Abstract
Oestrogen action is mediated via specific receptors that act as ligand-activated transcription factors. A monoclonal antibody specific to the C-terminus of human oestrogen receptor beta has been characterized and the prevalence of expression of oestrogen receptor beta protein investigated in a well defined set of breast cancers. Reverse transcription-polymerase chain reaction analysis of RNA from tissue biopsies detected oestrogen receptor beta in all samples examined. The anti-oestrogen receptor beta antibody cross reacted specifically with both long (∼59 Kd) and short (∼53 Kd) forms of recombinant oestrogen receptor beta. Western blot analysis of breast tumours contained both forms of oestrogen receptor beta protein although in some samples lower molecular weight species (32–45 Kd) were identified. Fifty-one breast cancer biopsies were examined using immunohistochemistry; 41 (80%) were immunopositive for oestrogen receptor alpha, 48 (94%) were immunopositive for oestrogen receptor beta and 38 (74.5%) co-expressed both receptors. Expression of oestrogen receptor beta was exclusively nuclear and occurred in multiple cell types. There was no quantitative relationship between staining for the two ERs although in tumours in which both receptors were present immunoexpression of oestrogen receptor alpha was invariably more intense. The significance of oestrogen receptor beta protein expression in breast cancers to therapy remains to be determined but the availability of a well characterized antibody capable of detecting oestrogen receptor beta in archive material will facilitate the process.
British Journal of Cancer (2002) 86, 250–256. DOI: 10.1038/sj/bjc/6600035 www.bjcancer.com
© 2002 The Cancer Research Campaign
Keywords: oestrogen receptor, ERβ, monoclonal, breast
Until recently it was accepted that the major effects of oestrogen on the growth and development of the breast and its tumours was mediated through a single oestrogen receptor (ERα, Green et al, 1986). Ligand binding assays and immunohistochemical studies indicated that most breast tumours possessed such receptors and their presence was associated with the likelihood of response to endocrine therapy (McGuire et al, 1982; Jordan et al, 1988; Miller, 1996). However in 1996 an additional ER isotype, usually known as ERβ, was identified in rat (Kuiper et al, 1996) and human (Mosselman et al, 1996). Both receptors share significant sequence homology within their DNA and ligand binding domains but are encoded on different chromosomes (Enmark et al, 1997). Studies in vitro have demonstrated that although both ERα and ERβ bind oestradiol with equal affinity (Kuiper et al, 1997) these receptors may have differential responses to some oestrogen agonists and antagonists (Watanabe et al, 1997; Barkhem et al, 1998; Jones et al, 1999; Sun et al, 1999). Notably ERβ appears to have a higher affinity for phytoestrogens, including genestein, than does ERα (Kuiper et al, 1997). When present within in the same cell, ERα and ERβ have the capacity to form either homo- or heterodimers (Pace et al, 1997) and the proportions of the different isotypes may be critical to modulation of gene expression (Hall and McDonnell, 1999). Studies in mammary tissues of the rat have suggested that one role of ERβ may be to antagonize ERα-mediated actions in epithelial cells (Saji et al, 2000), a function supported by data from in vitro cell transfections (Hall and McDonnell, 1999).
To date studies demonstrating the expression of ERβ in breast cancer tissues have largely been confined to the demonstration of expression of ERβ mRNA (Dotzlaw et al, 1997; Leygue et al, 1998; Speirs et al, 1999; Vladusic et al, 2000). Messenger RNAs encoding variant forms of both ERα (Bollig and Miksicek, 2000) and ERβ (Lu et al, 1998) have been identified in breast cancers and in breast cancer cell lines and there has been considerable debate over the role of such variants in cancer progression (Balleine et al, 1999; Huang et al, 1999).
The present investigation was designed to characterize the expression of ERβ and ERα proteins in a series of 51 breast cancers; some samples were also subjected to analysis for mRNAs by RT–PCR. We have made use of specific monoclonal antibodies and used both immunohistochemistry on well-fixed tissues in which the cellular architecture has been preserved as well as Western analysis of tissue extracts. These investigations have demonstrated wide spread expression of ERβ protein and provide new information important for further exploration of the relationship between the co-expression of ERβ and ERα and the in response of breast cancers to endocrine therapies.
MATERIALS AND METHODS
Patients and tissue samples
Samples of breast were obtained from 51 consecutive patients presenting to the Edinburgh Breast Unit with diagnosis of breast cancer who had given informed consent for tissue to be used for research purposes. Samples were snap frozen to provide material for extraction of RNA or protein, or fixed in 10% neutral buffered formaldehyde for 16 to 24 h then stored in 70% (w v−1) ethanol prior to processing into paraffin wax at the Department of Pathology using standard procedures.
Detection of ERα and ERβ by reverse transcription-polymerase chain reaction (RT–PCR)
RNA was extracted using the Tri-reagent system according to the manufacturer's instructions (Sigma, Poole, Dorset, UK), dissolved in RNase-free water and stored at −70°C. One microgram of RNA was reverse transcribed for 1 h at 42°C in a 20-μl reaction using the Superscript system (Gibco-BRL, Paisley, Scotland, UK). Upon completion of the incubation, the sample cDNAs were each diluted to a final volume of 60 μl, and 20 μl used in individual PCR reactions containing primers specific for ERα, ERβ or alpha-actin (positive control). The primers employed were as follows: human ERα (Green et al, 1986), forward 5′-GGCCAGTACCAATGACAAGGGAAG-3′ (nucleotides 787–811); ERα, reverse 5′-CCAGCAAGCATGTCGAAGATCTCC-3′ (nucleotides 1558–1580); human ERβ (Ogawa et al, 1998a), forward 5′-GTTGCGCCAGCCCTGTTAC-3′ (nucleotides 493–512); ERβ, reverse 5′-CTCGTCGGCACTTCTCTGTCTC-3′ (nucleotides 788–809); alpha-actin forward, 5′-GGAGCAATGATCTTGATCTT-3′; alpha-actin reverse, 5′-CCTTCCTGGGCATGGAGTCCT-3′. The primers used to amplify the oestrogen receptor cDNAs were chosen to span regions separated by two intronic regions. PCR reactions were carried out using ‘Hot start’ Taq polymerase (Qiagen, Crawley, West Sussex, UK) and the following cycling conditions; 96°C for 30 s, 56°C for 1 min, 72°C for 1 min, repeated for 30 cycles for ERα, similar conditions were used for ERβ except that the annealing temperature was 52°C. The expected sizes of the amplified bands were; ERα, 793 bp; ERβ, 316 bp; alpha actin 120 bp. Nine samples were analyzed.
Antibodies
The anti-hERα mouse monoclonal antibody (code 1D5) was obtained from DAKO (Cambridge, UK). A peptide located at the C-terminus of hERβ (Mosselman et al, 1996) (CSPAEDSKSKEGSQNPQSQ) was used to prepare a monoclonal antibody in mice according to standard methods and positive clones were identified by ELISA using recombinant human ERβ (P2466, PanVera, Madison, WI, USA) (Saunders et al, 2000). This antibody has been used previously to demonstrate expression of ERβ using human ovarian tissue sections (Saunders et al, 2000).
Western analysis
Two forms of recombinant human ERβ1 were obtained from Pan Vera (Madison, WI, USA). These were hERβ1 ‘short’, a ∼53 Kd form of the receptor (βs) synthesized from a cDNA (Mosselman et al, 1996) lacking the first potential start site for translation (Ogawa et al, 1998a), and hERβ1 ‘long’ (βL) the larger protein (∼59 Kd) synthesized from the full length cDNA (Ogawa et al, 1998a). Recombinant hERα (∼66 Kd) was also obtained from Pan Vera. Gel analysis and blotting were carried out as described previously (Saunders et al, 2000). Briefly, proteins were extracted from frozen biopsy specimens by rapid homogenization of tissue in denaturing/loading buffer (50 mM Tris-HCl pH 6.8, 100 mM DTT, 2% SDS, 0.1% bromophenol blue, 10% glycerol, all from Sigma). Recombinant proteins (0.5 μg lane−1), tissue extracts (30–50 μg total protein) and prestained protein molecular weight markers (BioRad) were separated on denaturing minigels containing an acrylamide gradient from 4 to 20% (w v−1) polyacrylamide (Novex, San Diego, CA, USA). Membranes were incubated overnight with the mouse monoclonal anti hERβ1 (code M9) at 1 in 500 or mouse monoclonal anti-hERα (code1D5) at 1 in 100; both the antibodies were diluted in TBST containing 5% normal donkey serum. Bound antibodies were detected using rabbit anti-mouse IgG and the ECL visualization system (Amersham, Bucks, UK) according to the manufacturer's instructions.
Immunohistochemistry
Sections (4 μm) were mounted on Superfrost coated slides (BDH, Poole, Dorset, UK) dewaxed and rehydrated in gradient alcohols and distilled water. Endogenous peroxidases were blocked with 3% hydrogen peroxide for 10 min and sections were subjected to heat-induced antigen retrieval in 0.01 m citrate buffer, pH 6.0 (Norton et al, 1994) before staining with specific antibodies as outlined below.
Anti-ERα
All staining for ERα was carried out in the Pathology Department of the Western General Hospital. An endogenous biotin block was carried out by applying 100 μl egg white blocking solution for 30 min. Anti-ERα, (Dako) was diluted 1 in 50 in biotin diluent for primary antibodies (PBS, goat serum and d-biotin), and incubated in the sections for 60 min at room temperature. The secondary antibody, biotinylated anti-mouse Ig(Vector Laboratories) was diluted 1 : 2000, in ‘background reducing diluent’ (Dako) and applied to sections for 30 min at room temperature. The tertiary system (ABC-HRP, Dako) was applied as per manufacturer's instructions for 30 min at room temperature. The tissue was visualized by immersing sections in 3,3′-diaminobenzidine tetra-hydrochloride (DAB) for 5 min. Sections were counterstained using Mayers haematoxylin (Sigma-Aldrich, Poole, Dorset), dehydrated through gradient alcohols and mounted.
Anti-ERβ
Immunolocalization was undertaken as described in detail in Saunders et al (2000), Sections were blocked for 30 min in normal rabbit serum (NRS, Diagnostics Scotland, Carluke) diluted 1 : 4 in TBS containing 5% BSA (NRS/TBS/BSA), rinsed briefly in TBS and an avidin biotin block performed using reagents from Vector (Peterborough, UK). Anti-ERβ antibody was diluted 1 : 40 in NRS/TBS and incubated on sections overnight at 4°C. Sections were washed twice for 5 min each time in TBS and incubated with rabbit anti mouse, (Dako, Cambridge, UK) diluted 1 : 500 in NRS/TBS/BSA. Thereafter, bound antibodies were visualized by incubation with 3,3′-diaminobenzidine tetra-hydrochloride (liquid DAB cat K3468, DAKO). Sections were counterstained with haematoxylin.
Images were captured using an Olympus Provis microscope (Olympus Optical Co, London, UK) equipped with a Kodak DCS330 camera (Eastman Kodak Co., Rochester, NY, USA), stored on a Macintosh PowerPC computer and assembled using Photoshop 5.5 (Adobe, Mountain View, CA, USA).
Quantitation of immunohistochemical staining
Quantitation was based on a scoring system reported in detail previously (Allred et al, 1998; Leake et al, 2000). This method is based on a composite additive score of intensity 0–3 and proportion of malignant epithelial cells staining 0–5. This gives a range from 0–8 for each tissue. Samples were analyzed using the SPSS package (version 10 for Macintosh; SPSS Inc, Chicago, IL, USA) and plotted as a box and whisker plot. No correlation between ERα and ERβ scores was detected.
RESULTS
Detection of mRNAs for ERα and ERβ in breast cancer samples
All samples tested (n=9) were positive for ERβ following RT–PCR (Figure 1). This signal always appeared greater than those for ERα and was present in both ERα positive and negative samples. Actin was amplified from all samples although the efficiency of the reaction was variable.
Figure 1.
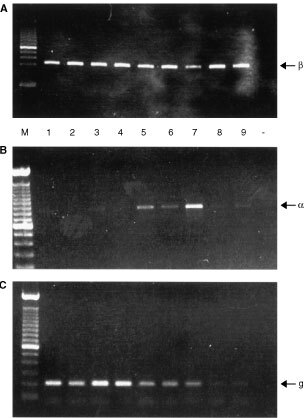
Detection of oestrogen receptor mRNAs by RT–PCR. (A) ERβ, (B) ERα, (C) Alpha-actin. In all panels, lane M 100 bp ladders, lanes 1–9 breast tumour samples, the negative control lane (−) contained a sample prepared without reverse transcriptase. Note that although a cDNA specific for ERβ was amplified from all samples, the amount of ERα cDNA amplified from the same sample set was highly variable.
Specificity of antisera and extraction of ER proteins from breast cancer biopsies
On Western blots (Figure 2) antibodies directed against ERα and ERβ bound to either recombinant ERα or recombinant ERβ protein depending upon the isotype to which they were directed. These results were consistent with previously published data (Saunders et al, 2000); no binding of the ERβ specific monoclonal to ERα was observed (Figure 2, lower panel, lane α). The anti-hERβ monoclonal that was directed against a peptide at the C-terminus of hERβ bound to both short (Mosselman et al, 1996) and long (Ogawa et al, 1998a) forms of ERβ. This result is consistent with data that has demonstrated that the difference in size of the long and short forms of ERβ is due to use of alternative start sites for translation within the full length mRNA and that the C-termini of both proteins are identical.
Figure 2.
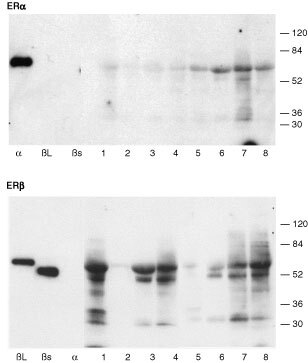
Western analysis of proteins extracted from breast cancer samples. Proteins were separated, blotted and incubated with antibodies directed against ERα (upper panel) or ERβ (lower panel). The anti-ERα antibody bound to recombinant hERα but not to recombinant hERβ (βs, βL). The anti-ERβ1 antibody bound to both long (βL) and short (βs) forms of recombinant hERβ but not to recombinant hERα (α). Proteins migrating with the same apparent molecular size as recombinant ERα (α, upper panel, arrowhead) were detected in all breast samples (lanes 1 to 8, note identical samples were used for both gels and are loaded in the same order). In sample numbers 6 and 7 additional lower molecular weight forms of ERα were present. Variable amounts of ERβ proteins were detected in the same samples. Proteins migrating with the same apparent molecular size as both long and short forms of ERβ proteins (arrowheads) were detected in breast sample numbers 1, 3, 4, 6, 7, 8; additional lower molecular weight variants were present in these same extracts but samples 2 and 5 lacked significant levels of ERβ.
Tissue biopsied from eight tumours, that were histologically shown to be cancers, were also examined. The predominant form of the ERα protein (Figure 2, upper panel) extracted from all biopsies migrated with an apparent molecular size (∼66 Kd) identical to recombinant ERα run in a parallel lane (α). In only two samples (lanes 6 and 7) did we see evidence of expression of shorter/variant ERα proteins.
The amount of ERβ protein detected in extracts from cancer biopsies was highly variable (Figure 2 lower panel). It was notable that in six of the eight samples proteins migrating with apparent molecular sizes corresponding to both long (∼59 Kd) and short (53 Kd) ERβ were present. We have found that this antibody recognizes ERβ protein extracted from human ovary, prostate (Saunders et al, 2000) endometrium and testis and human cell lines (MCF-7, Ishikawa, unpublished observations). In breast tumour samples that appeared to contain high levels of expression of full length ERβ (numbers 1, 3, 4, 7, 8) several lower molecular weight protein species with apparent molecular weights from 32 to 45 Kd were detected.
Immunolocalization of oestrogen receptors
Typical examples of immunostaining for ERα and ERβ are shown in Figures 3 and 4 respectively. Staining for ERα (Figure 3) was predominantly nuclear and almost exclusively restricted to malignant epithelium (insets A′ and B′) in this tissue series. Note that the malignant tissues illustrated in Figure 4A,B are the same as those in Figure 3A,B (codes 5580 and 5667 respectively) and clearly illustrate that ERα expression (Figure 3) can occur in the presence (Figure 4A) or absence (Figure 4B) of ERβ. Expression of ERβ was almost exclusively nuclear and often appeared granular and heterogeneous (Figure 4A′). Expression of ERβ was noted in a wider range of cells than was ERα and was found in non-malignant components of the tumour including normal glandular elements (Figure 4D arrows), blood vessels, adipose tissue and stromal cells (asterisks) as well as in non-invasive intraduct cancers (Figure 4C).
Figure 3.
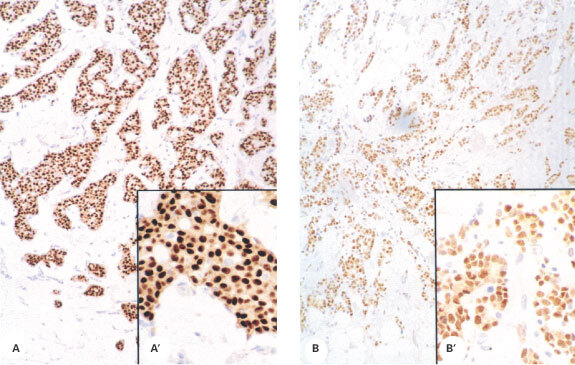
Immunoexpression of ERα in human breast cancers. Nuclear expression of ERα was largely confined to malignant epithelium in the 40 samples in which it was detected; intensity was variable. (A) example of intense immunostaining (sample code 5580); (B) sample code 5667, magnification ×20, insets A′ and B′ show higher power magnification of the same tissue samples.
Figure 4.
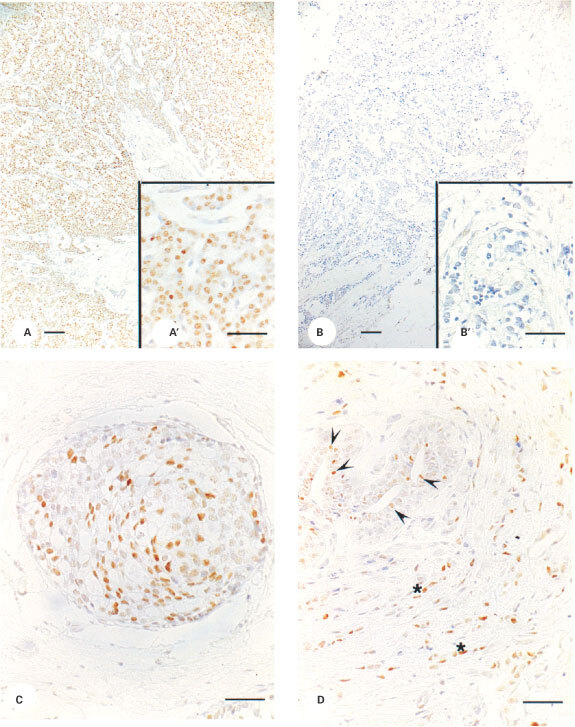
Immunoexpression of ERβ in human breast tissues. Nuclear expression of ERβ protein was detected in 94% of the samples examined. (A,B) show examples of immunopositive (A, code 5580) and immunonegative (B, code 5667) staining of malignant tissue. Expression of ERβ was also noted in non-invasive ductal cancer (C) and in epithelial (D, arrowheads) and stromal (D, asterisks) cells in areas of breast not associated with malignant growth. (A,B), Magnification ×10, bar=100 μM, insets A′ and B′ are from the same tissues as A and B, magnification ×40, bar=50 μM. (C, D) Magnification ×40, bar=50 μM.
Quantitation of immunohistochemical staining
Most of the tumours (48 out of 51) displayed staining for ERβ in malignant epithelium with a range of scoring between 2 and 7 (median score 4.5). ERα staining was found in 41 out of 50 tumours with a range of scoring between 6 and 8 (median score 7.5). Quantitatively it was possible to identify ERα-positive, ERβ-positive tumours (38 out of 51, Figures 3A and 4A) as well as ERα-positive, ERβ-negative tumours (3 out of 51, Figure 3B compared with Figure 4B; 2 out of 51). ERα-negative, ERβ-positive tumours were detected (10 out of 51) but we observed no double negatives. There was no quantitative relationship between immunohistochemical scores for ERα and ERβ (Figure 5).
Figure 5.
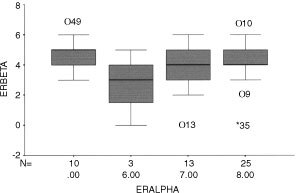
Quantification of immunoexpression for ER isotypes. Box and whisker plot summarising the relationship between score for ERα (x-axis) and ERβ (y-axis) for each sample. Solid horizontal line shows the median for the data, the top of the box the 25th percentile, the bottom the 75th percentile and the additional lines the range of the data. Note that there were no samples with an ERα score of 1 to 5.
DISCUSSION
Many breast cancers, like the normal tissue from which they are derived, appear sensitive to oestrogens. The major action of oestrogen appears to be mediated by specific receptor proteins that act as nuclear transcription factors. Until recently, studies have concentrated on the ERα member of the family and these have clearly demonstrated the involvement of the protein in maintaining the growth of hormone sensitive tumours. As a consequence ERα measurements have been used to select patients for endocrine therapy and the protein has become a therapeutic target by which to treat patients with breast cancer. Nevertheless there have been paradoxical observations such as tumours regressing following endocrine deprivation therapy in apparently ERα negative disease. Oestrogen responses in ERα knockout mice and the differential effects of anti-oestrogens in tissues and tumours were also unexplained.
Our ability to correlate ER status with outcome of therapy has been complicated by the finding of a second oestrogen receptor (ERβ) which can bind oestrogens including oestradiol and tamoxifen with high affinity (Kuiper et al, 1996, 1997; Mosselman et al, 1996). As a result there has been a major effort to delineate the role of ERβ in the natural history of breast cancer. Many papers have reported that the mRNAs for both ERα and ERβ are expressed in breast cancer cell lines (Watanabe et al, 1997; Moore et al, 1998; Vladusic et al, 2000), in breast cancer tissue (Dotzlaw et al, 1997) and in the normal human and rodent mammary gland (Moore et al, 1998; Saji et al, 2000). Studies that have compared levels of expression of the mRNAs encoding the two receptors have reported that the amount of ERβ mRNA does not appear to be correlated with that of ERα (Dotzlaw et al, 1997; Iwao et al, 2000; Vladusic et al, 2000) consistent with expression of the receptors by different genes (Enmark et al, 1997). Some studies have reported that up-regulation/over expression of ERβ mRNA may be correlated with development of oestrogen-independent tumour growth and a poor prognosis (Speirs et al, 1999; Iwao et al, 2000).
Modelling studies using ERα have defined the amino acids within the protein which interact with natural as well as synthetic oestrogens and anti-oestrogens (Ekena et al, 1997). The major determinants of ligand binding are conserved between ERα and ERβ consistent with their ability of both to bind oestradiol (Kuiper et al, 1997). Barkhem et al (1998) have used cell lines stabily transfected with either ERα or ERβ to test the affinity and potency of widely used anti-oestrogens including tamoxifen, raloxifine and ICI 164,384 and concluded that the ligand binding cavity of ERβ is more different to that of ERα than can be anticipated from the primary sequence. Recently novel non-steroidal ligands that show subtype specific binding affinity and transcriptional potency have been identified (Sun et al, 1999) and ligand-dependent differences in the ability of ERα and ERβ to recruit co-activators following exposure to xenoestrogens described (Routledge et al, 2000). ER-driven gene activation can be determined by the formation of homo- or hetero-dimers, the cell type, and whether the ligand-activated receptors bind to a promotor containing ERE or an AP-1 site (Watanabe et al, 1997; Jones et al, 1999). Furthermore the experience with studies on ERα has been that mRNA is not necessarily translated into protein make it essential that assays for ERβ are performed at the level of protein.
The monoclonal antibody used to detect ERβ in the present study was raised against a peptide at the C-terminus of human ERβ1 (Mosselman et al, 1996; Moore et al, 1998). This peptide is not conserved in any of the ERβ variants formed by alternative splicing of the F domain of the protein (Moore et al, 1998; Ogawa et al, 1998b) and does not recognize recombinant ERβ2/βcx on Western blots (unpublished observations). Similarly Western blotting indicated that the monoclonal antibody identified ERβ but not ERα in breast cancers. Most of the ERβ1 protein detected in the extracts from the breast cancers migrated with the same apparent size as the ‘long’ and ‘short’ forms of recombinant ERβ1, which are formed by translation from different ATGs in the mRNA (Mosselman et al, 1996; Ogawa et al, 1998a). We did not detect proteins corresponding in size to those that could be translated from mRNAs deleted in exons 5 or 6 (Lu et al, 1998; Brandenberger et al, 1999) predicted to be 16.8 and 13 Kd respectively. The most prominent proteins other than full length ERβ1 migrated between 30 and 36 Kd these could represent use of alternative start sites, translation from an exon 2 deleted mRNA (∼35 Kd) or translation of protein from mRNA deleted for both exons 5 and 6 (AF074599) which is predicted to be ∼43 Kd (short) or ∼49 Kd (long) from the mRNA sequence. It is notable that mRNAs corresponding to alternatively spliced forms of ERβ have been detected in breast cancer tissues and cell lines (Lu et al, 1998; Moore et al, 1998; Vladusic et al, 1998; Iwao et al, 2000) as well as in normal human tissues (Ogawa et al, 1998b; Scobie et al, 2001). Furthermore, monoclonal antibodies directed against the N terminus of ERβ have detected expression of proteins other than full length ERβ in breast cancer cell lines (Fuqua et al, 1999) which might have been formed by translation of alternatively spliced mRNAs. During the course of the present study we found that recombinant ERβ proteins (both from commercial sources and prepared in house) degrade if subjected to a single freeze-thaw cycle or following prolonged storage even at low temperatures (−70°C). Therefore although considerable attention was paid to extraction of the breast tumour samples and to the storage of extracts we believe that the most likely explanation for the lower molecular weight bands identified in samples containing the highest levels of ERβ1 is that these are breakdown products of the full length protein which have formed during handling of the protein extracts.
We have used our ERβ1 specific monoclonal antibody to immuno-localize ERβ1 in a series of breast cancers as well as in other human and primate tissues (Saunders et al, 2000; Scobie et al, 2001). The present study has demonstrated the presence of ERβ1 in cell nuclei not only the malignant epithelium but also non-malignant elements of most breast cancers. The qualitative and quantitative expression of ERβ was independent of that of ERα. We have observed that ERβ1 was also expressed in multiple types of non-cancer cells within the breast tissue and this will therefore further complicate the assessment of ERβ status. For example, methods such as RT–PCR or Western blotting which use tissue extracts may contain a contribution from cells other than those derived from the malignant component of the tumour. It will therefore be important to quantify expression in different compartments of the breast separately. This precludes the simple use of Western and Northern blotting together with other technologies in which tissue is homogenized and extracted.
Whilst our studies were being written up three reports describing immunolocalization of ERβ to breast cancer samples were published. Mann et al (2001) used a rabbit polyclonal antibody directed against the N-terminus of human ERβ on formalin fixed samples; on the Western blot shown in their article multiple bands are shown, the most prominent of which appeared shorter than the recombinant standard and this may reflect degradation of protein in their extracts or non-specific reactivity of the antibody used. In their paper immunopositive staining of human breast cancer for ERβ was present in 66 and 70% of the two sets of samples reported but no mention was made of immunopositive staining of cells other than those of the malignancy. The authors mentioned the potential cross-reactivity of their antibody with isoforms of ERβ including ERβcx (Ogawa et al, 1998b) which will not occur with the antibody used in the current study. It is notable that the polyclonal rabbit antibody used by Omoto et al (2001) is raised to an identical part of the ERβ1 protein to our monoclonal and we would therefore expect similar results to our own. In their study they used frozen sections of tissue and found that only 59% (52 out of 88) were positive for ERβ, with only 38% of the ERα negative samples expressing the ERβ subtype. This proportion is much lower than in the current study or in the tissue set studied by Jarvinen et al (2000) who used frozen sections fixed briefly with Zamboni's, and found 60% of cancers contained ERβ1 positive cells using a commercial polyclonal antibody raised to the same region of the protein. The need to use frozen sections clearly limits the utility of these antibodies and highlights an important difference with the reagent used in the present study which appears capable of identifying ERβ1 in material fixed by formalin, methacarn (unpublished observations) or Bouins (Saunders et al, 2000). In studies using fixed samples from human tissues including ovary, placenta, vas deferens, testis and endometrium we have used monoclonal and polyclonal antibodies to localize ERβ proteins (Saunders et al, 2000; Critchley et al, 2001; Scobie et al, 2001). In all cases we find the protein to be nuclear in location in agreement with the findings using fixed tissues of human breast (present study) the only exceptions being dividing cells, and some myoid cell types where background staining of the cytoplasm associated with the secondary antibodies was a problem. We have detected cytoplasmic staining using some commercial anti ERβ antibodies especially those that have not been affinity purified and with some secondary antibodies especially those raised in goats (unpublished observations). These findings may explain some of the cytoplasmic staining seen in the figures published by others (Jarvinen et al, 2000; Mann et al, 2001; Omoto et al, 2001).
In conclusion, we believe that to assess the responsiveness of breast cancers to oestrogenic and anti-oestrogenic stimuli it will be necessary to measure both ERα and ERβ at the level of protein. The presence of ERβ in both malignant and non-malignant components of breast tumours means that assessments in individual compartments may also be required. This approach is being utilized in our ongoing studies.
Acknowledgments
We thank Dr Graeme Scobie for useful discussion and Dr D Stewart Irvine for assistance with statistical analysis.
References
- AllredDCHarveyJMBerardoMClarkGM1998Prognostic and predictive factors in breast cancer by immunohistochemical analysis Mod Pathol 11155168 [PubMed] [Google Scholar]
- BalleineRLHuntSMNClarkeCL1999Coexpression of alternatively spliced estrogen and progesterone receptor transcripts in human breast cancer J Clin Endocrinol Metab 8413701377 [DOI] [PubMed] [Google Scholar]
- BarkhemTCarlssonBNilssonYEnmarkEGustafssonJNilssonS1998Differential response of estrogen receptor alpha and estrogen receptor beta to partial estrogen receptor agonists/antagonists Mol Pharmacol 54105112 [DOI] [PubMed] [Google Scholar]
- BolligAMiksicekRJ2000An estrogen receptor-a splicing variant mediates both positive and negative effects on gene transcription Mol Endocrinol 14634649 [DOI] [PubMed] [Google Scholar]
- BrandenbergerAWLebovicDITeeMKRyanIPTsengJFJaffeRBTaylorRN1999Oestrogen receptor (ER)-alpha and ER-beta isoforms in normal endometrial and endometriosis-derived stromal cells Mol Hum Reprod 5651655 [DOI] [PubMed] [Google Scholar]
- CritchleyHODBrennerRMDrudyTAWilliamsKANayakNRMillarMRSaundersPTK2001Estrogen receptor beta, but not estrogen receptor alpha, is present in the vascular endothelium of the human and nonhuman primate endometrium J Clin Endocrinol Metab 8613701378 [DOI] [PubMed] [Google Scholar]
- DotzlawHLeygueEWatsonPHMurphyLC1997Expression of estrogen receptor-beta in human breast tumors J Clin Endocrinol Metab 8223712374 [DOI] [PubMed] [Google Scholar]
- EkenaKWeisKEKatzenellenbogenJAKatzenellenbogenBS1997Different residues of the human estrogen receptor are involved in the recognition of structurally diverse estrogens and anti-estrogens J Biol Chem 27250695075 [DOI] [PubMed] [Google Scholar]
- EnmarkEPelto-HuikkoMGrandienKLagercrantzSLagercrantzJFriedGNordenskjoldMGustafssonJ-A1997Human estrogen receptor β-gene structure, chromosomal localization, and expression pattern J Clin Endocrinol Metab 8242584265 [DOI] [PubMed] [Google Scholar]
- FuquaSASchiffRParraIFriedrichsWESuJLMcKeeDDSlentz-KeslerKMooreLBWillsonTMMooreJT1999Expression of wild-type estrogen receptor beta and variant isoforms in human breast cancer Cancer Res 5954255428 [PubMed] [Google Scholar]
- GreenSWalterPKumarVKrustABornertJ-MArgosPChambonP1986Human oestrogen receptor cDNA: sequence, expression and homology to v-erb-A Nature 320134139 [DOI] [PubMed] [Google Scholar]
- HallJMMcDonnellDP1999The estrogen receptor β-isoform (ERβ) of the human estrogen receptor modulates ERα transcriptional activity and is a key regulator of the cellular response to estrogens and antiestrogens Endocrinology 14055665578 [DOI] [PubMed] [Google Scholar]
- HuangALeygueEDotzlawHMurphyLCWatsonPH1999Influence of estrogen variants on the determination of ER status in human breast cancer Breast Cancer Res Treat 58219225 [DOI] [PubMed] [Google Scholar]
- IwaoKMiyoshiYEgawaCIkedaNTsukamotoFNoguchiS2000Quantitative analysis of estrogen receptor-alpha and -beta messenger RNA expression in breast carcinoma by real-time polymerase chain reaction Cancer 1517321738 [DOI] [PubMed] [Google Scholar]
- JarvinenTAHPelto-HuikkoMHolliKIsolaJ2000Estrogen receptor β is co-expressed with ERa, and PR and associated with nodal status, grade and proliferation rate in breast cancer Am J Pathol 1562935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- JonesPSParrottEWhiteINH1999Activation of transcription by estrogen receptor a and β is cell type- and promoter-dependent J Biol Chem 2743200832014 [DOI] [PubMed] [Google Scholar]
- JordanVCWolfMFMireckiDMWhitfordDAWelshonsWV1988Hormone receptor assays: clinical usefulness in the management of carcinoma of the breast CRC Crit Rev Clin Lab Sci 2697151 [DOI] [PubMed] [Google Scholar]
- KuiperGGJMCarlssonBGrandienKEnmarkEHaggbladJNilssonSGustafssonJ-A1997Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta Endocrinology 138863870 [DOI] [PubMed] [Google Scholar]
- KuiperGGJMEnmarkEPelto-HukkoMNilssonSGustafssonJ-A1996Cloning of a novel estrogen receptor expressed in rat prostate Proc Natl Acad Sci USA 9359255930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeakeRBarnesDPinderSEllisIAndersonLAndersonTAdamsonRRhodesTMillerKWalkerR2000Immunohistochemical detection of steroid receptors in breast cancer: a working protocol J Clin Pathol 53634635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeygueEDotzlawHWatsonPHMurphyLC1998Altered estrogen receptor a and β messenger RNA expression during human breast tumorigenesis Cancer Res 5831973201 [PubMed] [Google Scholar]
- LuBLeygueEDotzlawHMurphyLCWatsonPH1998Estrogen receptor-β mRNA variants in human and murine tissues Mol Cell Endocrinol 138199203 [DOI] [PubMed] [Google Scholar]
- MannSLauciricaRCarlsonNYounesPSAliNYounesALiYYounesM2001Estrogen receptor beta expression in invasive breast cancer Human Pathol 32113118 [DOI] [PubMed] [Google Scholar]
- McGuireWLOsborneCKClarkGMKnightWA1982Steroid hormone receptors and carcinoma of the breast Am J Physiol 243E99E102 [DOI] [PubMed] [Google Scholar]
- MillerWR1996Prediction of estrogen sensitivity/dependenceInEstrogen and Breast Cancer,Miller WR (ed.)pp 151169Austin, Texas: Landes, RG [Google Scholar]
- MooreJTMcKeeDDSlentz-KeslerKMooreLBJonesSAHorneELSuJLKliewerSALeymannJMWillsonTM1998Cloning and characterisation of human estrogen receptor beta isoforms Biochem Biophys Res Commun 2477578 [DOI] [PubMed] [Google Scholar]
- MosselmanSPolmanJDijkemaR1996ERbeta: identification and characterization of a novel human estrogen receptor FEBS Lett 3924953 [DOI] [PubMed] [Google Scholar]
- NortonAJJordanSYeomansP1994Brief, high-temperature heat denaturation (pressure cooking): a simple and effective method of antigen retrieval for routinely processed tissues J Pathol 173371379 [DOI] [PubMed] [Google Scholar]
- OgawaSInoueSWatanabeTHiroiHOrimoAHosoiTOuchiYMuramatsuM1998aThe complete primary structure of human estrogen receptor beta (hER beta) and its heterodimerization with ER alpha in vivo and in vitro Biochem Biophys Res Commun 243129132 [DOI] [PubMed] [Google Scholar]
- OgawaSInoueSWatanabeTOrimoAHosoiTOuchiYMuramatsuM1998bMolecular cloning and characterization of human estrogen receptor bcx: a potential inhibitor of estrogen action in human Nucl Acids Res 2635053512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- OmotoYInoueSOgawaSToyamaTYamashitaHMuramatsuMKobayashiSIwaseH2001Clinical value of the wild type estrogen receptor β expression in breast cancer Cancer Lett 163207212 [DOI] [PubMed] [Google Scholar]
- PacePTaylorJSuntharalingamSCoombesRCAliS1997Human estrogen receptor beta binds DNA in a manner similar to, and dimerizes with, estrogen receptor alpha J Biol Chem 2722583225838 [DOI] [PubMed] [Google Scholar]
- RoutledgeEJWhiteRParkerMRSumpterJP2000Differential effects of xenoestrogens on coactivator recruitment by estrogen receptor (ER) α and ERβ J Biol Chem 2758598685993 [DOI] [PubMed] [Google Scholar]
- SajiSJensenEVNilssonSRylanderTWarnerMGustafssonJ-A2000Estrogen receptors α and β in the rodent mammary gland Proc Natl Acad Sci USA 97337342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SaundersPTKMillarMRMacphersonSHarkissDAndersonRAOrrBGroomeNPWilliamsKScobieGFraserHM2000Differential expression of estrogen receptor-alpha and -beta and androgen receptor in the ovaries in marmoset and human Biol Reprod 6310981105 [DOI] [PubMed] [Google Scholar]
- ScobieGSMacphersonSMillarMRGroomeNPRomanaPGSaundersPTK2001Human estrogen receptors: differential expression of ERalpha and beta and the identification of ERbeta variants Steroids(in press)
- SpeirsVParkesATKerinMJWaltonDSCarletonPJFoxJNAtkinSL1999Coexpression of estrogen receptor a and β: poor prognostic factors in human breast cancer Cancer Res 59525528 [PubMed] [Google Scholar]
- SunJMeyersMJFinkBERajendranRKatzenellenbogenJAKatzenellenbogenBS1999Novel ligands that function as selective estrogens or antiestrogens for estrogen receptor-a or estrogen receptor-β Endocrinology 140800804 [DOI] [PubMed] [Google Scholar]
- VladusicEAHornbyAEGuerra-VladusicFKLakinsJLupuR2000Expression and regulation of estrogen receptor beta in human breast tumors and cell lines Oncol Rep 7157167 [DOI] [PubMed] [Google Scholar]
- VladusicEAHornbyAEGuerra-VladusicFKLupuR1998Expression of estrogen receptor beta messenger RNA variant in breast cancer Cancer Res 58210214 [PubMed] [Google Scholar]
- WatanabeTInoueSOgawaSIshiiYHiroiHIkedaKOrimoAMuramatsuM1997Agonistic effect of tamoxifen is dependent upon cell type, ERE-promoter context, and estrogen receptor subtype: functional difference between estrogen receptors α and β Biochem Biophys Res Commun 236140145 [DOI] [PubMed] [Google Scholar]


