Short abstract
Ligand-protein mapping was found to follow a power law and the preferential attachment principle, leading to the identification of the molecules, mostly nucleotide-containing compounds, that are likely to have evolved earliest.
Abstract
Background
Extant life depends greatly on the binding of small molecules (such as ligands) with macromolecules (such as proteins), and one ligand can bind multiple proteins. However, little is known about the global patterns of ligand-protein mapping.
Results
By examining 2,186 well-defined small-molecule ligands and thousands of protein domains derived from a database of druggable binding sites, we show that a few ligands bind tens of protein domains or folds, whereas most ligands bind only one, which indicates that ligand-protein mapping follows a power law. Through assigning the protein-binding orders (early or late) for bio-ligands, we demonstrate that the preferential attachment principle still holds for the power-law relation between ligands and proteins. We also found that polar molecular surface area, H-bond acceptor counts, H-bond donor counts and partition coefficient are potential factors to discriminate ligands from ordinary molecules and to differentiate super ligands (shared by three or more folds) from others.
Conclusion
These findings have significant implications for evolution and drug discovery. First, the chronology of ligand-protein binding can be inferred by the power-law feature of ligand-protein mapping. Some nucleotide-containing ligands, such as ATP, ADP, GDP, NAD, FAD, dihydro-nicotinamide-adenine-dinucleotide phosphate (NDP), nicotinamide-adenine-dinucleotide phosphate (NAP), flavin mononucleotide (FMN) and AMP, are found to be the earliest cofactors bound to proteins, agreeing with the current understanding of evolutionary history. Second, the finding that about 30% of ligands are shared by two or more domains will help with drug discovery, such as in finding new functions from old drugs, developing promiscuous drugs and depending more on natural products.
Background
Life is essentially a molecular network, not only in the individual sense but also at the ecosystem level [1,2]. The network depends greatly on the binding of small molecules (for example, ligands and cofactors) with macromolecules (for example, proteins). Small-molecule ligands not only participate in many basic enzymatic reactions (as coenzymes or substrates) to build metabolic networks, but also act as extra- and intra-cellular signals to help construct regulation networks [3-9]. The great potential of small-molecule ligands to make links between different proteins means that one ligand can bind to diverse targets [10-13]. In fact, some ligands are extremely powerful in contacting proteins, which are termed hubs of biochemical networks [14-17]. However, little is known about the global patterns of ligand-protein mapping, which stimulated our interest to do a comprehensive analysis and explore the biological and chemical bases underlying the mapping patterns. Since ligand-protein binding is one of the most basic biochemical processes, the present study has significant implications for tracing the important events in the origin of life and as well as for understanding the new paradigms in drug discovery.
Results
Distribution patterns of ligands in the protein universe
Although considerable efforts have been devoted to constructing ligand databases [18-26], it is still a great challenge to select clearly defined ligands from them. Thanks to the endeavor of Rognan and co-workers, a well-defined ligand database, the Annotated Database of Druggable Binding Sites from the PDB (sc-PDB), was released recently [27]. For this database, the ligands were collected according to the following criteria: only host proteins with high-resolution (<2.5 Å) crystal structures were considered; water molecule, metal ions and other 'unwanted molecules' (for example, solvents, detergents and covalently bound ligands) were removed; only small-molecular-weight ligands (ranging from 70 to 800 Da for heavy atoms) were selected; and only ligands with a limited solvent-exposed surface (that is, less than 50% of their surface exposed to the solvent) were picked. In addition, the corresponding binding sites were also extracted and were defined by all of the protein residues with at least one atom within 6.5 Å of any ligand atom. Taken together, the clear definition for the ligands in sc-PDB guarantees the repeatability of the present analysis, which gives sc-PDB an advantage over other ligand databases.
Through searching sc-PDB, 2,186 small-molecule ligands were selected, which are bound by 5,740 domains (the domains were counted at a non-redundant level and constituted domain space; Additional data file 1). According to SCOP 1.69 [28,29], these domains were classified into 591 folds. As one fold may cover multiple domains and bind more than one ligand, the fold occurrences amounted to 3,224, which constituted the fold universe.
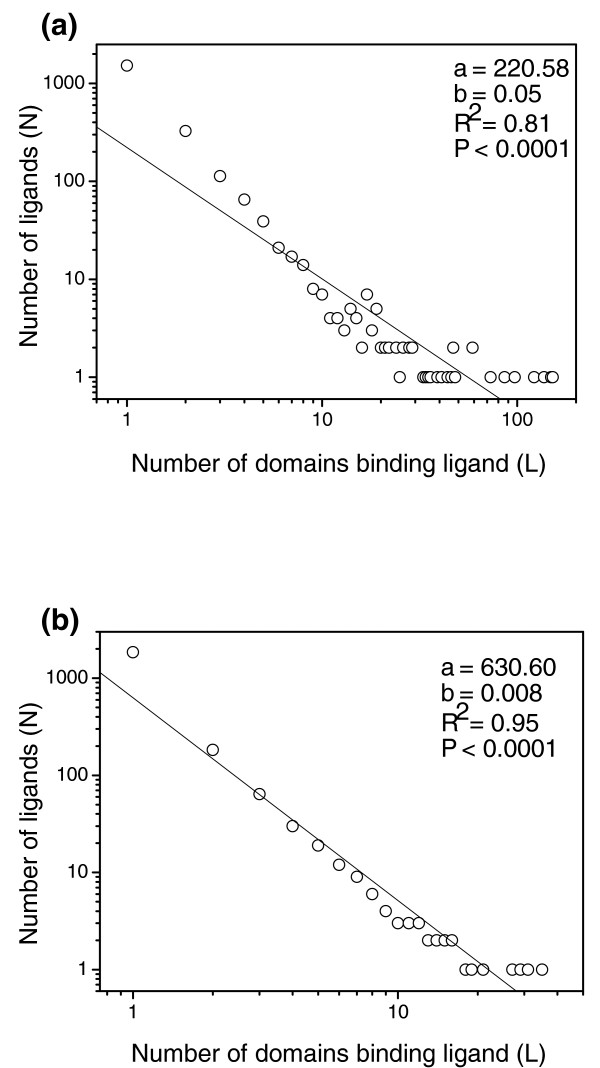
As shown in Additional data file 1, ligands do not distribute evenly in the domain space. A few ligands cover 100+ domains, 681 ligands (31.2%) are shared by 2 or more domains and 1,505 (68.8%) bind only one. Moreover, ligands also populate unevenly in the protein architecture universe. For instance, 1,833 ligands (83.9%) are bound by only one fold, 185 (8.5%) by two, while 24 ligands (1.1%) are bound by 10+ folds (Additional data file 1). The most common ligand, ATP (adenosine-5'-triphosphate), is shared by 35 folds. As illustrated in Figure 1, the number of ligands (N) decays with increasing number (L) of domains and folds that bind the ligand and follows the power law N = aL-b (P < 0.0001). It is interesting to note that most of the widely shared ligands (such as those shared by 15+ folds; Additional data file 1) are hubs of metabolic networks [14-16] and are vital to metabolism (especially energy metabolism).
Figure 1.
Power-law behaviors of ligand-protein binding. The number of ligands (N) decays with an increase in the number (L) of (a) domains and (b) folds that bind the ligand and follows the equation N = aL-b. The figure illustrates that a few ligands cover tens of protein domains or folds, while most ligands bind only one domain or fold.
Biological basis underlying the power-law behaviors of ligand-protein binding
Although power law is a central concept in network sciences and has been implicated in most biological networks [14-16], it is a challenge to elucidate the mechanisms underlying the rule. The most popular theoretical models resort to preferential attachment principle, which attributes the different connections of nodes to their different emerging orders, that is to say, the more connected nodes originated earlier than the less connected nodes [30]. Although the preferential attachment principle has been justified for protein networks [31-33], it remains unclear whether it can be applied to protein-ligand binding.
As a large part of the sc-PDB-derived ligands are synthetic, to explore the applicability of the preferential attachment principle to protein-ligand binding, we extracted bio-ligands from the ligand dataset. To do this, the MetaCyc database (9.5; a metabolic-pathway database that contains 5,253 metabolites) [34] was employed to filter the non-metabolic ligands. As a result, 128 bio-ligands were obtained, which bind to 1,662 domains (counted at a non-redundant level). According to SCOP 1.69 [28,29], these domains were classified into 207 folds. As one fold may cover multiple domains and bind more than one ligand, the fold occurrences amounted to 574. Although these ligands are only metabolism-relevant, they also follow power-law distribution in the protein universe (Additional data file 2).
As the quantity of bio-ligands is limited, to guarantee statistical significance, the 128 bio-ligands were classified into only two categories: first, 70 early ligands, which are owned by both prokaryotic (Escherichia coli) and eukaryotic (yeast or higher) species; and second, 54 late ligands, which are owned only by eukaryotic (yeast or higher) species (4 ligands failed in age assignment) (Additional data file 3). It is interesting to note that early ligands cover 7.1 folds on average, in contrast to late ligands, which cover only 1.2 folds on average, and that all (100%) super ligands (shared by 3+ folds) originated early, while most (64.8%) ordinary ligands (bind to 3 or less folds) appeared late. All of these findings strongly suggest that the preferential attachment principle still holds for ligand-protein binding to a large extent.
Chemical basis underlying the power-law behaviors of ligand-protein binding
It has been widely accepted that protein folds are among the most conserved elements of life [35-37]. However, the present analysis indicates that 353 ligands (16.1%) are shared by 2 or more folds and 104 ligands (4.8%) can cover 3+ folds, which suggests that ligand binding is not constrained by the global architecture of proteins. This finding is consistent with a recent concept that the local structures around an active site are more basic than folds to describe a protein's biological space (binding site for potential ligands) [38]. This phenomenon can be elucidated, at least in part, in terms of the structure-function relationships of proteins. First, binding sites and ligands are quite flexible and plastic [39-41], and therefore, binding-site selection is, to certain extent, ligand dependent [42-44]. Second, ligand binding is governed by a few conserved residues and, thus, is a local rather than a global property of proteins [10,11]. However, the structural factors underlying the strong protein-binding ability of the super ligands still remain unknown. In addition, it is also of interest to explore the structural features discriminating ligands from ordinary molecules. Therefore, the chemical space consisting of ligands and ordinary molecules was charted to reveal the relationship between the ligand distribution patterns in the protein universe and in the chemical space.
The chemical space is composed of 2,176 ligands derived from sc-PDB (due to the lack of atomic parameters, 10 of the 2,186 ligands failed to go through the descriptor calculations) and 2,184 small molecules randomly selected from ACD-SC (Available Chemicals Directory-Screening Compounds, Version 2005.1, Molecular Design Ltd. Information Systems Inc., San Leardo, CA, USA; which collects chemicals that are commercially available and is broadly regarded as a source of ordinary molecules [45]). Seventy descriptors characterizing the structural features of these molecules were calculated, of which 13 were calculated by Sybyl (Tripos Inc., St Louis, Missouri, USA [46]), 49 by Cerius2 (Version 4.10L, Accelrys Inc., San Diego, CA, USA [47]) and 8 by an in-house program written in Perl (Table 1).
Table 1.
Descriptors of chemical space consisting of sc-PDB-derived ligands and ACD-SC-derived ordinary molecules and corresponding loadings (Varimax normalized) for the first two factors*
| Descriptors | Characterization | Factor loadings | Software | |
| 1 | 2 | |||
| AREA | Total molecular surface area | 0.974 | 0.103 | Sybyl |
| PSA | Polar molecular surface area | 0.255 | 0.892 | |
| PV | Polar molecular volume | 0.501 | 0.741 | |
| VOL | Total molecular volume | 0.991 | 0.062 | |
| MOLWEIGHT | Molecular weight | 0.958 | 0.206 | |
| Acceptor | H-bond acceptor counts | 0.464 | 0.799 | |
| Donor | H-bond donor counts | 0.376 | 0.817 | |
| BondCount | Total bond counts | 0.972 | 0.060 | |
| Chiral | Counts of chiral center | 0.367 | 0.617 | |
| Hydrophobe | Hydrophobic fragment counts | 0.767 | -0.417 | |
| RingCount | Ring counts | 0.686 | -0.069 | |
| RotBonds | Number of rotatable bonds | 0.630 | 0.428 | |
| HeavyAtoms | Number of non-H atoms | 0.978 | 0.149 | |
| Carbons | Number of carbons atoms | 0.943 | -0.228 | Perl |
| Oxygens | Number of oxygen atoms | 0.425 | 0.793 | |
| Nitrogens | Number of nitrogen atoms | 0.475 | 0.324 | |
| Sulfurs | Number of sulfur atoms | 0.141 | -0.009 | |
| Phosphorus | Number of phosphorus atoms | 0.162 | 0.617 | |
| Halides | Number of halide atoms | 0.076 | -0.170 | |
| DoubleBonds | Number of double bonds | 0.527 | 0.378 | |
| TripleBonds | Number of triple bonds | -0.009 | -0.109 | |
| RadOfGyration | Radius of gyration | 0.888 | 0.004 | Cerius 2 |
| ShadowXY | Surface area projections | 0.967 | 0.076 | |
| ShadowXZ | 0.951 | 0.053 | ||
| ShadowYZ | 0.877 | 0.093 | ||
| ShadowXYfrac | -0.610 | -0.027 | ||
| ShadowXZfrac | -0.421 | -0.002 | ||
| ShadowYZfrac | -0.289 | 0.039 | ||
| Shadownu | 0.268 | -0.117 | ||
| ShadowXlength | 0.849 | -0.008 | ||
| ShadowYlength | 0.798 | 0.075 | ||
| ShadowZlength | 0.756 | 0.059 | ||
| Density | Density | -0.089 | 0.354 | |
| PMImag | Principal moment of inertia | 0.819 | 0.134 | |
| AlogP | Log of the partition coefficient using Ghose and Crippen's method. | 0.425 | -0.727 | |
| AlogP98 | Log of the partition coefficient, atom-type value, using latest parameters. | 0.365 | -0.852 | |
| Fh2o | Desolvation free energy for water | -0.479 | -0.762 | |
| Foct | Desolvation free energy for octanol | -0.578 | -0.617 | |
| LogP | Log of the partition coefficient. | -0.022 | -0.892 | |
| MR | Molar refractivity using Hopfinger's method. | 0.835 | -0.110 | |
| MolRef | Molar refractivity using linear additive method based on AlogP atom types | 0.986 | -0.033 | |
| JX | Balaban indices | -0.567 | 0.027 | |
| Kappa1 | Kappa topological indices | 0.969 | 0.189 | |
| Kappa2 | 0.926 | 0.026 | ||
| Kappa3 | 0.691 | 0.033 | ||
| Kappa1AM | 0.958 | 0.220 | ||
| Kappa2AM | 0.901 | 0.050 | ||
| Kappa3AM | 0.630 | 0.046 | ||
| PHI | Molecular flexibility index | 0.800 | 0.078 | |
| SC0 | Subgraph topological counts | 0.980 | 0.147 | |
| SC1 | 0.973 | 0.125 | ||
| SC2 | 0.943 | 0.186 | ||
| SC3P | 0.904 | 0.141 | ||
| SC3C | 0.749 | 0.389 | ||
| SC3CH | 0.016 | -0.086 | ||
| CHI0 | Kier and Hall Chi connectivity indices | 0.974 | 0.190 | |
| CHI1 | 0.983 | 0.115 | ||
| CHI2 | 0.958 | 0.210 | ||
| CHI3P | 0.939 | 0.136 | ||
| CHI3C | 0.655 | 0.484 | ||
| CHI3CH | 0.015 | -0.087 | ||
| CHIV0 | 0.990 | 0.076 | ||
| CHIV1 | 0.971 | 0.120 | ||
| CHIV2 | 0.913 | 0.137 | ||
| CHIV3P | 0.838 | 0.096 | ||
| CHIV3C | 0.476 | 0.148 | ||
| CHIV3CH | 0.016 | -0.088 | ||
| Wiener | Wiener topological index | 0.854 | 0.186 | |
| logZ | Logarithm of Hosoya topological index | -0.220 | -0.131 | |
| Zagreb | Zagreb topological index | 0.958 | 0.162 | |
*The first factor explains 52.8% of the variance and the second explains 12.7%. Factors with high loadings (>0.9 for first factors and >0.8 for second factors) are shown in bold.
We used factor analysis to visualize the diversity of the molecules. Factor analysis is widely used to study the patterns of relationship among many dependent variables, with the goal of discovering something about the nature of the independent variables (called factors) that affect them [48,49]. In the present analysis, two factors, which can explain 65.5% of the variance, were extracted by principal component analysis and rotated by the Varimax method [50] to chart the two-dimensional chemical space of small molecules. The factor loadings (Varimax normalized) are listed in Table 1.
From the factor loadings, we see that the first factor, explaining 52.8% of the variance, contains high loadings (>0.9; shown in bold in Table 1) from constitutional properties (such as total molecular surface area, total molecular volume, molecular weight, total bond counts, number of non-hydrogen atoms and number of carbons atoms) and topological properties (such as Kappa topological indices, subgraph topological counts, Kier and Hall Chi connectivity indices and Zagreb topological Index). In comparison, the second factor, explaining 12.7% of the variance, contains important contributions (with loadings of higher than 0.8; shown in bold in Table 1) from electronic properties, such as polar molecular surface area, H-bond acceptor counts (whose loading is 0.799), H-bond donor counts and partition coefficient (measured by AlogP98 and LogP).
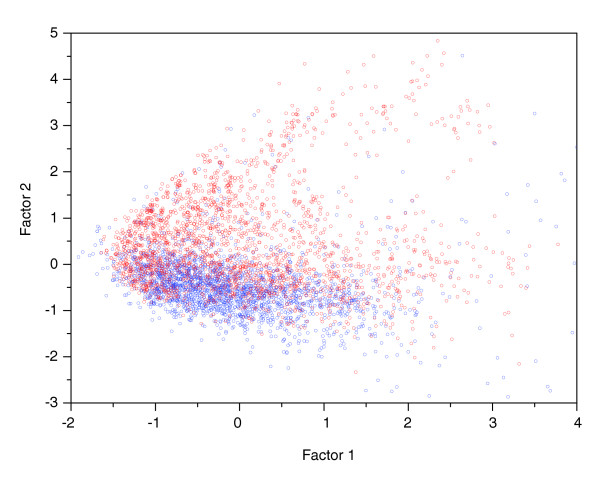
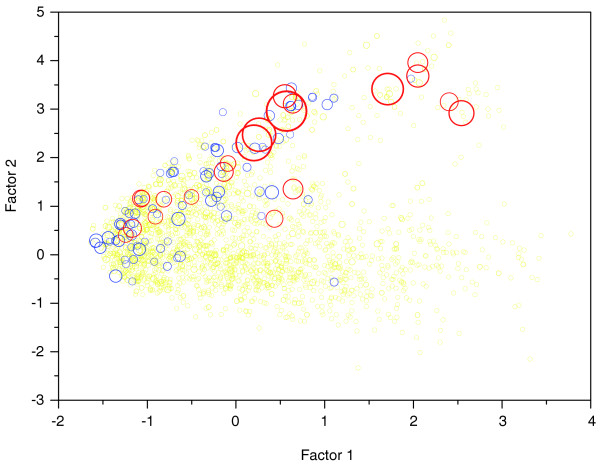
In the chemical space formed by the two factors (Figure 2), one can find some differences between the distribution patterns of ligands and ordinary molecules. That is, ligands (in red) occupy the relatively upper part of the space, while ordinary molecules (in blue) hold the relatively lower part, which implies that it is the second factor that discriminates ligands from ordinary molecules. As a consequence, it can be deduced that polar molecular surface area, H-bond donor counts, H-bond acceptor counts and partition coefficient are likely responsible for the differences between ligands and ordinary molecules, which agrees well with the current understanding of the chemical basis of ligand-protein binding that electrostatic interactions (including H-bond) and hydrophobic interactions make major contributions to the binding. More interestingly, as shown in Figure 3, super ligands (in blue and red) do not distribute randomly in the chemical space, but concentrate in the relatively upper part of the space, which suggests that polar molecular surface area, H-bond donor counts, H-bond acceptor counts and partition coefficient are also key factors discriminating super ligands from others.
Figure 2.
Chemical space consisting of ligands (derived from sc-PDB) and ordinary molecules (randomly selected from ACD-SC), defined by the first two factors derived from 70 descriptors. The figure illustrates that ligands (in red) occupy the relatively upper part of the space, while ordinary molecules (in blue) occupy the relatively lower part, which means that it is the second factor that discriminates ligands from ordinary molecules. From the loadings of the second factor, it can be deduced that polar molecular surface area, H-bond donor counts, H-bond acceptor counts and partition coefficient are likely responsible for the differences between ligands and ordinary molecules, which is supported by the different average values of the four kinds of parameters for ligands and ordinary molecules (Table 2).
Figure 3.
Chemical space consisting of sc-PDB-derived ligands, defined by the first two factors derived from 70 descriptors. The figure illustrates that super ligands (shared by 3+ folds; in blue), especially those that are shared by 10+ folds (in red), concentrate in the relatively upper part of the space (the area of the circle is directly proportional to the number of folds that bind the ligand), which suggests that polar molecular surface area, H-bond donor counts, H-bond acceptor counts and partition coefficient are responsible for the strong protein-binding potential of the super ligands, which is supported by the different average values of the four kinds of parameters for ligands with different protein-binding potentials (Table 2).
To shed more light on the above findings, the average values of descriptors characterizing polar molecular surface area, H-bond donors, H-bond acceptors and partition coefficient were calculated for ordinary molecules, ligands and super ligands. From Table 2, it can be seen that there indeed exist correlations between protein-binding ability and the four kinds of parameters. The protein-binding potential of ligands is positively correlated with polar molecular surface area, H-bond donor and acceptor counts, and negatively correlated with partition coefficient (measured by AlogP98 and LogP).
Table 2.
Average values of descriptors characterizing polar molecular surface area, H-bond donors, H-bond acceptors, partition coefficient and rotatable bonds for ordinary molecules, ligands and ligands with different protein-binding potentials
| Descriptor* | Small molecules† | Average values | Standard error | Number of molecules |
| PSA | Molecules | 111.81 | 1.79 | 2,184 |
| Ligands | 230.59 | 2.79 | 2,176 | |
| Ligands (≤ 3) | 225.71 | 2.80 | 2,072 | |
| Ligands (4-9) | 304.28 | 15.67 | 80 | |
| Ligands (≥ 10) | 406.83 | 33.10 | 24 | |
| Donor | Molecules | 1.51 | 0.04 | 2,184 |
| Ligands | 3.97 | 0.07 | 2,176 | |
| Ligands (≤ 3) | 3.87 | 0.07 | 2,072 | |
| Ligands (4-9) | 5.24 | 0.43 | 80 | |
| Ligands (≥ 10) | 8.21 | 0.90 | 24 | |
| Acceptor | Molecules | 3.35 | 0.05 | 2,184 |
| Ligands | 5.87 | 0.09 | 2,176 | |
| Ligands (≤ 3) | 5.74 | 0.09 | 2,072 | |
| Ligands (4-9) | 7.69 | 0.53 | 80 | |
| Ligands (≥ 10) | 11.00 | 1.18 | 24 | |
| AlogP98 | Molecules | 2.87 | 0.05 | 2,184 |
| Ligands | 0.81 | 0.06 | 2,176 | |
| Ligands (≤ 3) | 0.92 | 0.06 | 2,072 | |
| Ligands (4-9) | -1.33 | 0.25 | 80 | |
| Ligands (≥ 10) | -1.80 | 0.38 | 24 | |
| LogP | Molecules | 0.77 | 0.08 | 2,184 |
| Ligands | -2.27 | 0.10 | 2,176 | |
| Ligands (≤ 3) | -2.10 | 0.10 | 2,072 | |
| Ligands (4-9) | -5.06 | 0.50 | 80 | |
| Ligands (≥ 10) | -8.11 | 0.96 | 24 | |
| RotBond | Molecules | 4.88 | 0.09 | 2,184 |
| Ligands | 7.49 | 0.11 | 2,176 | |
| Ligands (≤ 3) | 7.43 | 0.11 | 2072 | |
| Ligands (4-9) | 8.00 | 0.50 | 80 | |
| Ligands (≥ 10) | 11.33 | 1.19 | 24 |
* PSA, polar molecular surface area; Donor, H-bond donor counts; Acceptor, H-bond acceptor counts; AlogP98, log of the partition coefficient, atom-type value, using latest parameters; LogP, log of the partition coefficient; RotBond, number of rotatable bonds. †Molecules, ACD-SC-derived ordinary molecules; Ligands, sc-PDB-derived ligands; Ligands (≤ 3), ligands covering ≤ 3 folds; Ligands (4-9), ligands covering 4-9 folds; Ligands (≥ 10), ligands covering ≥ 10 folds.
Recently, through examining the conformational diversity of some very common ligands (that is, ATP, NAD and FAD) bound to proteins, Stockwell and Thornton [41] suggested that molecular flexibility is important for ligands to bind diverse proteins. This opinion is partially supported by the present analysis. Although the contribution from the number of rotatable bonds (RotBonds) to the second factor is not very strong (the loading is 0.428; Table 1), there is a correlation between the protein-binding ability of ligands and index RotBonds. As listed in Table 2, the average RotBonds for ligands is significantly higher than that for ordinary molecules (independent samples t-test shows that P < 0.0001), and it is clear that the more folds the ligands cover, the higher the average RotBonds are for the ligands.
Discussion
Since ligand-protein binding is one of the most basic biochemical processes, the present findings have broad biological and medical implications.
Implications for tracing the chronology of ligand binding to proteins
The most challenging issue in life sciences may be elucidating how organisms originated from inorganic scratches (gases, water and clays), during which one of the most important missions is to establish the chronology of the important biological events. Thanks to the continuing efforts of chemists and biologists, the chronologies of the evolution of amino acids and proteins have been established in principle [37,51-55]. However, as many proteins bind ligands that are essential for their functions and the ligands are likely to have originated independently of proteins [56-59], the binding of ligands with primordial proteins would also be a critical step in the origin of life. Thus, it is intriguing to explore the chronology of ligand-protein binding and answer the following questions: which ligand was first recognized by a protein and what kind of architecture did the host protein have. Nevertheless, since there is no fossil of the last universal common ancestor, let alone the more ancestral organisms, it is a great challenge to trace the protein-binding history of early ligands.
As stated above, through determining the protein-binding ages of ligands, a rough temporal order (early or late) for ligand-protein binding can be inferred (as shown in Additional data file 3). However, considering the fact that fold distribution pattern in the sequence universe helps greatly to reveal the chronology of the evolution of protein architecture [37,53,54], we speculate that the power-law distribution of ligands in the protein universe may implicate a more explicit temporal order for ligand-protein binding. In fact, the preferential attachment principle underlying the power-law behavior of ligand-protein mapping suggests that the more widely a ligand is shared, the earlier it bound to proteins. As protein architecture is more conserved than sequence [35-37], the fold-based inference is believed to be more robust than the domain-based one. Therefore, the nine bio-ligands that are most popular in the fold universe (covering 15+ folds; Table 3) are considered to have bound their host proteins relatively earlier than others and to follow the order (from early to late): ATP, ADP (adenosine-5'-diphosphate), GDP (guanosine-5'-diphosphate), NAD (nicotinamide-adenine-dinucleotide), FAD (flavin-adenine dinucleotide), NDP (dihydro-nicotinamide-adenine-dinucleotide phosphate), NAP (nicotinamide-adenine-dinucleotide phosphate), FMN (flavin mononucleotide) and AMP (adenosine monophosphate).
Table 3.
The most prevalent bio-ligands in the fold universe (shared by 15+ folds) and the most common folds used by host proteins of each ligand
| Ligands | Number of folds | Most common folds |
| Adenosine-5'-triphosphate (ATP) | 35 | P-loop containing nucleoside triphosphate hydrolases (c.37) |
| Adenosine-5'-diphosphate (ADP) | 31 | P-loop containing nucleoside triphosphate hydrolases (c.37) |
| Guanosine-5'-diphosphate (GDP) | 29 | P-loop containing nucleoside triphosphate hydrolases (c.37) |
| Nicotinamide-adenine-dinucleotide (NAD) | 27 | NAD(P)-binding Rossmann-fold domains (c.2) |
| Flavin-adenine dinucleotide (FAD) | 21 | FAD/NAD(P)-binding domain (c.3) |
| Dihydro-nicotinamide-adenine-dinucleotide phosphate (NDP) | 18 | NAD(P)-binding Rossmann-fold domains (c.2) |
| Nicotinamide-adenine-dinucleotide phosphate (NAP) | 16 | NAD(P)-binding Rossmann-fold domains (c.2) |
| Flavin mononucleotide (FMN) | 16 | Flavodoxin-like (c.23) |
| Adenosine monophosphate (AMP) | 15 | Adenine nucleotide alpha hydrolase-like (c.26) |
A close inspection of ATP's host proteins reveals that although ATP covers 35 folds and 97 domains, most domains belong to a small group of folds, indicating that power law is still effective (Additional data file 4). According to the preferential attachment principle of fold usage [37], it is reasonable to infer that the most prevalent fold, P-loop hydrolase (c.37), was employed by ATP's first host (Table 3). Interestingly, c.37 is the most ancient fold predicted by a phylogenomic analysis of protein architectures [37,53,54]. Similar analyses allowed us to deduce the most ancestral host proteins of the other eight early ligands (Additional data file 4, Table 3). It is interesting to note that the predicted earliest hosts for the nine bio-ligands appeared in roughly the same order as the protein structures deduced by a phylogenomic analysis (that is, c.37 is the earliest, followed by c.2, c.23, c.3 and c.26, all of which belong to the α/β class) [37,53,54]. Although no consensus has been reached on the exact temporal order of protein architectures, α/β is generally considered to be the most ancient protein class [37,53,54,60-62]. In addition, based on an extensive analysis of sequences and structures of numerous proteins, Trifonov and co-workers [63-65] also inferred that some P-loop ATP-binding domains represent the most ancient proteins. Recently, through a phylogenomic analysis on protein architectures of modern metabolic networks, Caetano-Anollés and co-workers [66] indicated that enzymes with the P-loop hydrolase fold engaged in nucleotide (especially purine) metabolism may be the most primitive members of metabolic systems. Through examining the structures and functions of these members, we found that most (approximately 80%) of them need ATP to work normally. Therefore, the present speculations on the chronology of ligand-protein binding are self-consistent and are in line with the up-to-date knowledge on protein evolutionary history.
To get a deeper insight into the evolutionary features of ligands, the building block usage of 128 bio-ligands was analyzed. As shown in Additional data file 5, nucleic acid bases are the most frequently used building blocks, followed by carbohydrates and amino acids, which is in accordance with Nobeli et al.'s [67] finding that nucleic acid bases are the most common fragments of metabolites. More interestingly, many early bio-ligands (45.0%) contain nucleic acid bases; in particular, the nine earliest bio-ligands all contain one or more bases. In contrast, carbohydrates or amino acids are contained by only a small proportion of early bio-ligands (25.0% and 7.5%, respectively). This provides further evidence to support the notion that early ligands are vestiges of the RNA world [56].
As mentioned above, the presently revealed chronology of early ligands' host proteins is roughly in line with the previously deduced evolutionary history of protein architectures [37,53,54]. Thus, it is interesting to ask: is the accordance between both events fortuitous? Our answer is maybe not. Considering the prevalent ligand-induced protein folding [68-72], we conjecture that early ligands might have facilitated protein formation as catalysts (to assemble amino acids or peptide segments), as molecular chaperons (to help protein folding) and/or as selectors (because of the important functions of the early ligands), which naturally resulted in the accordance between both events. This conjecture implicates that the origin of primitive proteins benefited from ligand binding, which is reasonable in terms of the thermodynamics of ligand binding and protein folding.
It has been found that some early ligands, such as ADP and GDP, can bind proteins related to the very old P-loop hydrolase fold (for example, preprotein translocase SecA (1M74), ADP-ribosylation factor-like protein 3 (1FZQ) and GTP-binding protein (1A4R)) with an affinity (free energy) of 10-15 kcal/mol [73], which is just in the range of the free energy loss (10-20 kcal/mol) during protein folding [74,75]. Thus, the free energy release during ligand binding may meet the free energy demand during protein folding. It is tempting to examine the conjecture of ligand-induced formation and/or folding of primordial proteins through experimentation. To do that, in vitro selection may be an appropriate methodology [76]. It is interesting to note that in vitro selection of proteins (consisting of 80 residues) targeted to bind ATP has been performed [77]. The randomly generated proteins indeed belong to the α/β class, but are not related to P-loop hydrolases fold [78]. However, considering the fact that the shortest protein sequence for the P-loop hydrolase fold contains 94 residues (according to the Protein Databank), we suggest that to explore whether the formation of the most ancient proteins was induced by ATP, one should adopt longer protein sequences in the in vitro selection experiments and use small amino acids as building blocks, because in the primordial world only these amino acids were available [51,55].
Implications for understanding the new paradigms in drug discovery
Nowadays, the pharmaceutical industry is facing an unprecedented challenge. Global research funding has doubled since 1991, whereas the number of approved new drugs has fallen by 50% [79,80]. To meet the more-investment-less-outcome challenge, some novel drug discovery strategies have appeared in recent years, which include finding new functions from old drugs, developing promiscuous drugs rather than selective agents and depending more on natural products than on combinatorial libraries of synthetic compounds to derive drug leads. Since the essence of drug action is the binding between drugs and target biomolecules (most of which are proteins), the ligand-protein binding features revealed in the present study have important implications for understanding these new drug discovery strategies.
As indicated above, approximately 30% of ligands are bound by two or more domains (this number gets ~15%, if counted on fold level), which suggests that if a ligand can bind to a protein, it has great potential to bind to others. Considering the fact that the US Food and Drug Administration (FDA) has approved approximately 2,000 drugs (chemical entities) and there exist only 2,000-3,000 druggable genes and 600-1,500 drug targets [81,82], it is truly possible to find new functions from these old 'safe' drugs, which supports an increasingly shared notion in drug development that the most fruitful basis for the discovery of a new drug is to start with an old drug [83-85].
Since most human diseases, such as cancer, diabetes, heart disease, arthritis and neurodegenerative diseases, involve multiple pathogenetic factors, the more-investment-less-outcome predicament is attributed in part to the limitations of the current one-drug-one-target paradigm in drug discovery [79,86]. Therefore, more and more efforts are devoted to finding new therapeutics aimed at multiple targets [86], which is becoming a new paradigm in drug discovery. To hit the multiple targets implicated in complex diseases, two strategies are conceivable. One is called the multicomponent therapeutic strategy, which incorporates two or more active ingredients in one drug [86-89], as was applied in some traditional medicines (in China and many other countries) and in recently developed drug cocktails. The other is to hit the multiple targets with a single component, which is termed the one-ligand-multiple-targets strategy or promiscuous drug strategy [89-99]. Compared with the former strategy, the latter might take advantage of lower risks of drug-drug interactions and more predictable pharmacokinetic behaviors [91,92] and thus has been paid more and more attention. The feasibility of the one-ligand-multiple-targets strategy is supported by the present findings, because a certain proportion of ligands do indeed bind to two or more domains (even folds). In addition, the presently revealed structural features of super ligands are of significance for selecting and/or designing multipotent agents. Of course, the new strategy should be treated with wariness, because of the potential side effects of the promiscuous ligands.
Another feature of the recent drug discovery paradigm shift is that more attention has been given to natural-product repositories rather than combinatorial libraries of synthetic compounds for finding novel drug leads [100,101]. Due to their biosynthetic origin, natural products are natively bound to proteins (synthases). In light of the present findings, one can conclude that natural products have more potential than synthetic compounds to bind proteins, including those of human, which helps to understand the natural product-based drug discovery strategy. In addition, it can be inferred that it is rather easy to build a protein-ligand network on the basis of naturally occurring small-molecule ligands, which definitely benefits the birth of networked life and facilitates the formation of links within different species.
Materials and methods
Data selection/collection
Until June 2006, 2,721 ligands had been recorded in sc-PDB. As our interest was focused on non-peptide ligands, 433 peptides were eliminated. After removing 102 repeated ligands (which have the same structures to others but were given different release names), 2,186 small-molecule ligands remained (Additional data file 1), which bind to 5,740 non-redundant domains (to remove the redundancy of domains, only one domain was chosen from each species). Domain is defined as an independently folded unit within a protein, often joined by a flexible segment of the polypeptide chain [102]. For a small proportion of ligands that are shared by two domains, both domains were counted. According to SCOP 1.69 [28,29], these domains were classified into 591 folds. As one fold may cover multiple domains and hold more than one ligand, the fold occurrences amounted to 3,224.
Since sc-PDB is a subset of the PDB, one may be concerned about the robustness of the conclusions derived when using it. However, considering the facts that the present inferences were made mainly on the level of protein fold and that folds are much more conserved than domains, and thus fold increase is much slower than that of domains in the PDB [103], it is believed that the present conclusions are solid. In fact, even if the latest data of the sc-PDB (containing 396 new ligands and 827 new domains, which were kindly provided by Dr Rognan and have not been uploaded on the website) are considered, all of the present conclusions still hold.
Descriptor calculation
Seventy descriptors characterizing the structural features of 2,186 ligands selected from the sc-PDB and 2,184 small molecules randomly selected from ACD-SC were calculated by Sybyl (13 descriptors) [46], Cerius 2 (49 descriptors) [47] and an in-house program written in Perl (8 descriptors). Then, the calculated data were linked together with Perl for further analysis. Because of the lack of atomic parameters for ten ligands (that is, 2,3,4,5,6-pentafluorobenzyl alcohol, 2-amino-4-oxo-4,7-dihydro-3h-pyrrolo [2,3-d] pyrimidine-5-carbonitrile, 3,5,3',5'-tetraiodo-l-thyronine, 6,7-dinitroquinoxaline-2,3-dione, 9-hydroxy aristolochic acid, 3,5,7-trihydroxy-2-(4-hydroxyphenyl)-4h-chromen-4-one, 5-hydroxy-2-(4-hydroxyphenyl)-1-benzofuran-7-carbonitrile, 3,3',5,5'-tetraiodothyroacetic acid, 3,5,7,3',4'-pentahydroxyflavone and radicicol), some descriptors could not be calculated for these molecules. Hence, only 2,176 ligands went through the calculation. However, as each of the ten ligands covers only one fold, their absence has no impact on the conclusion of the present study.
Factor analysis
SPSS 13.0 (SPSS Inc., Chicago, IL, USA) was employed to do the factor analysis. The factors were extracted by means of principal component analysis [48,49] and the parameter settings were as follows: a correlation matrix was used; and two factors were extracted to visualize the two-dimensional chemical space of ligands and ordinary molecules. In order to simplify the interpretation of the extracted factors, factor rotation was performed, during which the most popular orthogonal rotation method, Varimax, developed by Kaiser [50], was employed. For other variables, default parameters were adopted.
Age assignment for bio-ligands
An early bio-ligand is defined as that owned by both prokaryotic (E. coli) and eukaryotic (yeast or higher) species, while a late bio-ligand is defined as that owned only by eukaryotic (yeast or higher) species. As there is no direct information on ligand ownership, we used the information of their host proteins to deduce their ages. That is, a ligand is early, provided that at least one of its host proteins is owned by both E. coli and yeast (or higher species); and a ligand is late if none of its host proteins is owned by E. coli but at least one is owned by yeast or higher species. During the age-assigning process, not only the host proteins recorded in sc-PDB were checked, but also the corresponding homologous proteins retrieved from Swiss-Prot [104] were considered.
Abbreviations
ACD-SC, Available Chemicals Directory-Screening Compounds; RotBond, rotatable bond; sc-PDB, Annotated Database of Druggable Binding Sites from the PDB.
Authors' contributions
H.-Y.Z. designed the study. H.-F.J., D.-X.K. and L.S. collected the data and performed the calculation. All authors analyzed the data. H.-Y.Z., H.-F.J. and L.-L.C. wrote the paper.
Additional data files
The following additional data are available with the online version of this paper. Additional data file 1 lists ligands and the numbers of domains and folds that bind them. Additional data file 2 illustrates the power-law behaviors of metabolism-relevant ligands. Additional data file 3 provides building blocks and ownerships of metabolism-relevant ligands. Additional data file 4 illustrates the power-law behaviors of folds for proteins binding ATP, ADP and NAD. Additional data file 5 illustrates the building block usage of bio-ligands.
Supplementary Material
Ligands and the numbers of domains and folds that bind them.
Power-law behaviors of metabolism-relevant ligands.
Building blocks and ownerships of metabolism-relevant ligands.
Power-law behaviors of folds for proteins binding ATP, ADP and NAD.
Building block usage of bio-ligands.
Acknowledgments
Acknowledgements
We thank Prof. Antonio Lazcano and Dr Xin-Min Li for fruitful discussions and Dr Didier Rognan for generously providing the sdf file of sc-PDB and the latest data. This work was partially supported by National Basic Research Program of China (2003CB114400) and National Natural Science Foundation of China (30570383 and 30600119).
Contributor Information
Hong-Fang Ji, Email: jhf@sdut.edu.cn.
De-Xin Kong, Email: dxkong@163.com.
Liang Shen, Email: shen@sdut.edu.cn.
Ling-Ling Chen, Email: llchen@sdut.edu.cn.
Bin-Guang Ma, Email: mbgmbg@163.com.
Hong-Yu Zhang, Email: zhanghy@sdut.edu.cn.
References
- Barabási AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Fewell JH. Social insect networks. Science. 2003;301:1867–1870. doi: 10.1126/science.1088945. [DOI] [PubMed] [Google Scholar]
- Whittaker RH, Feeny PP. Allelochemics: chemical interactions between species. Science. 1971;171:757–770. doi: 10.1126/science.171.3973.757. [DOI] [PubMed] [Google Scholar]
- Dixon RA. Natural products and plant disease resistance. Nature. 2001;411:843–847. doi: 10.1038/35081178. [DOI] [PubMed] [Google Scholar]
- Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin IT, Halitschke R, Paschold A, von Dahl CC, Preston CA. Volatile signaling in plant-plant interactions: 'talking trees' in the genomics era. Science. 2006;311:812–815. doi: 10.1126/science.1118446. [DOI] [PubMed] [Google Scholar]
- Keller L, Surette MG. Communication in bacteria: an ecological and evolutionary perspective. Nat Rev Microbiol. 2006;4:249–258. doi: 10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Ladurner AG. Rheostat control of gene expression by metabolites. Mol Cell. 2006;24:1–11. doi: 10.1016/j.molcel.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Cappello V, Tramontano A, Koch U. Classification of proteins based on the properties of the ligand-binding site: The case of adenine-binding proteins. Proteins. 2002;47:106–115. doi: 10.1002/prot.10070. [DOI] [PubMed] [Google Scholar]
- Denessiouk KA, Johnson MS. When fold is not important: A common structural framework for adenine and AMP binding in 12 unrelated protein families. Proteins. 2000;38:310–326. doi: 10.1002/(SICI)1097-0134(20000215)38:3<310::AID-PROT7>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L, Koonin EV. Emergence of diverse biochemical activities in evolutionarily conserved structural scaffolds of proteins. Curr Opin Chem Biol. 2003;7:12–20. doi: 10.1016/S1367-5931(02)00018-2. [DOI] [PubMed] [Google Scholar]
- Russell RB, Sasieni PD, Sternberg MJE. Supersites within superfolds. Binding site similarity in the absence of homology. J Mol Biol. 1998;282:903–918. doi: 10.1006/jmbi.1998.2043. [DOI] [PubMed] [Google Scholar]
- Jeong H, Tombor B, Albert R, Oltvai ZN, Barabási AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- Wagner A, Fell DA. The small world inside large metabolic networks. Proc R Soc Lond Ser B. 2001;268:1803–1810. doi: 10.1098/rspb.2001.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma HW, Zeng AP. Reconstruction of metabolic networks from genome data and analysis of their global structure for various organisms. Bioinformatics. 2003;19:270–277. doi: 10.1093/bioinformatics/19.2.270. [DOI] [PubMed] [Google Scholar]
- Arita M. The metabolic world of Escherichia coli is not small. Proc Natl Acad Sci USA. 2004;101:1543–1547. doi: 10.1073/pnas.0306458101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalsky E, Dunkel M, Goede A, Preissner R. SuperLigands-a database of ligand structures derived from the Protein Data Bank. BMC Bioinformatics. 2005;6:122. doi: 10.1186/1471-2105-6-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendlich M. Databases for protein-ligand complexes. Acta Crystallogr. 1998;D54:1178–1182. doi: 10.1107/s0907444998007124. [DOI] [PubMed] [Google Scholar]
- Goto S, Nishioka T, Kanehisa M. LIGAND: chemical database for enzyme reactions. Bioinformatics. 1998;14:591–599. doi: 10.1093/bioinformatics/14.7.591. [DOI] [PubMed] [Google Scholar]
- Shin JM, Cho DH. PDB-Ligand: a ligand database based on PDB for the automated and customized classification of ligand-binding structures. Nucleic Acids Res. 2005;33:D238–D241. doi: 10.1093/nar/gki059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold ND, Jackson RM. A searchable database for comparing protein-ligand binding sites for the analysis of structure-function relationships. J Chem Inf Model. 2006;46:736–742. doi: 10.1021/ci050359c. [DOI] [PubMed] [Google Scholar]
- Feldmana HJ, Snydera KA, Ticolla A, Pintiliea G, Hogue CWV. A complete small molecule dataset from the protein data bank. FEBS Lett. 2006;580:1649–1653. doi: 10.1016/j.febslet.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Gold ND, Jackson RM. SitesBase: a database for structure-based protein-ligand binding site comparisons. Nucleic Acids Res. 2006;34:D231–D234. doi: 10.1093/nar/gkj062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block P, Sotriffer CA, Dramburg I, Klebe G. AffinDB: a freely accessible database of affinities for protein-ligand complexes from the PDB. Nucleic Acids Res. 2006;34:D522–D526. doi: 10.1093/nar/gkj039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovin A, Dimitropoulos D, Oldfeld T, Rachedi A, Henrick K. MSDsite: A database search and retrieval system for the analysis and viewing of bound ligands and active sites. Proteins. 2005;58:190–199. doi: 10.1002/prot.20288. [DOI] [PubMed] [Google Scholar]
- Kellenberger E, Muller P, Schalon C, Bret G, Foata N, Rognan D. sc-PDB: an annotated database of druggable binding sites from the Protein Data Bank. J Chem Inf Model. 2006;46:717–727. doi: 10.1021/ci050372x. [DOI] [PubMed] [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Andreeva A, Howorth D, Brenner SE, Hubbard TJP, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acid Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY. Preferential attachment in the protein network evolution. Phys Rev Lett. 2003;91:138701–138704. doi: 10.1103/PhysRevLett.91.138701. [DOI] [PubMed] [Google Scholar]
- Ekman D, Light S, Björklund ÅK, Elofsson A. What properties characterize the hub proteins of the protein-protein interaction network of Saccharomyces cerevisiae? Genome Biol. 2006;7:R45. doi: 10.1186/gb-2006-7-6-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prachumwat A, Li WH. Protein function, connectivity, and duplicability in yeast. Mol Biol Evol. 2006;23:30–39. doi: 10.1093/molbev/msi249. [DOI] [PubMed] [Google Scholar]
- Caspi R, Foerster H, Fulcher CA, Hopkinson R, Ingraham J, Kaipa P, Krummenacker M, Paley S, Pick J, Rhee SY, et al. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 2006;34:D511–D514. doi: 10.1093/nar/gkj128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Luscombe NM, Gerstein M. Protein family and fold occurrence in genomes: power-law behaviour and evolutionary model. J Mol Biol. 2001;313:673–681. doi: 10.1006/jmbi.2001.5079. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI, Karev GP. The structure of the protein universe and genome evolution. Nature. 2002;420:218–223. doi: 10.1038/nature01256. [DOI] [PubMed] [Google Scholar]
- Caetano-Anollés G, Caetano-Anollés D. An evolutionarily structured universe of protein architecture. Genome Res. 2003;13:1563–1571. doi: 10.1101/gr.1161903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArdle BM, Quinn RJ. Identification of protein fold topology shared between different folds inhibited by natural products. ChemBioChem. 2007;8:788–798. doi: 10.1002/cbic.200700035. [DOI] [PubMed] [Google Scholar]
- Todd AE, Orengo CA, Thornton JM. Plasticity of enzyme active sites. Trends Biochem Sci. 2002;27:419–426. doi: 10.1016/S0968-0004(02)02158-8. [DOI] [PubMed] [Google Scholar]
- Macchiarulo A, Nobeli I, Thornton JM. Ligand selectivity and competition between enzymes in silico. Nat Biotechnol. 2004;22:1039–1045. doi: 10.1038/nbt999. [DOI] [PubMed] [Google Scholar]
- Stockwell GR, Thornton JM. Conformational diversity of ligands bound to proteins. J Mol Biol. 2006;356:928–944. doi: 10.1016/j.jmb.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Van Regenmortel MHV. Molecular recognition in the post-reductionist era. J Mol Recognit. 1999;12:1–2. doi: 10.1002/(SICI)1099-1352(199901/02)12:1<1::AID-JMR449>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Ma B, Kumar S, Tsai CJ, Nussinov R. Folding funnels and binding mechanisms. Protein Eng. 1999;12:713–720. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]
- Ma B, Shatsky M, Wolfson HJ, Nussinov R. Multiple diverse ligands binding at a single protein site: a matter of pre-existing populations. Protein Sci. 2002;11:184–197. doi: 10.1110/ps.21302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Available Chemicals Directory-Screening Compounds http://www.akosgmbh.eu/acd-sc.htm
- SYBYL 7.0 http://www.tripos.com/index.php?family=modules,SimplePage,,,&page=comp_informatics
- Cerius2 http://www.accelrys.com/products/cerius2/
- Kim JO, Mueller CW. Factor Analysis: Statistical Methods and Practical Issues. Thousand Oaks, CA: Sage Publications; 1978. [Google Scholar]
- Reyment RA, Joreskog KG. Applied Factor Analysis in the Natural Sciences. Cambridge: Cambridge University Press; 1993. [Google Scholar]
- Kaiser HF. The varimax criterion for analytic rotation in factor analysis. Psychometrika. 1958;23:187–200. doi: 10.1007/BF02289233. [DOI] [Google Scholar]
- Trifonov EN, Gabdank I, Barash D, Sobolevsky Y. Primordia vita deconvolution from modern sequences. Orig Life Evol Biosph. 2006;36:559–565. doi: 10.1007/s11084-006-9042-5. [DOI] [PubMed] [Google Scholar]
- Wong JT-F. Coevolutionary theory of the genetic code at age thirty. BioEssays. 2005;27:416–425. doi: 10.1002/bies.20208. [DOI] [PubMed] [Google Scholar]
- Caetano-Anollés G, Caetano-Anollés D. Universal sharing patterns in proteomes and evolution of protein fold architecture and life. J Mol Evol. 2005;60:484–498. doi: 10.1007/s00239-004-0221-6. [DOI] [PubMed] [Google Scholar]
- Wang M, Boca SM, Kalelkar R, Mittenthal JE, Caetano-Anollés G. Phylogenomic reconstruction of the protein world based on a genomic census of protein fold architecture. Complexity. 2006;12:27–40. doi: 10.1002/cplx.20141. [DOI] [Google Scholar]
- Zhang H-Y. Exploring the evolution of standard amino-acid alphabet: when genomics meets thermodynamics. Biochem Biophys Res Commun. 2007;359:403–405. doi: 10.1016/j.bbrc.2007.05.115. [DOI] [PubMed] [Google Scholar]
- White HB. Coenzymes as fossils of an earlier metabolic state. J Mol Evol. 1976;7:101–104. doi: 10.1007/BF01732468. [DOI] [PubMed] [Google Scholar]
- Miller SL, Schlesinger G. Prebiotic syntheses of vitamin coenzymes: I. Cysteamine and 2-mercaptoethanesulfonic acid (coenzyme M). J Mol Evol. 1993;36:302–307. doi: 10.1007/BF00182177. [DOI] [PubMed] [Google Scholar]
- Miller SL, Schlesinger G. Prebiotic syntheses of vitamin coenzymes: II. Pantoic acid, pantothenic acid, and the composition of coenzyme A. J Mol Evol. 1993;36:308–314. doi: 10.1007/BF00182178. [DOI] [PubMed] [Google Scholar]
- Huang F, Bugg CW, Yarus M. RNA-catalyzed CoA, NAD, and FAD synthesis from phosphopantetheine, NMN, and FMN. Biochemistry. 2000;39:15548–15555. doi: 10.1021/bi002061f. [DOI] [PubMed] [Google Scholar]
- Winstanley HF, Abeln S, Deane CM. How old is your fold? Bioinformatics. 2005;21(Suppl 1):i449–i458. doi: 10.1093/bioinformatics/bti1008. [DOI] [PubMed] [Google Scholar]
- Abeln S, Deane CM. Fold usage on genomes and protein fold evolution. Proteins. 2005;60:690–700. doi: 10.1002/prot.20506. [DOI] [PubMed] [Google Scholar]
- Ji H-F, Zhang H-Y. Protein architecture chronology deduced from structures of amino acid synthases. J Biomol Struct Dyn. 2007;24:321–323. doi: 10.1080/07391102.2007.10507122. [DOI] [PubMed] [Google Scholar]
- Berezovsky IN, Kirzhner VM, Kirzhner A, Rosenfeld VR, Trifonov EN. Protein sequences yield a proteomic code. J Biomol Struct Dyn. 2003;21:317–325. doi: 10.1080/07391102.2003.10506928. [DOI] [PubMed] [Google Scholar]
- Berezovsky IN, Kirzhner A, Kirzhner VM, Trifonov EN. Spelling protein structure. J Biomol Struct Dyn. 2003;21:327–339. doi: 10.1080/07391102.2003.10506929. [DOI] [PubMed] [Google Scholar]
- Trifonov EN. Early molecular evolution. Israel J Ecol Evol. 2006. pp. 375–387.
- Caetano-Anollés G, Kim HS, Mittenthal JE. The origin of modern metabolic networks inferred from phylogenomic analysis of protein architecture. Proc Natl Acad Sci USA. 2007;104:9358–9363. doi: 10.1073/pnas.0701214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobeli I, Ponstingl H, Krissinel EB, Thornton JM. A structure-based anatomy of the E. coli metabolome. J Mol Biol. 2003;334:697–719. doi: 10.1016/j.jmb.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/S0959-440X(02)00289-0. [DOI] [PubMed] [Google Scholar]
- Fink AL. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Grandori R, Schwarzinger S, Müller N. Cloning, overexpression and characterization of micro-myoglobin, a minimal heme-binding fragment. Eur J Biochem. 2000;267:1168–1172. doi: 10.1046/j.1432-1327.2000.01114.x. [DOI] [PubMed] [Google Scholar]
- Wang RX, Fang XL, Lu YP, Yang CY, Wang SM. The PDBbind database: methodologies and updates. J Med Chem. 2005;48:4111–4119. doi: 10.1021/jm048957q. [DOI] [PubMed] [Google Scholar]
- Dobson CM, Šali A, Karplus M. Protein folding: a perspective from theory and experiment. Angew Chem Int Ed. 1998;37:868–893. doi: 10.1002/(SICI)1521-3773(19980420)37:7<868::AID-ANIE868>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. New York: Freeman; 1999. [Google Scholar]
- Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Ann Rev Biochem. 1999;68:611–648. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- Keefe AD, Szostak JW. Functional proteins from a random-sequence library. Nature. 2001;410:715–718. doi: 10.1038/35070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Surdo P, Walsh MA, Sollazzo M. A novel ADP- and zinc-binding fold from function-directed in vitro evolution. Nat Struct Mol Biol. 2004;11:382–383. doi: 10.1038/nsmb745. [DOI] [PubMed] [Google Scholar]
- Buehler LK. Advancing drug discovery-beyond design. PharmaGenomics. 2004;4:24–26. [Google Scholar]
- Ruffolo RR. Why has R&D productivity declined in the pharmaceutical industry? Expert Opin Drug Discov. 2006;1:99–102. doi: 10.1517/17460441.1.2.99. [DOI] [PubMed] [Google Scholar]
- Russ AP, Lampel S. The druggable genome: an update. Drug Discov Today. 2005;10:1607–1610. doi: 10.1016/S1359-6446(05)03666-4. [DOI] [PubMed] [Google Scholar]
- Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- Wermuth CG. Selective optimization of side activities: another way for drug discovery. J Med Chem. 2004;47:1303–1314. doi: 10.1021/jm030480f. [DOI] [PubMed] [Google Scholar]
- Lipinski C, Hopkins A. Navigating chemical space for biology and medicine. Nature. 2004;432:855–861. doi: 10.1038/nature03193. [DOI] [PubMed] [Google Scholar]
- O'Connor KA, Roth BL. Finding new tricks for old drugs: an efficient route for public-sector drug discovery. Nat Rev Drug Discov. 2005;4:1005–1014. doi: 10.1038/nrd1900. [DOI] [PubMed] [Google Scholar]
- Fitzgerald JB, Schoeberl B, Nielsen UB, Sorger PK. Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol. 2006;2:458–466. doi: 10.1038/nchembio817. [DOI] [PubMed] [Google Scholar]
- Zimmermann GR, Lehár J, Keith CT. Multi-target therapeutics: when the whole is greater than the sum of the parts. Drug Discov Today. 2007;12:34–42. doi: 10.1016/j.drudis.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Kitano H. A robustness-based approach to systems-oriented drug design. Nat Rev Drug Discov. 2007;6:202–210. doi: 10.1038/nrd2195. [DOI] [PubMed] [Google Scholar]
- Keith CT, Borisy AA, Stockwell BR. Multicomponent therapeutics for networked systems. Nat Rev Drug Discov. 2005;4:71–78. doi: 10.1038/nrd1609. [DOI] [PubMed] [Google Scholar]
- Mencher SK, Wang LG. Promiscuous drugs compared to selective drugs (promiscuity can be a virtue). BMC Clin Pharmacol. 2005;5:3. doi: 10.1186/1472-6904-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morphy R, Kay C, Rankovic Z. From magic bullets to designed multiple ligands. Drug Discov Today. 2004;9:641–651. doi: 10.1016/S1359-6446(04)03163-0. [DOI] [PubMed] [Google Scholar]
- Morphy R, Rankovic Z. Designed multiple ligands. An emerging drug discovery paradigm. J Med Chem. 2005;48:6523–6543. doi: 10.1021/jm058225d. [DOI] [PubMed] [Google Scholar]
- Zhang H-Y. One-compound-multiple-targets strategy to combat Alzheimer's disease. FEBS Lett. 2005;579:5260–5264. doi: 10.1016/j.febslet.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Frantz S. Drug discovery:Playing dirty. Nature. 2005;437:942–943. doi: 10.1038/437942a. [DOI] [PubMed] [Google Scholar]
- Hopkins AL, Mason JS, Overington JP. Can we rationally design promiscuous drugs? Curr Opin Struct Biol. 2006;16:127–136. doi: 10.1016/j.sbi.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Roth BL, Sheffler DJ, Kroeze WK. Magic shotguns versus magic bullets: selectively non-selective drugs for mood disorders and schizophrenia. Nat Rev Drug Discov. 2004;3:353–359. doi: 10.1038/nrd1346. [DOI] [PubMed] [Google Scholar]
- Hampton T. 'Promiscuous' anticancer drugs that hit multiple targets may thwart resistance. J Am Med Assoc. 2004;292:419–422. doi: 10.1001/jama.292.4.419. [DOI] [PubMed] [Google Scholar]
- Zhang H-Y, Yang D-P, Tang G-Y. Multifunctional antioxidants: from screening to design. Drug Discov Today. 2006;11:749–754. doi: 10.1016/j.drudis.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Morphy R, Rankovic Z. Fragments, network biology and designing multiple ligands. Drug Discov Today. 2007;12:156–160. doi: 10.1016/j.drudis.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Breinbauer R, Vetter IR, Waldmann H. From protein domains to drug candidates-natural products as guiding principles in the design and synthesis of compound libraries. Angew Chem Int Ed. 2002;41:2878–2890. doi: 10.1002/1521-3773(20020816)41:16<2878::AID-ANIE2878>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Paterson I, Anderson EA. The renaissance of natural products as drug candidates. Science. 2005;310:451–453. doi: 10.1126/science.1116364. [DOI] [PubMed] [Google Scholar]
- Rose GD. Hierarchic organization of domains in globular proteins. J Mol Biol. 1979;134:447–470. doi: 10.1016/0022-2836(79)90363-2. [DOI] [PubMed] [Google Scholar]
- Levitt M. Growth of novel protein structural data. Proc Natl Acad Sci USA. 2007;104:3183–3188. doi: 10.1073/pnas.0611678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence data bank and its supplement TrEMBL in 1999. Nucleic Acids Res. 1999;27:49–54. doi: 10.1093/nar/27.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ligands and the numbers of domains and folds that bind them.
Power-law behaviors of metabolism-relevant ligands.
Building blocks and ownerships of metabolism-relevant ligands.
Power-law behaviors of folds for proteins binding ATP, ADP and NAD.
Building block usage of bio-ligands.