mRNA splicing. In: Berg P, editor. Genetic Chemistry: the Molecular Basis of Heredity. Houston, TX: The Robert A Welch Foundation; 1985. pp. 353–382. [Conferences on Chemical Research, vol.XXIX] [Google Scholar]
- Zhuang Y, Weiner AM. The conserved dinucleotide AG of the 3' splice site may be recognized twice during in vitro splicing of mammalian mRNA precursors. Gene. 1990;90:263–269. doi: 10.1016/0378-1119(90)90189-X. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M, Sherlock G, Binkley G, Jin H, Matese JC, Hernandez-Boussard T, Rees CA, Cherry JM, Botstein D, Brown PO, Alizadeh AA. SOURCE: a unified genomic resource of functional annotations, ontologies, and gene expression data. Nucleic Acids Res. 2003;31:219–223. doi: 10.1093/nar/gkg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth M, Huse K, Reichwald K, Taudien S, Hampe J, Rosenstiel P, Birkenmeier G, Schreiber S, Platzer M. Method for preparing single-stranded DNA templates for pyrosequencing using vector ligation and universal biotinylated primers. Anal Biochem. 2006;356:194–201. doi: 10.1016/j.ab.2006.04.043. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 lists the putative unusual splice sites evident from EST-to-genome alignments that failed the quality checks. Supplementary Table 2 provides data about the comprehensive analysis of putative 3' TG splice sites suggested by spliced alignments of RefSeq transcripts. Supplementary Table 3 contains all primer sequences. Supplementary Figure S1 shows the distance-dependent occurrence of TG-AG and AG-AG splice acceptor tandems. Supplementary Figure S2 shows a LOGO representation of the TG 3' splice site sequence context.