Abstract
Mutant presenilins have been found to cause Alzheimer disease. Here, we describe the identification and characterization of HOP-1, a Caenorhabditis elegans presenilin that displays much more lower sequence identity with human presenilins than does the other C. elegans presenilin, SEL-12. Despite considerable divergence, HOP-1 appears to be a bona fide presenilin, because HOP-1 can rescue the egg-laying defect caused by mutations in sel-12 when hop-1 is expressed under the control of sel-12 regulatory sequences. HOP-1 also has the essential topological characteristics of the other presenilins. Reducing hop-1 activity in a sel-12 mutant background causes synthetic lethality and terminal phenotypes associated with reducing the function of the C. elegans lin-12 and glp-1 genes. These observations suggest that hop-1 is functionally redundant with sel-12 and underscore the intimate connection between presenilin activity and LIN-12/Notch activity inferred from genetic studies in C. elegans and mammals.
Genetic linkage studies have identified a number of loci associated with familial Alzheimer disease (1). Two of these loci encode related multipass transmembrane proteins, presenilins 1 and 2 (PS1 and PS2). Mutations in the genes encoding PS1 and PS2 loci are dominant and fully penetrant for early onset Alzheimer disease (2–4). The presenilins are ubiquitously expressed (3, 4) and found in conjunction with intracellular membranes (5). However, the normal role of presenilins, and the mechanism by which mutant presenilins cause Alzheimer disease, are not known.
Genetic studies in simple organisms offer a powerful approach to understanding the normal role of presenilins. The Caenorhabditis elegans sel-12 gene encodes a protein that displays about 50% amino acid sequence identity to PS1 and PS2 (6). Genetic analysis established that reducing or eliminating sel-12 activity causes an egg-laying defective (Egl) phenotype, and that sel-12 activity facilitates the activity of LIN-12 and GLP-1, two receptors of the LIN-12/Notch family (6). SEL-12 appears to be a bona fide presenilin, because human PS1 and PS2 can rescue the Egl phenotype of a sel-12 mutant (7). Furthermore, the membrane topology of SEL-12 and PS1 appears to be similar (8–10). In addition to the functional and structural similarities, expression studies indicate that SEL-12 and human presenilins are expressed throughout development in many different cell types (3, 4, 7).
We have identified another candidate C. elegans presenilin based on predicted amino acid sequence by searching the genomic sequence database (11, 12). Here, we show that this gene, which we have named hop-1 (hop = homolog of presenilin), encodes a functional presenilin, by demonstrating that HOP-1 can rescue the egg-laying defect of a sel-12 mutant. We also show that HOP-1 has characteristic features of presenilin membrane topology. Finally, we show that reducing hop-1 activity in a sel-12 mutant background results in novel phenotypes, suggesting that hop-1 and sel-12 are functionally redundant.
MATERIALS AND METHODS
Genetic Methods.
Methods for handling and culturing C. elegans have been described by Brenner (13). The wild-type parent for all strains used was C. elegans var. Bristol strain N2 (13). Rescue and RNA-mediated interference experiments were performed at 20°C. Strains for topological analysis were grown at 25°C, a temperature that maximizes β-galactosidase activity.
hop-1 cDNA.
hop-1 cDNA was isolated by PCRs using a cDNA library provided by R. Barstead (14) as the template. A major portion of hop-1 cDNA was amplified with primers from within the exons of Genefinder (12) predicted gene now called C18E3.8. The forward primer, C18LXJ1-5′-CGGGATCCTTTGCATGTTGTTCGTCGCG, harbors amino acids of Cys-Met-Leu-Phe-Val-Ala in the amino terminal region of C18E3.8. The reverse primer, C18LXJ2-5′-CGGGATCCAAATTAGCTGTGAGGTGC, is complementary to amino acids of Thr-Ser-Gln-Leu-Ile in the carboxyl-terminal region of C18E3.8. The amplified product was subcloned and sequenced to verify the exon/intron boundary predicted by genefinder. The sequence of cosmid C18E3 has been deposited by the genome sequencing consortium (Genbank accession no. AF000265).
The 5′ and 3′ ends of the transcript were determined using rapid amplification of cDNA ends (RACE). For 5′ RACE, primers with the sequence of either spliced leader SL1 (15) or SL2 (16) were used for the amplification. A specific product was only obtained with SL1 as one of the primers after a second round of amplification, using a nested primer inside the first amplification product. For 3′ RACE, dT-adaptor, 5′-GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT, was used for the amplification. After a second round of amplification using a nested primer inside the first amplification product, a specific product was obtained and subcloned into pBluescript.
Full-length hop-1 cDNA was obtained by joining these three cDNA fragments with appropriate restriction endonucleases.
Plasmids.
pLSX (9) is an expression vector containing 2.8 kb of sel-12 5′ flanking region, a polylinker, and 755 bp of 3′ noncoding region including a polyadenylation sequence from the unc-54 gene (17). cDNAs containing initiation codons are inserted into the polylinker site and expression is driven by sel-12 5′ flanking region. This vector leads to good expression in a subset of cells that express sel-12 (X.L., D. Levitan, and I.G., unpublished observations) and served as the parent plasmid for the rescue and topology experiments. The full-length hop-1 cDNA or portions of hop-1 cDNA fused in-frame to the cDNAs encoding LacZ or TM::LacZ proteins (17) were placed into pLSX.
Transgenic Lines.
Transgenic lines were established by microinjection of plasmid mixtures into the hermaphrodite germ line to create extrachromosomal arrays (18).
For the rescue experiments, pLSX::hop-1 or pLSX::sel-12 was injected at a concentration of 20 μg/ml into recipient sel-12(ar171) unc-1(e538) hermaphrodites along with 100 μg/ml of pRF4, a plasmid containing the cloned dominant rol-6(su1006) gene (18), as a cotransformation marker. F1 Roller progeny were picked, and F2 Roller progeny used to establish lines. To assess rescue of sel-12(ar171), approximately 30 L4 Rol progeny from 5–7 independent lines were picked individually and scored daily for the ability to lay eggs. sel-12(ar171) never lays eggs (6), and transgenic hermaphrodites were scored as “Egl+” if they displayed robust egg-laying, characteristic of wild-type hermaphrodites, for 2 days as adults.
For the topology experiments, plasmids were injected at a concentration of 20 μg/ml into recipient N2 hermaphrodites along with 100 μg/ml of pRF4, a plasmid containing the cloned dominant rol-6(su1006) gene (18) as a cotransformation marker. F1 Roller progeny were picked and F2 Roller progeny were used to establish lines. Larvae were fixed using an acetone fixation protocol described by Fire (19) and stained for β-galactosidase activity overnight at room temperature.
RNA-Mediated Interference Experiments.
Plasmids harboring hop-1 and sel-12 cDNAs were linearized with appropriate restriction endonucleases and served as templates for production of antisense RNAs in vitro (Stratagene). Transcripts were treated with DNase, followed by phenol–chloroform extractions and isopropanol precipitation. After washing once with 70% ethanol, pellets were resuspended in diethylpyrocarbonate (DEPC)-treated distilled water. Wild-type (N2), sel-12(ar131), or sel-12(ar171) unc-1(e538) young adult hermaphrodites were injected with RNAs in both distal gonad arms at an estimated concentration of 50–250 μg/ml. Injected worms were plated individually at 20°C and were transferred to a new plate every day. Their progeny were counted and examined under the dissecting microscope. Embryonic arrest (dead eggs) was assessed 1 day after the eggs were laid.
At least 12, and as many as 30 hermaphrodites were injected with antisense RNAs for each set of injections into recipient hermaphrodites of a given genotype. The injection protocol was repeated two independent times for hop-1 or sel-12 RNA, or DEPC-dH20 into wild-type N2 hermaphrodites, and two independent times for sel-12 RNA or DEPC-dH20 into sel-12 mutant hermaphrodites. We did not observe novel mutant phenotypes or reduced viability of progeny in any of these experiments. Injection of hop-1 RNA into sel-12(ar131) or sel-12(ar171) unc-1 hermaphrodites was repeated four independent times for each mutant genotype. Offspring displaying Glp-1 and Lag phenotypes were seen in all four sets of injections into each mutant. We also often observed many inviable embryos (Emb phenotype) among the progeny of injected hermaphrodites; we do not know if this reflects the combination of reduced maternal and zygotic glp-1 and lin-12 activity or the involvement of hop-1 in other processes. sel-12 mutant hermaphrodites injected with DEPC-dH20 did not produce offspring displaying Glp-1 or Lag phenotypes or a significant increase in the number of inviable offspring.
It is difficult to quantify precisely the different phenotypic classes obtained among the progeny of injected sel-12 Egl hermaphrodites. However, for injections into sel-12(ar131), in some experiments we quantified the proportion of mutant offspring among the progeny of injected Egl+ hermaphrodites. For example, in one set of injections in which hop-1 RNA (at approximately 200 μg/ml) was injected into sel-12(ar131) hermaphrodites, we observed 56 viable adult hermaphrodites and 103 inviable offspring (65% inviable); 40/56 (71%) viable adult hermaphrodites were sterile, and all sterile hermaphrodites examined had defects characteristic of reducing glp-1 activity in the germ line. A high proportion of sterile and inviable offspring was reproducibly seen in all injections involving hop-1 RNA (data not shown).
RESULTS
Identification and Sequence of a Candidate Presenilin.
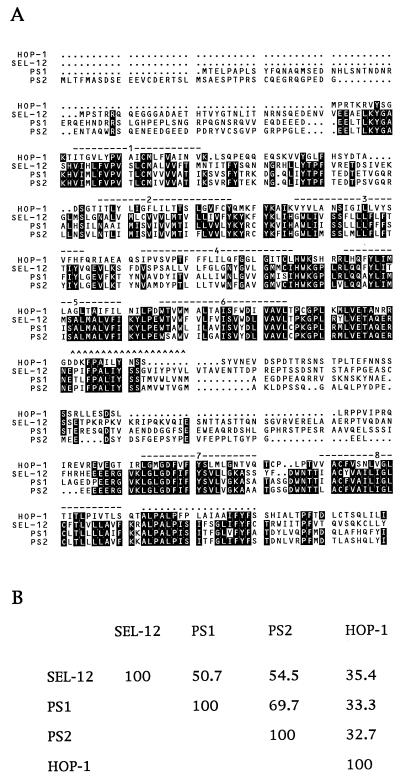
We searched the C. elegans genome sequence database (11, 12) with the SEL-12 amino acid sequence and identified a second potential presenilin gene, C18E3.8. We obtained and sequenced cDNAs for C18E3.8 (GenBank accession number AF021905). The predicted protein encoded by this cDNA is also shown aligned with SEL-12, PS1, and PS2 in Fig. 1. Because the functional and structural studies described below suggest that C18E3.8 is a bona fide presenilin, we have named C18E3.8 hop-1, for homolog of presenilin.
Figure 1.
(A) Predicted protein sequence of HOP-1 and its alignment with the predicted protein sequences of C. elegans SEL-12, human PS1, and human PS2. The pileup program of the GCG–Wisconsin package was used to create this alignment (30). Amino acids that are identical between at least three of the four proteins are highlighted in black. The predicted transmembrane domains based on topological studies of SEL-12 (9) are overlined. Two additional features referred to in the Discussion are marked: a hydrophobic region found in SEL-12 (“the seventh hydrophobic region”), PS1, and PS2 that does not span the membrane is overlined with carets (^), and a hydrophobic region found in all four presenilins (the “tenth hydrophobic region” of SEL-12) that is not membrane spanning is overlined with dots. The sequence of SEL-12 is from ref. 6, with a minor correction as described in ref. 7. The PS1 sequence is from ref. 4, and the PS2 sequence is from ref. 3. (B) Amino acid identity among the C. elegans and human presenilins. The gap program of the GCG–Wisconsin package was used to calculate the percentage of amino acid identity (30).
hop-1 Can Rescue the Egg-Laying Defect of a sel-12 Presenilin Mutant.
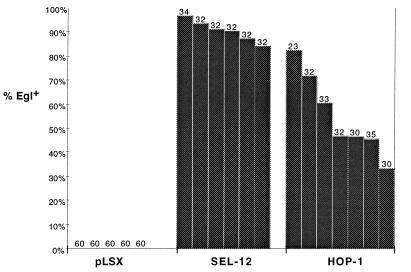
Human PS1 and PS2 can efficiently rescue the distinctive Egl phenotype caused by a mutation that reduces sel-12 activity (7). We used the same basic approach to assess the presenilin activity of hop-1. The sel-12(ar171) mutation strongly reduces sel-12 activity, so that all sel-12(ar171) hermaphrodites are Egl. We created transgenes expressing hop-1 by microinjecting a plasmid containing the hop-1 cDNA under the control of 2.8 kb of sel-12 5′ flanking region into sel-12(ar171) hermaphrodites (see Materials and Methods). We found that hop-1 can efficiently rescue the Egl defect of sel-12(ar171) hermaphrodites (Fig. 2), indicating that HOP-1 is a functional homolog of SEL-12, PS1 and PS2.
Figure 2.
Rescue of the sel-12 Egl and abnormal vulva phenotypes by HOP-1. The data are shown for transgenic lines generated by injecting a construct that places a sel-12(+) cDNA or a hop-1(+) cDNA under the control of sel-12 5′ flanking sequence at a concentration of 20 μg/ml. Each line in the histogram represents data for an independent transgenic line; the number of hermaphrodites scored is shown above each line. The transgene is indicated on the horizontal axis. The percentage of Egl+ hermaphrodites is indicated on the vertical axis. For pLSX control lines, Egl+ hermaphrodites were never seen. See Materials and Methods for details about generating and scoring transgenic lines.
HOP-1 Has Similar Topology to SEL-12 Presenilin.
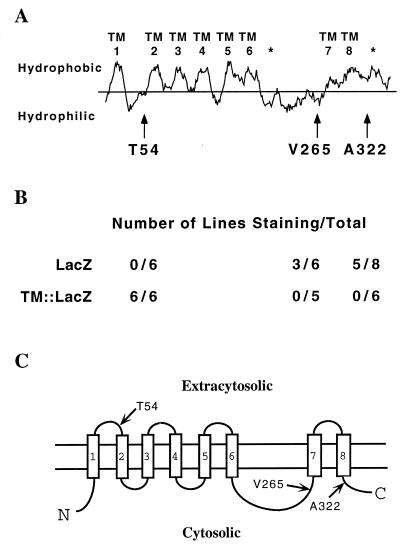
The amino and carboxyl termini of SEL-12 and PS1 are cytosolic, as is a large intracellular loop located between the sixth and seventh transmembrane domains (8, 9). We have determined that HOP-1 has these essential features of presenilin topology by using the LacZ hybrid protein approach that we used to deduce the topology of SEL-12 presenilin (9). If LacZ is fused at different points in a membrane protein, its location in the cytosol or in an extracytosolic compartment, and hence the topology of the membrane protein, can be deduced by its activity, because β-galactosidase is active within the cytosol but not in the extracytosolic compartment (20, 21).
We constructed transgenes encoding hybrid HOP-1::LacZ proteins in which LacZ or TM::LacZ [TM denotes a short hydrophilic spacer followed by a synthetic transmembrane domain (17)] was placed after the first, sixth, or eighth putative transmembrane domains (see Fig. 3). Several independent transgenic C. elegans lines expressing individual LacZ hybrid proteins were established (18) and assayed for β-galactosidase activity (19). Transgenic lines expressing individual hybrid proteins should consistently stain positively for β-galactosidase activity if the LacZ moiety is located in the cytosol, but should not stain if LacZ is located extracytosolically; for each pair of hybrid proteins one hybrid should be active, whereas the other should not be active. Our results suggest that the amino terminus, loop, and carboxyl terminus of HOP-1 are all cytosolic (Fig. 3), as they are in other presenilins (8, 9).
Figure 3.
HOP-1 topology. (A) Hydrophobicity plot of HOP-1, generated using the Kyte–Doolittle algorithm (31) (window size = 15). Transmembrane domains based on topological studies of SEL-12 (9) are numbered. The location of hydrophobic regions of SEL-12 that do not span the membrane are indicated by asterisks. Arrows indicate the position of LacZ and TM::LacZ fusions. (B) β-Galactosidase activity of HOP-1::LacZ and HOP-1::TM::LacZ fusion proteins. (C) Inferred topology of HOP-1, based on the data in B and sequence similarities with other presenilins.
Reducing hop-1 Activity Causes Novel Phenotypes in a sel-12 Mutant Background.
RNA-mediated interference is a widely used method for reducing gene activity in C. elegans and has been shown in many cases to give a phenotype similar to null alleles (22, 23). Both antisense and sense RNAs appear to have the same effect (22). RNA-mediated interference appears to cause targeted suppression of the corresponding gene in the germ line of injected hermaphrodites, so that progeny display phenotypes associated with loss of gene function (22, 23). Although the penetrance of these phenotypes is incomplete, the expressivity can be high. Maternal gene activities seem to be more sensitive to RNA-mediated interference than are zygotic gene activities.
We injected the germ lines of wild-type and sel-12 mutant hermaphrodites with antisense RNA from sel-12 or hop-1 (Table 1). When wild-type hermaphrodites were injected with sel-12 or hop-1 RNA, no mutant offspring were seen. However, when sel-12 mutant hermaphrodites were injected with hop-1 RNA, there were many dead embryos, arrested larvae, and sterile hermaphrodites among the progeny. These phenotypes are not seen in sel-12 mutants (6) or in sel-12 mutant hermaphrodites injected with sel-12 RNA or water (Table 1).
Table 1.
Summary of novel phenotypes obtained by RNA interference
| RNA injected | Recipient genotype
|
||
|---|---|---|---|
| Wild type | sel-12(ar131) | sel-12(ar171)* | |
| DEPC-dH2O | None | None | None |
| sel-12 | None | None | None |
| hop-1 | None | Glp-1, Lag, Emb | Glp-1, Lag, Emb |
See Materials and Methods for a detailed description of the RNA interference experiments and Results for more information about mutant phenotypes. Glp-1, phenotypes associated with the reduction of glp-1 activity: defective germ line proliferation and premature entry of germ cells into meiosis, or missing anterior pharynx (25, 26). Lag, phenotypes associated with the concomitant reduction of lin-12 and glp-1 activity: missing rectum and/or excretory cell (27). Emb, embryonic lethal (cellular anatomy not analyzed). DEPC-dH2O, diethyl pyrocarbonate-treated distilled water.
*Complete genotype: sel-12(ar171) unc-1(e538).
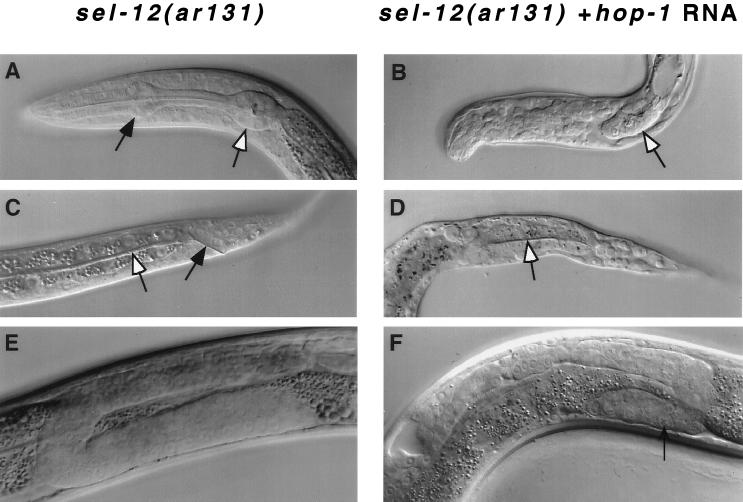
Previous work established that sel-12 facilitates lin-12 and glp-1 activity (6). Characteristic anatomical defects due to abnormal cell fate specification are observed when lin-12 or glp-1 activity is reduced; additional defects are observed when the activities of both genes are reduced concomitantly (24–27). Many of these various defects were frequently observed among the progeny of sel-12 mothers that had been injected with hop-1 RNA (Table 1, Fig. 4). (i) All of the sterile hermaphrodites examined had a severely reduced number of germ cells and premature differentiation of germ cells as sperm, the hallmarks of reduced glp-1 activity in the germ line (25, 26). (ii) Some arrested larvae lacked an anterior pharynx, a defect associated with reduced maternal glp-1 activity (25, 26). (iii) Some arrested larvae lacked a rectum and an excretory cell, the hallmarks of the “Lag” phenotype associated with concomitantly reduced lin-12 and glp-1 activity and, to a lesser extent, with reduced lin-12 activity (27) .
Figure 4.
Nomarski photomicrographs of sel-12(ar131) hermaphrodites. (A, C, and E) sel-12(ar131) hermaphrodites. (B, D, and F) Progeny of sel-12(ar131) hermaphrodites that were injected with hop-1 antisense RNA (see Materials and Methods). (A and B) The head region. The white arrow indicates the position of the posterior bulb; the black arrow indicates the position of the anterior bulb of the pharynx, which is often missing in progeny of hop-1 RNA-injected parents, as in glp-1 mutants (25, 26). (C and D) The tail region. The white arrow indicates the intestine; the black arrow indicates the rectum, which is often missing in progeny of hop-1 RNA-injected parents, as in lin-12 glp-1 double mutants (27). (E and F) The gonad of L4 hermaphrodites. In F , germ line proliferation is reduced, and the arrow indicates a region of the germ line undergoing premature spermatogenesis; these phenotypes are characteristic of glp-1 mutants (25, 26).
Although many dead embryos and arrested larvae display defects known to result from reduced lin-12 and/or glp-1 activity, they may also have additional anatomical defects (data not shown). Further analysis of embryos depleted in the activities of both hop-1 and sel-12 will be facilitated by the isolation of conventional hop-1 mutations and the construction of hop-1(−);sel-12(−) double mutants.
DISCUSSION
Despite considerable sequence differences from other presenilins, HOP-1 appears to be a bona fide presenilin. HOP-1 can rescue the egg-laying defect caused by mutations in sel-12 when hop-1 is expressed under the control of sel-12 regulatory sequences. Furthermore, HOP-1 has the essential topological characteristics of the other presenilins. Finally, hop-1 and sel-12 appear to be functionally redundant. It is curious that both C. elegans and humans have at least two functional presenilins, but the meaning of this observation is unclear.
Sequence Comparison Between HOP-1 and Other Presenilins.
SEL-12, PS1, and PS2 are highly similar in all eight transmembrane domains as well as two other hydrophobic regions that do not appear to span the membrane (9). Most of the sequence divergence is found in two cytosolic regions, one at the amino terminus and the other in the loop between the sixth and seventh transmembrane domains. This region of the loop has features reminiscent of a PEST [region rich in proline (P), glutamate (E), serine (S), and threonine (T)] protein destabilization sequence (9). HOP-1 is similar to the other presenilins in terms of the overall length and spacing of the transmembrane domains, the presence of a PEST sequence between the sixth and seventh hydrophobic domains, and the presence of a hydrophobic region following the eighth transmembrane domain. However, HOP-1 is considerably more diverged from SEL-12, PS1, and PS2 at the primary amino acid sequence level (Fig. 1).
The divergence of HOP-1 from the other presenilins is instructive in view of the efficient rescue of sel-12 mutants by expression of HOP-1. There is a surprising amount of divergence between HOP-1 and the other presenilins in the eight transmembrane domains. HOP-1 is different from the human presenilins even in transmembrane domains that are highly conserved between SEL-12 and the human presenilins (compare, for example, TM1 or TM5 in the four proteins; Fig. 1). In general, however, the substitutions appear to be fairly conservative, preserving the hydrophobic character of the transmembrane domains.
SEL-12, PS1, and PS2 have two additional hydrophobic regions that do not span the membrane (8, 9) (see Fig. 3). The carboxyl-terminal hydrophobic region is strikingly conserved in sequence and hydrophobicity in HOP-1 as well. However, the internal hydrophobic region of the cytosolic loop of SEL-12 is not appreciably hydrophobic in HOP-1 (see Fig. 3). Nevertheless, there is a notable region of conservation in the cytosolic loop (SEL-12 T232 through S259 and HOP-1 T188 through S215). These observations suggest that, rather than mediating an association with the membrane, this segment may mediate a protein–protein or other molecular interaction necessary for presenilin function (9). It is curious that there is a splice junction conserved in HOP-1, SEL-12, and PS1 just at the end of the conserved region (HOP-1 S215) (there are no available genomic sequence data for PS2).
Functional Redundancy of hop-1 and sel-12.
RNA-mediated interference is a method for reducing endogenous gene activity in C. elegans (22, 23). By using RNA-mediated interference to reduce hop-1 activity, we have found evidence for functional redundancy of hop-1 and sel-12. When hop-1 activity is reduced in a sel-12 mutant background, we observed phenotypes characteristic of glp-1 and lin-12 single mutants, and lin-12 glp-1 double mutants. These results suggest that hop-1, like sel-12 (6), facilitates lin-12 and glp-1 signaling.
The fact that we see hop-1 RNA-mediated interference phenotypes only when sel-12 activity is reduced suggests that hop-1 and sel-12 are functionally redundant. The simplest interpretation of this apparent functional redundancy is that, in some cases, HOP-1 and SEL-12 may be expressed and may function within the same cells. The cells in which HOP-1 and SEL-12 expression and function overlap might include at least some of the cells undergoing lin-12- and glp-1-mediated cell–cell interactions, so that reducing both hop-1 and sel-12 activities cause characteristic cell fate transformations. Although we have thus far been unable to determine the expression pattern of hop-1 directly, apparently because it is not highly expressed, we note that sel-12 is expressed in many cells and cell types during development (7) and appears to facilitate lin-12 signaling within the same cell (6).
Whereas hop-1 and sel-12 appear to be functionally redundant, we cannot conclude from our experiments that hop-1 does not have any unique roles, because the RNA-mediated interference method may not completely eliminate hop-1 gene activity in a wild-type genetic background. Nevertheless, it is possible that conventional hop-1 null alleles may not cause a significant visible phenotype. However, we expect that hop-1(−);sel-12(−) double mutants would display highly penetrant phenotypes associated with reducing lin-12 and/or glp-1 activity, and possibly additional defects as well.
Genetic analysis suggests that the hop-1 and sel-12 presenilins facilitate lin-12 and glp-1 activity (ref. 6; this work). Furthermore, targeted disruption of the mouse PS1 gene causes striking phenotypes associated with reduced Notch activity (28, 29). Although presenilins may be involved in other processes as well, these observations suggest that there is an intimate relationship between presenilin activity and Notch activity. Understanding the molecular basis for this relationship is an important goal that will be facilitated by further genetic analysis of sel-12 and hop-1, and of suppressors of mutant phenotypes associated with sel-12 and hop-1, in C. elegans.
Acknowledgments
We are grateful to Richard Ruiz and Denise Brousseau for technical assistance; Barth Grant, Jane Hubbard, and Diane Levitan for valuable advice; and Tom Cline, David Hirsh, and members of the laboratory for comments on this manuscript. This work was supported by National Institute of Health Grant NS35556. I.G. is an Associate Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- PS1
presenilin 1
- PS2
presenelin 2
- Egl
egg-laying defective
- RACE
rapid amplification of cDNA ends
References
- 1.Schellenberg G D. Proc Natl Acad Sci USA. 1995;92:8552–8559. doi: 10.1073/pnas.92.19.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levy-Lahad E, Wijsman E M, Nemens E, Andereson L, Goddard K A, Beber J L, Bird T D, Schellenberg G D. Science. 1995;269:970–973. doi: 10.1126/science.7638621. [DOI] [PubMed] [Google Scholar]
- 3.Rogaev E I, Sherrington R, Rogaeva E A, Levesque G, Ideda M, et al. Nature (London) 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 4.Sherrington R, Rogaev E I, Liang Y, Rogaeva E A, Levesque G, et al. Nature (London) 1995;375:754–760. [Google Scholar]
- 5.Kovacs D M, Fausett H J, Page K J, Kim T-W, Moir R D, Merriam D E, Hollister R D, Hallmark O G, Mancini R, Felsenstein K M, Hyman B T, Tanzi R E, Wasco W. Nat Med. 1996;2:224–229. doi: 10.1038/nm0296-224. [DOI] [PubMed] [Google Scholar]
- 6.Levitan D, Greenwald I. Nature (London) 1995;377:351–354. doi: 10.1038/377351a0. [DOI] [PubMed] [Google Scholar]
- 7.Levitan D, Doyle T G, Brousseau D, Lee M K, Thinakaran G, Slunt H H, Sisodia S S, Greenwald I. Proc Natl Acad Sci USA. 1996;93:14940–14944. doi: 10.1073/pnas.93.25.14940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doan A, Thinakaran G, Borchelt D R, Slunt H H, Ratovitsky T, Podlisny M, Selkoe D J, Seeger M, Gandy S E, Price D L, Sisodia S S. Neuron. 1996;17:1023–1030. doi: 10.1016/s0896-6273(00)80232-9. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Greenwald I. Neuron. 1996;17:1015–1021. doi: 10.1016/s0896-6273(00)80231-7. [DOI] [PubMed] [Google Scholar]
- 10.Thinakaran G, Borchelt D R, Lee M K, Slunt H H, Spitzer L, Kim G, Ratovitsky T, Davenport F, Nordstedt C, Seeger M, Hardy J, Levey A I, Gandy SE, Jenkins N A, Copeland N G, Price D L, Sisodia S S. Neuron. 1996;17:181–190. doi: 10.1016/s0896-6273(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 11.Waterston R H, Sulston J E, Coulson A R. In: C. elegans II. Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. Plainview, NY: Cold Spring Harbor Lab.; 1997. pp. 23–46. [Google Scholar]
- 12.Edgley M L, Turner C A, Riddle D L. In: C. elegans II. Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. Plainview, NY: Cold Spring Harbor Lab.; 1997. pp. 1059–1062. [Google Scholar]
- 13.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barstead R J, Waterston R H. J Biol Chem. 1989;264:10177–10185. [PubMed] [Google Scholar]
- 15.Krause M, Hirsh D. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang X-Y, Hirsh D. Proc Natl Acad Sci USA. 1989;86:8640–8644. doi: 10.1073/pnas.86.22.8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fire A, Harrison S W, Dixon D. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- 18.Mello C C, Kramer J M, Stinchcomb D T, Ambros V A. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fire, A. (1993) Genet. Anal. Tech. Appl. 151–158. [DOI] [PubMed]
- 20.Silhavy T J, Beckwith J R. Microbiol Rev. 1985;49:398–418. doi: 10.1128/mr.49.4.398-418.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Froshauer S, Green G N, Boyd D, McGovern K, Beckwith J. J Mol Biol. 1988;200:501–511. doi: 10.1016/0022-2836(88)90539-6. [DOI] [PubMed] [Google Scholar]
- 22.Guo S, Kemphues K J. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- 23.Rocheleau C, Downs W D, Lin R, Wittmann C, Bei Y, Cha Y-H, Ali M, Priess J R, Mello C C. Cell. 1997;90:707–716. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- 24.Greenwald I S, Sternberg P W, Horvitz H R. Cell. 1983;34:435–444. doi: 10.1016/0092-8674(83)90377-x. [DOI] [PubMed] [Google Scholar]
- 25.Austin J, Kimble J. Cell. 1987;51:589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- 26.Priess J R, Schnabel H, Schnabel R. Cell. 1987;51:601–611. doi: 10.1016/0092-8674(87)90129-2. [DOI] [PubMed] [Google Scholar]
- 27.Lambie E, Kimble J. Development (Cambridge, UK) 1991;112:231–240. doi: 10.1242/dev.112.1.231. [DOI] [PubMed] [Google Scholar]
- 28.Shen J, Bronson R T, Chen D F, Xia W, Selkoe D J, Tonegawa S. Cell. 1997;89:629–639. doi: 10.1016/s0092-8674(00)80244-5. [DOI] [PubMed] [Google Scholar]
- 29.Wong P C, Zheng H, Chen H, Becher M W, Sirinathsinghji D J S, Trumbauer M W, Chen H Y, Price D L, Van der Ploeg L H T, Sisodia S S. Nature (London) 1997;387:288–292. doi: 10.1038/387288a0. [DOI] [PubMed] [Google Scholar]
- 30.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kyte J, Doolittle R. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]