Abstract
Epipodophyllotoxins are associated with leukemias characterized by translocations of the MLL gene at chromosome band 11q23 and other translocations. Cytochrome P450 (CYP) 3A metabolizes epipodophyllotoxins and other chemotherapeutic agents. CYP3A metabolism generates epipodophyllotoxin catechol and quinone metabolites, which could damage DNA. There is a polymorphism in the 5′ promoter region of the CYP3A4 gene (CYP3A4-V) that might alter the metabolism of anticancer drugs. We examined 99 de novo and 30 treatment-related leukemias with a conformation-sensitive gel electrophoresis assay for the presence of the CYP3A4-V. In all treatment-related cases, there was prior exposure to one or more anticancer drugs metabolized by CYP3A. Nineteen of 99 de novo (19%) and 1 of 30 treatment-related (3%) leukemias carried the CYP3A4-V (P = 0.026; Fisher’s Exact Test, FET). Nine of 42 de novo leukemias with MLL gene translocations (21%), and 0 of 22 treatment-related leukemias with MLL gene translocations carried the CYP3A4-V (P = 0.016, FET). This relationship remained significant when 19 treatment-related leukemias with MLL gene translocations that followed epipodophyllotoxin exposure were compared with the same 42 de novo cases (P = 0.026, FET). These data suggest that individuals with CYP3A4-W genotype may be at increased risk for treatment-related leukemia and that epipodophyllotoxin metabolism by CYP3A4 may contribute to the secondary cancer risk. The CYP3A4-W genotype may increase production of potentially DNA-damaging reactive intermediates. The variant may decrease production of the epipodophyllotoxin catechol metabolite, which is the precursor of the potentially DNA-damaging quinone.
Second cancers are uncommon events occurring at a frequency of about 7% in survivors of primary malignant neoplasms (1). Although leukemias comprise a small fraction of second cancers (2), leukemias are the major second cancers that result from chemotherapy (3–6). There are two major forms of treatment-related leukemia, those with chromosome 5 and 7 monosomies induced by alkylating agents, and those with MLL gene translocations and other translocations related to DNA topoisomerase II inhibitors (7). Because only a minority of patients develop leukemia after chemotherapy, it has been suggested that differences in drug interactions with the host may be predisposing factors (8). Germ-line mutations in tumor-suppressor genes or genetic variation in drug metabolism are examples of host risk factors. Germ-line mutations in the NF-1 and p53 tumor-suppressor genes have been observed in alkylating agent-associated leukemias with chromosome 5 and 7 monosomies (9–12). Similar host risk factors for leukemias induced by DNA topoisomerase II inhibitors currently are unknown.
We explored genetic variation in drug metabolism as a potential host risk factor. Distinct phase I and phase II pathways of drug metabolism comprise a protective mechanism against environmental toxins (13–15). Phase I metabolism by cytochrome P450 (CYP) enzymes converts many compounds to reactive, electrophilic, water-soluble intermediates, some of which can damage DNA (14–19). The glutathione S-transferases and N-acetyltransferases, which are phase II enzymes, inactivate various toxic compounds, including compounds produced by phase I metabolism (13–15). Polymorphisms of potential relevance to chemical carcinogenesis have been described for various CYP, glutathione S-transferase, and N-acetyltransferase enzymes (13–15). A CYP2D6 polymorphism is associated with an increased risk of leukemia; it has been proposed that the poor-metabolizer phenotype may result in decreased ability to detoxify chemical carcinogens (20). An excess of the GSTT1 null genotype was observed in an adult white population with myelodysplastic syndrome, perhaps suggesting that resultant decreased detoxification of carcinogens may enhance susceptibility to myelodysplastic syndrome (21).
The epipodophyllotoxins, etoposide (VP16) and teniposide (VM26), as well as cyclophosphamide (CPM), ifosphamide (IFOS), vinblastine (VBL), and vindesine are substrates for metabolism by CYP3A (22), the most abundant component of the CYP system in the human liver (23). We identified a variant in the 5′ promoter region of the CYP3A4 gene (CYP3A4-V; ref. 24). The polymorphism is an agggcaagag → agggcaggag transition in the nifedipine-specific response element (24). In the present study, we examined de novo and treatment-related leukemias with and without MLL gene translocation for the presence of CYP3A4-V. The results suggest that the wild-type CYP3A4 genotype (CYP3A4-W) is significantly associated with epipodophyllotoxin-induced leukemogenesis.
METHODS
Subjects and Biosamples.
The Institutional Review Board of The Children’s Hospital of Philadelphia and The Committee for Research on Human Subjects at the University of Pennsylvania approved this research. Genomic DNAs and clinical information were obtained on patients with a diagnosis of leukemia. The patients were grouped according to whether the leukemia was de novo or followed anticancer treatment and whether the leukemia was characterized by translocation of the MLL gene at chromosome band 11q23. Genomic DNA was isolated from leukemic marrow or peripheral blood mononuclear cells as described, and Southern blot analysis was used to identify MLL gene rearrangements (25–27). Group 1 included 42 patients with de novo leukemias characterized by molecular translocation of the MLL gene (Table 1). Group 2 included 22 patients with treatment-related leukemias characterized by molecular translocation of the MLL gene, although in five cases this was not cytogenetically apparent (Table 2). All received prior chemotherapy with at least one agent metabolized by CYP3A4 (22). Exposures included VP16, VM26, CPM, IFOS, or VBL. In all the cases in group 1 and group 2, MLL gene rearrangement was within the breakpoint cluster region except in a single group 1 case, where the rearrangement mapped 5′ of the breakpoint cluster region between MLL intron 3 and exon 5 (27, 28).
Table 1.
Characteristics of 129 subjects with leukemia
|
De novo
|
Treatment-related
|
Total (129) | |||||
|---|---|---|---|---|---|---|---|
| Group 1 11q23 (42) | Group 3 non-11q23 (57) | Total (99) | Group 2 11q23 (22) | Group 4 non-11q23 (8) | Total (30) | ||
| Mean age at Dx in yr, (SD), {n} | 2.2, (5.1), {42} | 7.7, (5.4), {49} | 11.2, (4.7), {22} | 14.1, (8.8), {8} | |||
| Male | 16 (38%) | 37 (65%) | 53 (53%) | 13 (59%) | 3 (38%) | 16 (53%) | 69 (53%) |
| Female | 26 (62%) | 19 (33%) | 45 (45%) | 9 (41%) | 5 (63%) | 14 (47%) | 59 (46%) |
| NA | 0 | 1 (2%) | 1 (1%) | 0 | 0 | 0 | 1 (1%) |
| White | 37 (88%) | 36 (63%) | 73 (74%) | 19 (86%) | 6 (75%) | 25 (83%) | 98 (76%) |
| Black | 3 (7%) | 9 (16%) | 12 (12%) | 0 | 0 | 0 | 12 (9%) |
| Hispanic | 2 (5%) | 3 (5%) | 5 (6%) | 3 (14%) | 1 (13%) | 4 (13%) | 9 (7%) |
| Asian | 0 | 2 (4%) | 2 (2%) | 0 | 0 | 0 | 1 (2%) |
| NA | 0 | 7 (12%) | 7 (7%) | 0 | 1 (13%) | 1 (3%) | 8 (6%) |
| ALL | 21 (50%) | 57 (100%) | 78 (79%) | 3 (14%) | 0 | 3 (10%) | 81 (63%) |
| AML | 18 (43%) | 0 | 18 (18%) | 16 (73%) | 4 (50%) | 20 (67%) | 38 (29%) |
| Biphenotypic | 3 (7%) | 0 | 3 (3%) | 1 (5%) | 0 | 1 (3%) | 4 (3%) |
| MDS | 0 | 0 | 0 | 2 (9%) | 4 (50%) | 6 (20%) | 6 (5%) |
| FAB L1 | 14 (33%) | 33 (58%) | 47 (47%) | 3 (13%) | 0 | 3 (10%) | 50 (39%) |
| FAB L2 | 3 (7%) | 9 (16%) | 12 (12%) | 0 | 0 | 0 | 12 (9%) |
| FAB M1 | 2 (5%) | 0 | 2 (2%) | 1 (5%) | 0 | 1 (3%) | 3 (2%) |
| FAB M2 | 2 (5%) | 0 | 2 (2%) | 2 (9%) | 0 | 2 (7%) | 4 (3%) |
| FAB M4 | 4 (9%) | 0 | 4 (4%) | 11 (50%) | 2 (25%) | 13 (43%) | 17 (13%) |
| FAB M5 | 10 (24%) | 0 | 10 (10%) | 2 (9%) | 1 (13%) | 3 (10%) | 13 (10%) |
| FAB M6 | 0 | 0 | 0 | 0 | 1 (13%) | 1 (3%) | 1 (1%) |
| FAB M7 | 0 | 0 | 0 | 1 (5%) | 0 | 1 (3%) | 1 (1%) |
| RAEB | 0 | 0 | 0 | 0 | 1 (13%) | 1 (3%) | 1 (1%) |
| RAEB-t | 0 | 0 | 0 | 0 | 1 (13%) | 1 (3%) | 1 (1%) |
| NA | 7 (17%) | 15 (26%) | 22 (22%) | 2 (9%) | 2 (25%) | 4 (13%) | 26 (20%) |
| Mean survival in mo, [SD], (range), {n} | 20.4, [19.0], (0.7–71.1), {39} | 37.9, [30.3], (6–125), {49} | 10.4, [10.5], (0.1–42), {20} | 6.6, [5.8], (0.3–15), {8} | |||
| Mean interval in mo, [SD], (range), {n} | Not applicable | Not applicable | 40.5, [31.4], (11–132), {22} | 55.3, [34.8], (14–113), {8} | |||
NA, not available; ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; MDS, myelodysplastic syndrome; FAB, the French–American–British classification of morphology; RAEB, refractory anemia with excess blasts; RAEB-t, refractory anemia with excess blasts in transition; {n}, number of subjects for whom information was available. Survival is from diagnosis of de novo or treatment-related leukemia. Interval is from diagnosis (Dx) of primary cancer to diagnosis of treatment-related leukemia.
Table 2.
Features of group 2 and group 4 subjects with treatment-related leukemia
| Patient no. | Gender | Race | Primary cancer | Age at primary Dx, yr. mo | CYP3A4 substrate exposure | Secondary leukemia | FAB | Interval from primary Dx, mo | Karyotype | |
|---|---|---|---|---|---|---|---|---|---|---|
| Group 2 | 2 | F | W | ALL | 2.9 | VP16 | AML | M4 | 50 | 46,XX,t(11;19)(q23;p13.3)[10]/46,XX[10] |
| 3 | F | W | ALL | 1.7 | VP16, CPM | AML | M4 | 41 | 47,XX,+8,t(11;19)(q23;p13)[19]/48,XX,+8,+8,t(11;19)(q23;p13)[6] | |
| 4 | F | W | OS | 7 | VP16 | AML | M1 | 60 | 46,XX,t(9;11)(p22;q23)[27] | |
| 5 | M | W | HD | 15.8 | CPM, VLB | Biphenotypic | M4 | 16 | 46,XY,t(11q−;19p+)[25] | |
| 6 | M | W | NB | 3.6 | CPM, VM26 | AML | M4 | 118 | 46,XY,t(1;7)(q32;q32–34), inv(2)(p21q37),t(3;11)(q25;q23),t(7;?) (q22;?)[29] | |
| 8 | F | W | NHL | 9.2 | CPM | ALL | L1 | 21 | 47,XX,+del(6)(?q27)[1]/47,XX,+8,del(6)(q27),del(11)(q23)[29] | |
| 9 | M | W | PNET | 13.9 | CPM, IFOS, VP16 | AML | M5 | 11 | 46,XY,t(9;11)(p22;q23)[20] | |
| 10 | M | W | OS | 14 | IFOS, VP16 | ALL | L1 | 16 | 46,XY,t(4;11)(q21;q23)[7]/46,XY[18] | |
| 11 | F | H | ALL | 12.3 | VP16 | MDS | M7 | 33 | 47,XX,add(1)(p32),+mar[2]/46,XX[38] | |
| 12 | F | W | ALL | 1.9 | VP16 | AML | M4 | 41 | 46,XX,del(11)(q14),add(14)(p?11.2)[16]/46,XX[4] | |
| 13 | M | W | ERMS | 5.3 | CPM | AML | M4 | 60 | 46,XY[18] | |
| 21 | M | W | ALL | 4.0 | VP16 | AML | M4 | 23 | 46,XY,t(9;11)(p22;q23)[19]/46,XY[1] | |
| 23 | F | W | ALL | 10.9 | CPM, VP16 | AML | M2 | 132 | 46,XX,t(11;19)(q23;p13.3)[15]/46,XX[5] | |
| 24 | M | W | ERMS | 15.2 | CPM, IFOS, VP16 | AML | M4 | 24 | 46,XY,t(9;11)(p21;q23)[19] | |
| 33 | F | W | ARMS | 9.11 | CPM, IFOS, VP16 | ALL | L1 | 15 | 46,XX,t(4;11)(q21;q23)[4]/46,XX[3] | |
| 35 | M | W | WT | 7.4 | CPM, VP16 | AML | M5a | 39 | 46,XY,t(9;11)(p22;q23)[20] | |
| 36 | M | H | ARMS | 1.4 | IFOS, VP16 | AML | M4 | 60 | 46,XY | |
| 38 | M | W | ALL | 1.3 | CPM, VM26 | MDS | NA | 28 | 46,XX,t(11;19)(q23;p13.3) | |
| 39 | M | W | NB | 12.11 | CPM, VP16 | AML | M4 | 11 | 46,XY,del(11)(q23)[8]/46,XY[12] | |
| 40 | F | H | ALL | 8.9 | CPM, VP16 | AML | NA | 31 | 46,XX,t(11;17)(q23;q25) | |
| 44 | M | W | Ewing’s | 10.8 | CPM, IFOS, VP16 | AML | M2 | 34 | 46,XY[20] | |
| 47 | M | W | OS | 12.10 | IFOS | AML | M4 | 26 | 46,XY,t(9;11)(p22;q23)[18]/46,XY[2] | |
| Group 4 | 17 | M | W | ERMS | 1.11 | CPM, VP16 | MDS | RAEB-t | 65 | 45,XY,hsr(2)(q22),−5,der(7)del(7) (q11.23)hsr(7) (q11.23),der(12)t(12;19) (p11.2;q12),der(17)t(5;17) (p12;p11.2),−19,+mar1[19]/related nonclonal[1]/46,XY[2] |
| 19 | F | NA | ARMS | 13 | CPM, IFOS, VP16 | AML | M5a | 49 | 49,XX,+2,+12,+16[12]/51,XX,+2,+6,+12,+13,+16[7] | |
| 27 | F | W | HD | 19.0 | VLB | AML | NA | 96 | 46,XX,inv(3)(q21q26)[18]/46,idem,der(6)t(1;6)(q21;q21)[5] | |
| 32 | F | H | Ependymoma | 1.5 | CPM, VP16 | MDS | NA | 20 | 45,XX,−7[7]/46,XX[7] | |
| 34 | M | W | NHL | 14.11 | CPM, IFOS, VP16 | MDS | RAEB | 14 | 46,XY,t(11;20)(p15;q11.2)[17]/45,idem,−5[3] | |
| 43 | F | W | OS | 14.0 | IFOS, VP16 | MDS | M6 | 49 | 45,XX,del(5)?(q11.2q23),−7,−17,+mar [17]/46,XX[13] | |
| 45 | F | W | RMS | 0.11 | CPM, IFOS, VP16 | AML | M4 | 36 | 46,XX,5q− | |
| 51 | M | W | Undifferentiated sarcoma | 13.8 | CPM, IFOS | AML | M4 | 113 | 46,XY[6]/45,XY,−13[1]/45,XY,−15[1]/43,XY,−16,−21,−21[1] |
Dx, diagnosis; M, male; F, female; NA, not available; W, white; B, black; H, Hispanic; ALL, acute lymphoblastic leukemia; OS, osteosarcoma; RMS, rhabdomyosarcoma; ARMS, alveolar rhabdomyosarcoma; ERMS, embryonal rhabdomyosarcoma; HD, Hodgkin’s disease; NHL, non-Hodgkin’s lymphoma; PNET, peripheral neurectodermal tumor; WT, Wilm’s tumor; CPM, cyclophosphamide; IFOS, ifosphamide; VLB, vinblastine; VP16, etoposide; VM26, teniposide; AML, acute myeloid leukemia; MDS, myelodysplastic syndrome; FAB, the French–American–British classification of morphology. Features of patients 2–6, 8–13, 17, and 19 have been described in refs 11, 28, and 30, where patient numbers were the same.
The 57 patients in group 3 were diagnosed with de novo B lineage acute lymphoblastic leukemia and were studied as a control population with a common pediatric cancer. MLL gene rearrangement was excluded in all cases by Southern blot analysis of BamHI-digested DNA with the B859 cDNA probe from the MLL breakpoint cluster region (29). Group 4 included eight patients with treatment-related leukemia without cytogenetic and/or molecular evidence of translocation of chromosome band 11q23. All eight patients previously were diagnosed with cancer and had a history of exposure to at least one anticancer drug metabolized by CYP3A4.
Clinical and demographic features, karyotypes, and rearranged or germ-line MLL gene configuration have been described for 30 group 1 patients (25–27), 11 group 2 patients (11, 28, 30), and 2 group 4 patients (11, 28). Clinical and demographic features have been described for 27 group 3 patients (31).
CYP3A4 Genotype Determination.
Genomic DNA was available from bone marrow or peripheral blood either at diagnosis or relapse of leukemia. Genotypes were examined by PCR amplification of a 592-bp template from upstream of the CYP3A4 gene, extending into exon 1 (nucleotides −571 to +22), and analysis of the products in a conformation-sensitive gel electrophoresis assay (24). The sequences of the forward and reverse primers used for PCR were derived from the published sequence (32) and were 5′-AAC AGG CGT GGA AAC ACA AT-3′, and 5′-CTT TCC TGC CCT GCA CAG-3′, respectively (24). Each 50-μl PCR mixture contained ≈100 ng of genomic DNA, 4 units AmpliTaq DNA polymerase (Perkin–Elmer), 25 pmol of each primer, 200 μM each dNTP (Perkin–Elmer), 1.5 mM MgCl2, and PCR buffer II at 1× final concentration (Perkin–Elmer). After initial denaturation at 95°C for 5 min and incubation at 82°C for 1 min, we used a two-phase touchdown protocol for annealing. The first phase included 25 cycles at 94°C for 1 min, 66°C for 1 min (with a decrease of 0.5°C each cycle), and 72°C for 1 min, to reach a final annealing temperature of 54°C. The second phase included 15 cycles at 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min, and was followed by a final elongation at 72°C for 10 min. Control genomic DNA with known CYP3A4-V/V genotype was also amplified.
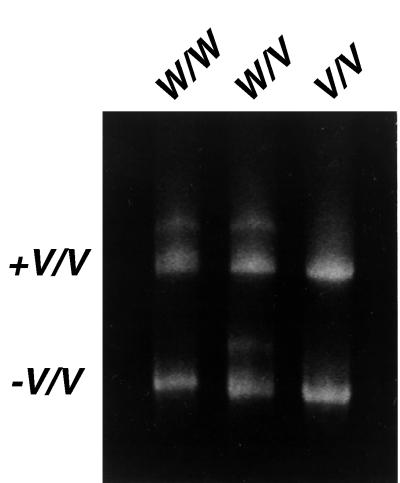
PCR products (6 μl) were admixed with 6 μl of PCR products with known CYP3A4-V/V genotype and electrophoresed in a 10% nondenaturing polyacrylamide gel in the same lane with 12 μl of the products without additional CYP3A4-V/V DNA. Differential migration of DNA heteroduplexes and homoduplexes was visualized by ethidium bromide staining, and CYP3A4 genotypes were classified as W/W, W/V or V/V. In CYP3A4 V/V subjects, a single homoduplex band would be observed with and without addition of V/V DNA. In W/V subjects, both homoduplex and heteroduplex bands would be observed without addition of V/V DNA, whereas addition of V/V DNA would have no effect on banding patterns. In contrast, for W/W subjects, a single homoduplex band would be observed without additional V/V DNA, but addition of V/V DNA would produce both homoduplex and heteroduplex patterns (Fig. 1). CYP3A4-V/V and CYP3A4-W/V were defined as CYP3A4-V (variant) genotypes.
Figure 1.
Migration patterns on conformation-sensitive gel electrophoresis analysis of PCR-amplified W/W, W/V, and V/V DNAs with (Upper) and without (Lower) addition of DNA with known V/V genotype, which changes the migration of only W/W DNA.
Statistical Methods.
Proportions in contingency tables were compared by nonparametric methods. Fisher’s Exact Test (FET) was used for analysis of contingency tables with less than five observations per cell. Odds ratios (OR) were estimated using logistic regression models for binary outcome data and were adjusted for age at diagnosis, race, and gender. All analyses were performed with SAS version 6.11 statistical software.
RESULTS
Descriptive Results.
Table 1 describes the molecular, demographic, and clinical characteristics of all four groups of subjects (n = 129). Among the four groups, there were significant differences in distribution of age at initial diagnosis (Kruskal–Wallis χ21 = 52.92; P < 0.0001) and gender (P = 0.034, FET) but not race (P = 0.170, FET).
Of the 22 subjects in group 2, 14 (64%) were exposed to CPM, 7 (32%) to IFOS, 1 (5%) to VBL, 2 (9%) to VM26, and 17 (77%) to VP16 (Table 2). The regimens contained one CYP3A4 substrate in seven cases and two or three CYP3A4 substrates in the other cases. Of the eight group 4 subjects, six (75%) were exposed to CPM, five (63%) to IFOS, one (13%) to VBL, none were exposed to VM26, and six (75%) were exposed to VP16 (Table 2). The chemotherapy included more than one drug metabolized by CYP3A4 in seven of eight cases.
The karyotypes in all treatment-related leukemias were examined for the presence of both copies of chromosome band 7q22, the genomic region encoding CYP3A4 (ref. 33; Table 2). The karyotypes suggested that both copies were present in a significant proportion of the cells and that both CYP3A4 alleles would be amplifiable from the leukemic samples.
Association of Case Group with CYP3A4 Genotype.
We observed significant differences in CYP3A4 genotype distribution among the four groups of subjects. As shown in Table 3, there was a significant deficit of CYP3A4-V genotypes among all treatment-related cases (3%) compared with all de novo cases (19%; P = 0.026, FET). The age-, gender-, and race-adjusted OR for this association was 0.07 (95% confidence interval (CI): 0.01–0.68). When we limited the analysis to leukemias with MLL gene translocations, we found no CYP3A4-V genotypes in the treatment-related cases in group 2, compared with 21% of the de novo cases in group 1 (P = 0.016, FET). Because none of the 22 group 2 subjects carried the CYP3A4-V, no OR could be estimated. Of the group 2 subjects, three did not have prior exposure to epipodophyllotoxin (Table 2). After removal of these cases and analysis of the group 2 subjects exposed to epipodophyllotoxin, the observation that CYP3A4-V was underrepresented in treatment-related leukemias with MLL gene translocations remained significant (P = 0.026, FET).
Table 3.
Association of CYP3A4 genotype with leukemia subsets
| Group* | Genotype frequency (row %)
|
|||
|---|---|---|---|---|
| CYP3A4-W | CYP3A4-V | FET P value | OR (95% CI)† | |
| All de novo cases (Groups 1, 3) | 80 (81%) | 19 (19%) | 0.026 | 0.09 (0.01–0.87) |
| All treatment-related cases (Groups 2, 4) | 29 (97%) | 1 (3%) | ||
| De novo 11q23 (Group 1) | 33 (79%) | 9 (21%) | 0.016 | ‡ |
| Treatment-related 11q23 (Group 2) | 22 (100%) | 0 (0%) | ||
| De novo 11q23 (Group 1) | 33 (79%) | 9 (21%) | 0.026 | ‡ |
| Epipodophyllotoxin-related 11q23 (Group 2 subset) | 19 (100%) | 0 (0%) | ||
| All 11q23 (Groups 1, 2) | 55 (86%) | 9 (14%) | 0.419 | 1.33 (0.42–4.20) |
| All non-11q23 (Groups 3, 4) | 54 (83%) | 11 (17%) | ||
| De novo non-11q23 (Group 3) | 47 (82%) | 10 (18%) | 0.592 | 0.37 (0.02–4.59) |
| Treatment-related non-11q23 (Group 4) | 7 (88%) | 1 (12%) | ||
| Treatment-related 11q23 (Group 2) | 22 (100%) | 0 (0%) | 0.267 | ‡ |
| Treatment-related non-11q23 (Group 4) | 7 (88%) | 1 (12%) | ||
Numbers correspond to four subject groups, as defined in Materials and Methods.
Adjusted by multiple logistic regression for age at diagnosis of leukemia, race, gender, and survival time.
Odds ratio and 95% CI are inestimable due to zero cells. The genotypes of the 9 group 1 subjects with CYP3A4-V were W/V in 8 cases and V/V in the other case; the genotypes of the 10 group 3 subjects with CYP3A4-V were W/V in 8 cases and V/V in 2 cases. The genotype of the single group 4 subject with CYP3A4-V was W/V.
To evaluate whether the genotype effect was specific to treatment-related leukemias with MLL gene translocations, we compared CYP3A4 genotype in patients with de novo and treatment-related leukemias without MLL gene translocations in groups 3 and 4. No difference was observed (P = 0.592, FET), but the sample size among these subjects was too small to make strong inferences (Table 3).
Because differences in survival times might affect the chances of developing treatment-related leukemia, we determined whether the mean survival time from diagnosis in patients with de novo leukemia in groups 1 and 3 was similar to the mean interval from the primary cancer diagnosis to the development of treatment-related leukemia in groups 2 and 4. The mean survival time in groups 1 and 3 was 30.1 months; the mean interval from the primary cancer diagnosis to the development of treatment-related leukemia in groups 2 and 4 was 44.4 months (Table 1). Because this difference was significant (F1,88 from ANOVA = 5.56; P = 0.02), we adjusted the association analyses for the total duration of follow-up. The resulting OR association comparing all de novo cases against all treatment-related cases (OR = 0.09, 95% CI: 0.01–0.87) was not substantially different from the analyses adjusted only for age at diagnosis, race, and gender, indicating that the effect of CYP3A4 genotype remained significant even after adjustment for different follow-up times among case groups.
DISCUSSION
DNA topoisomerase II inhibitors, especially the epipodophyllotoxins, are associated with treatment-related leukemias in about 2% of patients (28, 34–47). Molecular cancer epidemiology has not identified specific mutations that confer genetic susceptibility or mutagen sensitivity in this form of leukemia. Epipodophyllotoxins, CPM, IFOS, VBL, and vindesine all are substrates for CYP3A (22, 48). Previously, we observed an agggcaagag → agggcaggag polymorphism in the nifedipine-specific response element of the CYP3A4 promoter, which represents the CYP3A4-V (variant) genotype (24). The ethnic distributions of CYP3A4-V are 9% in whites, 53% in African Americans, and 0% in Taiwanese (49). In the present study, we observed a significant deficit of the CYP3A4-V among subjects who developed treatment-related leukemia after administration of chemotherapeutic agents that are metabolized by CYP3A.
These results suggest that CYP3A4-W is associated with the effects of chemotherapy, particularly the effects of epipodophyllotoxin, leading to leukemias with MLL gene translocations. Conversely, the findings suggest that after similar chemotherapy, CYP3A4-V is associated with DNA damage leading to leukemias with MLL gene translocation less frequently. We evaluated 19 subjects who had epipodophyllotoxin exposure and developed treatment-related leukemia with MLL gene translocation as a separate subgroup. A significant relationship was observed with CYP3A4 genotype in this patient subset. The chemotherapy included more than one CYP3A substrate in several of these patients. In addition, the deficit of CYP3A4-V was significant for all subjects with treatment-related leukemias, suggesting that the relationship of CYP3A4 genotype with alkylating agent-induced leukemia should be studied further.
CYP3A catalyzes O-demethylation of the epipodophyllotoxin dimethoxyphenol E ring to form the catechol metabolite (48). The catechol is the precursor to a quinone metabolite (50, 51), which potentially could produce depurinating N7-guanine adducts (52). Such adducts might result in the formation of abasic sites in DNA from the action of DNA glycosylases (53–56). Whereas enhanced DNA topoisomerase II cleavage is the mechanism of chromosomal breakage from epipodophyllotoxin parent drugs (57), abasic sites at critical positions in the DNA increase DNA topoisomerase II cleavage much more than the parent drugs (58–61). Unrepaired DNA adducts result in recruitment of recombinational repair when there is blockage of the replication fork (53). Redox cycling of the catechol and quinone metabolites generates reactive oxygen species and hydroxyl radicals that could cause oxidative damage to the DNA (51, 53). Oxidative DNA damage may generate interstrand crosslinks that recruit recombinational repair. Although this system is not well characterized in humans (53), recently it was shown that mammalian cells use homology-directed recombinational repair when there are double-strand breaks in DNA (62). Thus, epipodophyllotoxin catechol and quinone metabolites have potential genotoxic properties that may be of relevance to translocations.
We identified CYP3A4-V as a genotypic factor that modulates leukemogenic drug effects. It is biologically plausible that CYP3A metabolism is a host factor that modulates the risk of treatment-related leukemia. If CYP3A4-V, which disrupts a regulatory element 5′ to the CYP3A4 gene, is associated with decreased CYP3A4 expression or decreased activity of the enzyme, formation of genotoxic metabolites may be reduced in individuals carrying the CYP3A4-V. It is possible that CYP3A4-W is associated with increased metabolism of epipodophyllotoxin to the corresponding catechol compared with CYP3A4-V. The catechol can undergo redox cycling to form the potentially DNA-damaging quinone, and reactive oxygen species are produced (50, 51). Therefore, patients with CYP3A4-W would be more at risk.
Genetic polymorphism can account for large differences in the pharmacokinetics of chemotherapeutic agents, but metabolism of the majority is determined polygenetically and distributed unimodally. There is 5- to 20-fold interindividual variability in drug clearance, which is a consequence of both genetic and nongenetic factors (63). CYP3A-mediated first-pass metabolism occurs after oral drug administration and may contribute to the variability (63). CYP3A activity also can be modulated by inducers such as rifampin and anticonvulsants, inhibitors such as azole antifungal agents and macrolide antibiotics, by liver disease, or by aging (63).
The results of this analysis may indicate that interpatient differences in the pharmacokinetic profile are relevant to the leukemogenic effects of the epipodophyllotoxin anticancer drugs. However, in a population of children treated for primary acute lymphoblastic leukemia, Relling et al. found no differences between VP16 and VP16-catechol pharmacokinetics in patients who did and did not develop treatment-related acute myeloid leukemia (64). Whereas total VP16 and VP16–catechol were measured (64), epipodophyllotoxins are highly protein-bound (16). Therefore, it will be important to establish whether there are interindividual differences in protein binding of the catechol metabolite. Together with the observed association of CYP3A4-W genotype with treatment-related leukemia, these findings may indicate that pharmacokinetic studies of free epipodophyllotoxin and metabolites are warranted to investigate a role of the metabolites further. Alternatively, it is possible that CYP3A4-W confers another phenotype of relevance to leukemogenesis.
Additional investigation of the relationship of CYP3A4 genotype to myelodysplastic syndrome also may be warranted, because alkylating agents are associated with a distinct form of myelodysplastic syndrome and acute myeloid leukemia characterized by complete or partial deletion of chromosomes 5 and 7 (65, 66). CYP3A4 mediates 4-hydroxylation of IFOS, resulting in monooxygenation to a reactive intermediate (67). CYP3A4 also detoxifies IFOS by N-dechloroethylation (68). Although CYP3A4 is involved in CPM 4-hydroxylation, CPM is metabolized primarily by CYP2B6 (67). Therefore, the relationship of CYP3A4 genotype to DNA damage and the risk of treatment-related leukemia subsequent to exposure to CPM or IFOS may be more complex and more difficult to establish than the relationship of CYP3A4 genotype to epipodophyllotoxin-induced leukemia.
The patients studied represent a relatively large series of pediatric cancer subjects, given the paucity of treatment-related leukemias among survivors of primary childhood cancers, and they were characterized with respect to CYP3A4 genotype, chemotherapy exposures, and MLL gene translocations. Nonetheless, the sample size caused relatively small frequency counts and even “zero cell” observations in some of the comparisons. For example, although P values were significant, an OR effect of genotype could not be estimated for the 22 subjects with treatment-related leukemias characterized by MLL gene translocation, because there were no CYP3A4-V carriers in this group. Although the zero-cell observations are consistent with the finding that CYP3A4-V is underrepresented in patients with treatment-related leukemia, characterization of CYP3A4 genotype in additional patients may help to confirm this observation. Case–control studies of patients treated with the same CYP3A4 substrates also may be warranted to validate the findings. Ultimately, an understanding of the parameters that bring about variability in potentially leukemogenic genotoxicity may lead to the design of safer treatment regimens for high-risk individuals.
Acknowledgments
C.A.F. was supported by National Institutes of Health Grant 1R29CA66140-04, American Cancer Society Grant RPG-95-088-04-LBC, Leukemia Society of America Scholar Award (1996–2001), Children’s Cancer Group, National Leukemia Research Association Grant in Memory of Maria Bernabe Garcia, and The Children’s Hospital of Philadelphia High Risk High Impact Grant. P.C.N. was supported by National Institutes of Health Grant CA42232. I.A.B. was supported by National Institutes of Health Grant CA65878. T.R.R. was supported by Public Health Service Grants ES-08031, CA-73730, and CA-60798 and a grant from the University of Pennsylvania Cancer Center.
ABBREVIATIONS
- CI
confidence interval
- CPM
cyclophosphamide
- CYP
cytochrome P450
- FET
Fisher’s Exact Test
- IFOS
ifosphamide
- OR
odds ratio
- VBL
vinblastine
- VM26
teniposide
- VP16
etoposide
References
- 1. Flannery J T, Boice J D, Jr, Devesa S S, Kleinerman R A, Curtis R E, Fraumeni J F., Jr Natl Cancer Inst Monogr. 1985;68:13–24. [PubMed] [Google Scholar]
- 2.Kaldor J. Acta Oncol. 1990;29:647–655. doi: 10.3109/02841869009090070. [DOI] [PubMed] [Google Scholar]
- 3.Donaldson S S, Hancock S L. N Engl J Med. 1997;334:792–793. doi: 10.1056/NEJM199603213341210. [DOI] [PubMed] [Google Scholar]
- 4.Wolden S L, Lamborn K L, Cleary S F, Tate D J, Donaldson S. J Clin Oncol. 1998;16:536–544. doi: 10.1200/JCO.1998.16.2.536. [DOI] [PubMed] [Google Scholar]
- 5.Travis L B, Curtis R E, Boice J D, Jr, Platz C E, Hankey B F, Fraumeni J F., Jr Cancer Res. 1996;56:1564–1570. [PubMed] [Google Scholar]
- 6.Meadows A T, Baum E, Fossati-Bellani F, Green D, Jenkin R D T, Marsden B, Nesbit M, Newton W, Oberlin O, Sallan S G, et al. J Clin Oncol. 1985;3:532–538. doi: 10.1200/JCO.1985.3.4.532. [DOI] [PubMed] [Google Scholar]
- 7.Felix C. In: Multiple Primary Cancers: Incidence, Etiology, Diagnosis, and Prevention. Neugut A I, Meadows A T, Robinson E, editors. Baltimore: Williams & Wilkins; 1998. , in press. [Google Scholar]
- 8.Boice J, Jr, Travis L, Curtis L. Proc Am Assoc Cancer Res. 1997;38:645. (abstr.). [Google Scholar]
- 9.Maris J M, Wiersma S R, Mahgoub N, Thompson P, Geyer R J, Hurwitz C G H, Lange B J, Shannon K M. Cancer. 1997;79:1438–1446. [PubMed] [Google Scholar]
- 10.Side L, Taylor B, Cayouette M, Conner E, Thompson P, Luce M, Shannon K. N Engl J Med. 1997;336:1713–1720. doi: 10.1056/NEJM199706123362404. [DOI] [PubMed] [Google Scholar]
- 11.Felix C A, Hosler M R, Provisor D, Salhany K, Sexsmith E A, Slater D J, Cheung N-K V, Winick N J, Strauss E A, Heyn R, et al. Blood. 1996;87:4376–4381. [PubMed] [Google Scholar]
- 12.Panizo C, Patino A, Calasanz J, Rifon J, Sierrasesumaga L, Rocha E. Med Pediatr Oncol. 1998;30:165–169. doi: 10.1002/(sici)1096-911x(199803)30:3<165::aid-mpo7>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 13.Hayes J D, Pulford D J. Crit Rev Biochem Mol Biol. 1995;30:445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- 14.Smith G, Stanley L A, Sim E, Strange R C, Wolf C R. Cancer Surv. 1995;25:27–65. [PubMed] [Google Scholar]
- 15.Raunio H, Husgafvel-Pursianen K, Anttila S, Hietanen E, Hirvonen A, Pelkonen O. Gene. 1995;159:113–121. doi: 10.1016/0378-1119(94)00448-2. [DOI] [PubMed] [Google Scholar]
- 16.Hande K. Semin Oncol. 1992;19:3–9. [PubMed] [Google Scholar]
- 17.Sonnichsen D, Liu Q, Schuetz E, Schuetz J, Pappo A, Relling M. J Pharmacol Exp Ther. 1995;275:566–575. [PubMed] [Google Scholar]
- 18.Friche E, Danks M, Beck W. Cancer Res. 1992;52:5701–5706. [PubMed] [Google Scholar]
- 19.Relling M, Evans R, Dass C, Desiderio D, Nemec J. J Pharmacol Exp Ther. 1992;261:491–496. [PubMed] [Google Scholar]
- 20.Wolf C R, Smith C A D, Gough A C, Moss J E, Vallis K A, Howard G, Carey F J, Mills K, McNee W, Carmichael J, et al. Carcinogenesis. 1992;13:1035–1038. doi: 10.1093/carcin/13.6.1035. [DOI] [PubMed] [Google Scholar]
- 21.Chen H, Sandler D P, Taylor J A, Shore D L, Liu E, Bloomfield C D, Bell D A. Lancet. 1996;347:295–297. doi: 10.1016/s0140-6736(96)90468-7. [DOI] [PubMed] [Google Scholar]
- 22.Li A, Kaminski D, Rasmussen A. Toxicology. 1995;104:1–8. doi: 10.1016/0300-483x(95)03155-9. [DOI] [PubMed] [Google Scholar]
- 23.Wrighton S, Stevens J. Crit Rev Toxicol. 1992;22:1–21. doi: 10.3109/10408449209145319. [DOI] [PubMed] [Google Scholar]
- 24.Rebbeck T R, Jaffe J M, Walker A H, Wein A J, Malkowicz S B. J Natl Cancer Inst. 1998;90:1225–1229. doi: 10.1093/jnci/90.16.1225. [DOI] [PubMed] [Google Scholar]
- 25.Felix C, Kim C, Megonigal M, Slater D, Jones D, Spinner N, Stump T, Hosler M, Nowell P, Lange B, et al. Blood. 1997;90:4679–4686. [PubMed] [Google Scholar]
- 26.Megonigal M D, Rappaport E F, Jones D H, Williams T M, Lovett B D, Kelly K M, Lerou P H, Moulton T, Budarf M L, Felix C A. Proc Natl Acad Sci USA. 1998;95:6413–6418. doi: 10.1073/pnas.95.11.6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felix C A, Hosler M R, Slater D J, Parker R, Masterson M, Whitlock J A, Rebbeck T R, Nowell P C, Lange B J. J Pediatr Hematol Oncol. 1998;20:299–308. doi: 10.1097/00043426-199807000-00004. [DOI] [PubMed] [Google Scholar]
- 28.Felix C A, Hosler M R, Winick N J, Masterson M, Wilson A E, Lange B J. Blood. 1995;85:3250–3256. [PubMed] [Google Scholar]
- 29.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce C M, Canaani E. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 30.Megonigal M D, Rappaport E F, Jones D H, Kim C S, Nowell P C, Lange B J, Felix C A. Proc Natl Acad Sci USA. 1997;94:11583–11588. doi: 10.1073/pnas.94.21.11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felix C A, Poplack D G, Reaman G H, Steinberg S M, Cole D E, Taylor B J, Begley C G, Kirsch I R. J Clin Oncol. 1990;8:431–442. doi: 10.1200/JCO.1990.8.3.431. [DOI] [PubMed] [Google Scholar]
- 32.Hashimoto H, Toide K, Kitamura R, Fujita M, Tagawa S, Itoh S, Kamataki T. Eur J Biochem. 1993;218:585–595. doi: 10.1111/j.1432-1033.1993.tb18412.x. [DOI] [PubMed] [Google Scholar]
- 33.Inoue K, Inazawa J, Nakagawa H, Shimada T, Yamazaki H, Guengerich F P, Abe T. Jpn J Hum Genet. 1992;37:133–138. doi: 10.1007/BF01899734. [DOI] [PubMed] [Google Scholar]
- 34.Pui C-H, Hancock M L, Raimondi S C, Head D R, Thompson E, Wilimas J, Kun L E, Bowman L C, Crist W M, Pratt C B. Lancet. 1990;336:417–421. doi: 10.1016/0140-6736(90)91956-b. [DOI] [PubMed] [Google Scholar]
- 35.Pui C-H, Ribeiro R C, Hancock M L, Rivera G K, Evans W E, Raimondi S C, Head D R, Behm F G, Mahmoud M H, Sandlund J T, et al. N Engl J Med. 1991;325:1682–1687. doi: 10.1056/NEJM199112123252402. [DOI] [PubMed] [Google Scholar]
- 36.Winick N, McKenna R W, Shuster J J, Schneider N R, Borowitz M J, Bowman W P, Jacaruso D, Kamen B A, Buchanan G R. J Clin Oncol. 1993;11:209–217. doi: 10.1200/JCO.1993.11.2.209. [DOI] [PubMed] [Google Scholar]
- 37.Hunger S P, Tkachuk D C, Amylon M D, Link M P, Carroll A J, Welborn J L, Wilman C L, Cleary M L. Blood. 1993;81:3197–3203. [PubMed] [Google Scholar]
- 38.Broeker P L S, Super H G, Thirman M J, Pomykala H, Yonebayashi Y, Tanabe S, Zeleznik-Le N, Rowley J D. Blood. 1996;87:1912–1922. [PubMed] [Google Scholar]
- 39.Felix C A, Winick N J, Negrini M, Bowman W P, Croce C M, Lange B J. Cancer Res. 1993;53:2954–2956. [PubMed] [Google Scholar]
- 40.Pedersen-Bjergaard J. Leuk Res. 1992;16:61–65. doi: 10.1016/0145-2126(92)90102-d. [DOI] [PubMed] [Google Scholar]
- 41.Pedersen-Bjergaard J, Philip P. Blood. 1991;78:1147–1148. [PubMed] [Google Scholar]
- 42.Ratain M J, Kaminer L S, Bitran J D, Larson R A, Le Beau M M, Skosey C, Purl S, Hoffman P C, Wade J, Vardiman J W, et al. Blood. 1987;70:1412–1417. [PubMed] [Google Scholar]
- 43.Nucifora G, Rowley J. Blood. 1995;86:1–14. [PubMed] [Google Scholar]
- 44.Detourmignies L, Castaigne S, Stoppa A M, Harousseau J L, Sadoun A, Janvier M, Demory J L, Sanz M, Berger R, Bauters F, et al. J Clin Oncol. 1992;10:1430–1435. doi: 10.1200/JCO.1992.10.9.1430. [DOI] [PubMed] [Google Scholar]
- 45.Quensel B, Kantarjian H, Pedersen-Bjergaard J, Brault P, Estey E, Lai J, Tilly H, Stoppa A, Archimbaud E, Harousseau J, et al. J Clin Oncol. 1993;11:2370–2379. doi: 10.1200/JCO.1993.11.12.2370. [DOI] [PubMed] [Google Scholar]
- 46.Pedersen-Bjergaard J, Brondum-Nielsen K, Karle H, Johansson B. Leukemia. 1997;11:1571–1574. doi: 10.1038/sj.leu.2400769. [DOI] [PubMed] [Google Scholar]
- 47.Smith, M., Rubenstein, L., Anderson, J., Catalano, P., Freidlin, B., Heyn, R., Khayat, A., Krailo, M., Land, V., Miser, J., et al. (1998) J. Clin. Oncol., in press. [DOI] [PubMed]
- 48.Relling M. Am Soc Pharmacol Exp Ther. 1994;45:352–358. [Google Scholar]
- 49.Walker, A. H., Jaffe, J. M., Gunasegaram, S., Cummings, S. A., Huang, C.-S., Chern, H.-D., Olopade, O. I., Weber, B. L. & Rebbeck, T. R. (1998) Hum. Mutat., in press. [PubMed]
- 50.van Maanen J M, Retel J, de Vries J, Pinedo H M. J Natl Cancer Inst. 1988;80:1526–1533. doi: 10.1093/jnci/80.19.1526. [DOI] [PubMed] [Google Scholar]
- 51.Mans D, Retel J, van Maanen J, Lafleur M, van Schaik M, Pinedo H, Lankelma J. Br J Cancer. 1990;62:54–60. doi: 10.1038/bjc.1990.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cavalieri E L, Stack D E, Devanesan P D, Todorovic R, Dwivedy I, Higginbotham S, Johansson S L, Patil K D, Gross M L, Gooden J K, et al. Proc Natl Acad Sci USA. 1997;94:10937–10942. doi: 10.1073/pnas.94.20.10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Demple B, Harrison L. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 54.Sun B, Latham K A, Dodson M L, Lloyd R S. J Biol Chem. 1995;270:19501–19508. doi: 10.1074/jbc.270.33.19501. [DOI] [PubMed] [Google Scholar]
- 55.Singer B, Hang B. Chem Res Toxicol. 1997;10:713–732. doi: 10.1021/tx970011e. [DOI] [PubMed] [Google Scholar]
- 56.Krokan H E, Standal R, Slupphaug G. Biochem J. 1997;325:1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Corbett A, Osheroff N. Chem Res Toxicol. 1993;6:585–597. doi: 10.1021/tx00035a001. [DOI] [PubMed] [Google Scholar]
- 58.Kingma P, Corbett A, Burcham P, Marnett L, Osheroff N. J Biol Chem. 1995;270:21441–21444. doi: 10.1074/jbc.270.37.21441. [DOI] [PubMed] [Google Scholar]
- 59.Kingma P, Osheroff N. J Biol Chem. 1997;272:1148–1155. doi: 10.1074/jbc.272.2.1148. [DOI] [PubMed] [Google Scholar]
- 60.Kingma P, Osheroff N. J Biol Chem. 1997;272:7488–7493. doi: 10.1074/jbc.272.11.7488. [DOI] [PubMed] [Google Scholar]
- 61.Kingma P, Greider C, Osheroff N. Biochemistry. 1997;36:5934–5939. doi: 10.1021/bi970507v. [DOI] [PubMed] [Google Scholar]
- 62.Liang F, Han M, Romanienko P J, Jasin M. Proc Natl Acad Sci USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilkinson G R. J Pharmacokinet Biopharm. 1996;24:475–490. doi: 10.1007/BF02353475. [DOI] [PubMed] [Google Scholar]
- 64.Relling M V, Yanishevski Y, Nemec J, Evans W E, Boyett J M, Behm F G, Pui C-H. Leukemia. 1998;12:346–352. doi: 10.1038/sj.leu.2400928. [DOI] [PubMed] [Google Scholar]
- 65.Hawkins M M, Kinnier-Wilson L M, Stovall M A, Marsden H B, Potok M H N, Kingston J E, Chessells J M. Br Med J. 1992;304:951–958. doi: 10.1136/bmj.304.6832.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tucker M A, Meadows A T, Boice J D, Stovall M, Oberlin O, Stone B J, Birch J, Voute P A, Hoover R N, Fraumeni J F., Jr J Natl Cancer Inst. 1987;78:459–464. [PubMed] [Google Scholar]
- 67.Chang T, Weber G, Crespi C, Waxman D. Cancer Res. 1993;53:5629–5637. [PubMed] [Google Scholar]
- 68.Walker D, Flinois J P, Monkman S C, Beloc C, Boddy A V, Cholerton S, Daly A K, Lind M J, Pearson A D J, Beaune P H, et al. Biochem Pharmacol. 1994;47:1157–1163. doi: 10.1016/0006-2952(94)90387-5. [DOI] [PubMed] [Google Scholar]