Abstract
Abnormal fragile histidine triad transcripts were found in 20–30% of CIN2/3 lesions and 11% of normal cervical biopsies by RT–PCR. Bi-allelic loss of the fragile histidine triad gene and the loss of fragile histidine triad protein expression detectable by immunochemical staining with a polyclonal fragile histidine triad specific antibody was rare. The genomic changes showed no association with the presence of human papillomavirus types which carry high risk for cervical cancer (high risk human papillomavirus) as assessed by a type-specific multiplex PCR. The presence of abnormal fragile histidine triad transcripts in a subset of CIN2/3 lesions with no high risk human papillomavirus suggests that this could be an independent risk factor associated with an alternative carcinogenic pathway.
British Journal of Cancer (2002) 86, 376–381. DOI: 10.1038/sj/bjc/6600077 www.bjcancer.com
© 2002 The Cancer Research Campaign
Keywords: CIN, HPV, FHIT
Cervical cancer is the second most common malignancy in women worldwide. It is generally accepted that the development of cervical cancer from precancers (CIN2/3) involves multistep molecular changes and is therefore a preventable disease if precancers are detected and treated early.
The major risk factor for CIN2/3 and cancer is infection with high risk human papillomavirus (HR HPV) types which include HPV16, HPV18, HPV31, HPV33, HPV35, HPV45, HPV51, HPV52, HPV56, HPV58, HPV59 and HPV68. One or more of these virus types have been reported to be present in 90–95% of CIN2/3 lesions and in nearly all cancers (Lorincz et al, 1992; Bosch et al, 1995; Walboomers et al, 1996). However, infection with HR HPV does not invariably lead to the development of high grade cervical lesions and there is a clear requirement for other co-factors, presumably to advance the oncogenic process initiated by HR HPV (zur Hausen, 1999). The effects of co-factors are difficult to measure objectively because of the dominant effects of HR HPV infection but tobacco smoking has consistently been associated with cervical cancer (Szarewski et al, 1996) and has been found recently to induce genomic instability in head and neck cancers (Schantz et al, 2000). Other potential co-factors include oral contraceptive use and other sexually transmitted diseases.
Genomic instability has been found in cervical neoplasia and commonly involves the short arm of chromosome 3. Results reported so far are based on small numbers of samples and show no clear correlation between genomic changes and lesion grades (Butler et al, 2000; Yoshino et al, 2000). Allelic loss and microsatellite instability were found to occur in lesions with or without malignant potential in similar proportion (Wistuba et al, 1997; Chu et al, 1998; Yoshino et al, 1998; Butler et al, 2000; Lin et al, 2000). Inconsistent results were also obtained in the analysis of abnormal transcripts encoded by the fragile histidine triad (FHIT) gene at chromosome 3p14.2. These are also indicative of genomic instability (Sozzi et al, 1996) but the incidence reported varies from 30 to 80% in cervical cancer and 0 to 40% in normal tissue (Greenspan et al, 1997; Chu et al, 1998; Muller et al, 1998; Su et al, 1998; Yoshino et al, 1998; Nakagawa et al, 1999; Segawa et al, 1999). In only one study, which involved FHIT protein staining in 110 paraffin sections, was a reduction in FHIT gene expression observed to correlate with increasing lesion grade (Birrer et al, 1999). How genetic alterations in chromosome 3p interact with HR HPV is also not clear (Muller et al, 1998; Butler et al, 2000). Integration of HPV16 sequences at the FHIT locus has been described but this does not appear to be site specific (Wagatsuma et al, 1990; Wilke et al, 1995). HPV16 sequences were not present in abnormal FHIT transcripts (Muller et al, 1998) and the presence of HR HPV E6/E7 transcripts and abnormal FHIT transcripts were negatively correlated in cervical cancer (Segawa et al, 1999).
In this investigation, abnormal FHIT gene expression based on the detection of abnormal FHIT transcripts or a reduction of immunohistochemical staining for FHIT protein was studied in 281 and 114 biopsies respectively, of normal cervix and CIN lesions. We assessed whether these indicators of genomic instability at chromosome location 3p14.2 were present at an early stage, when the oncogenic potential of HR HPV is not yet fully expressed. The influence of HR HPV on genomic instability was also investigated. The potential implications of abnormal FHIT expression in the pathogenesis of CIN lesions are discussed.
MATERIALS AND METHODS
Specimens
All specimens were collected in accordance with the guidelines approved by the Whittington Hospital Trust UK ethics committee.
HPV and abnormal FHIT transcript status
Nucleic acid preparations from 281 biopsies of normal cervix and CIN lesions from a total of 91 women participating in a previous study of HPV E6 and E7 transcripts (Ho et al, 1994) were used. Five cancer biopsies from the same study acted as high grade lesion controls. Routine histology was reviewed by a consultant histopathologist. Nucleic acid extraction was carried out using guanidinium isothiocyanate at 60°C for 10 min to selectively lyse the epithelium and remove it from the underlying, more resistant, stromal tissue. After further extraction with hot phenol/chloroform and digestion with proteinase K (PK, 200 μg ml−1 (Sambrook et al, 1989)) total nucleic acids were precipitated and stored in 1 ml of ethanol containing 0.3 M sodium acetate (pH 5.2) at −70°C. HPV assays were carried out using 50 μl aliquots which were centrifuged at 14 000 g and 4°C for 15 min. The pellet was re-dissolved in 20 μl of TE (10 mM Tris, 1 mM EDTA, pH 8.0). Thirteen HR HPV types were assayed by a multiplex type specific PCR (mts–PCR) using primers that are completely homologous to the corresponding HPV genome sequence (Terry et al, 2001). The 3′ end of each primer was not homologous to any other region of any of the 13 genomes. The primers amplified DNA sequences in the E6 or E6/E7 genes, the region most likely to be retained after viral DNA integration.
FHIT transcripts were detected by semi-nested RT–PCR. Nucleic acids from a 500 μl aliquot in ethanol was pelleted, redissolved in TE, digested with RQ1 RNase free DNase I (Amersham Pharmacia Biotech) at 250 μg ml−1. The RNA was then phenol-chloroform extracted and re-precipitated with ethanol. FHIT cDNA was synthesised using a Reverse Transcription System kit (Promega) and subsequently amplified for 30 cycles (94°C for 30 s, 60°C for 1 min, 68°C for 2 min) with primers FH203S (forward) and FH1038 (reverse). The efficiency of reverse transcription was monitored using aldolase PCR primers (Chelly et al, 1988). The PCR product was diluted 1/10 and amplified in a semi-nested PCR for a further 30 cycles (cycling conditions as above) using primers FH203S (forward) and FH904S (reverse). The FHIT primers were as described by Fong et al (1997) but with the addition of 5′-leader sequences (nucleotides in lower case) to allow direct sequencing of the PCR products using available Cy5 labelled sequencing primers for rubella virus (in FH203S) or M13 (in FH904S).
Nucleic acid sequencing was carried out using an ALF-Express automatic sequencer.
HPV and FHIT protein status
Paraffin blocks from corresponding cases used for the FHIT transcript study were not available due to re-organizations in the National Health Service. For this analysis 114 biopsies of normal cervix and CIN lesions from a total of 42 women participating in two previous studies of cervical cancer risk factors (Mansell et al, 1994; Cuzick et al, 1996) were used. Nine cancer lesions (Cuzick et al, 1996) were included as high grade lesion controls. Routine histology was reviewed by a consultant histopathologist. Sections (3 μm thick) were mounted on coated slides (BDH). For HPV genotyping, individual sections were de-waxed with EZ-DeWax (InnoGenex) according to the instruction of the manufacturer, scraped from the slide, washed three times with ethanol, digested with PK and tested by mts–PCR as described above. For histological and immunohistochemical staining, sections were de-waxed with xylene and stained with either haematoxylin and eosin or FHIT antibody (Zymed). FHIT antibody was used at a dilution of 1/200 after epitope retrieval in accordance with the manufacturer's instructions and its presence detected by reaction with monoclonal alkaline phosphatase conjugated anti-rabbit IgG antibody (gamma chain specific) at a dilution of 1/100 (Sigma A2556) and 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (Sigma B5655) as substrate. Nuclei were stained with 0.1% nuclear fast red in 5% aluminium sulphate. Specific cytoplasmic immunostaining for FHIT protein in cervical lesions was assessed by comparison with normal epithelium (positive control, present in most sections) and with stromal cells (negative control, present in all sections).
RESULTS
The histological grades and the HR HPV status of the lesions used in this study, subdivided into those used for FHIT transcript or protein detection, are shown in Table 1. High grade lesions were predominantly associated with HPV16, HPV18, HPV31, HPV33, HPV35 and HPV58 but 20% (29 out of144) were negative for HR HPV. Seven lesions contained more than one HR HPV type. Cancer biopsies were included as positive controls for fully invasive disease and one of these was HR HPV negative. The association of high grade lesions with HR HPV was clearly evident (Ptrend<0.001 in the transcript study and Ptrend=0.012 in the protein study).
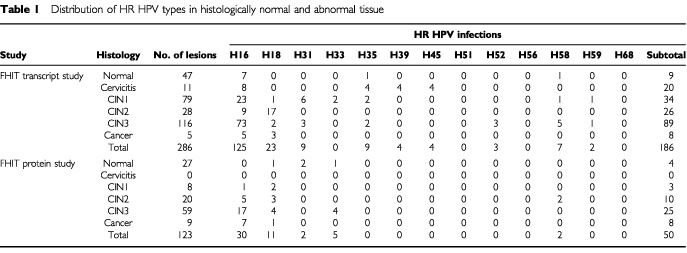
Table 1. Distribution of HR HPV types in histologically normal and abnormal tissue.
Representative semi-nested FHIT transcript PCR products are shown in Figure 1. Full length FHIT transcripts were not detectable in nine lesions (two cancers, five CIN3 and two CIN1). Abnormal splicing patterns similar to those previously reported (Sozzi et al, 1996) were found by nucleic acid sequencing to be responsible for size-shifted PCR fragments. These included (i) skipping of exons 4–8, exons 5–8, exons 5–6 with an alternative splice site within exon 7 or exons 6–9 with alternative splice sites within exons 5 and 10, (ii) replacement of exons 5–7 with 138 bases from intron 5 and (iii) an insertion of a 50 base sequence from intron 7 between exons 7 and 8 (Figure 2). After adjusting for HR HPV, abnormal FHIT transcript expression (aberrand and absent) was found to be associated with increasing CIN grades (Ptrend<0.001) (Table 2A).
Figure 1.
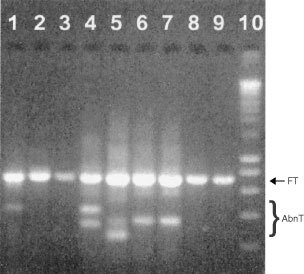
Agarose gel electrophoresis of semi-nested RT–PCR products (lanes 1–9). Size markers in lane 10 are 200, 400, 600, 800, 1000, 1500, 2000, 2500, 3000, 4000, 5000, 6000, 8000 and 10 000 bp from bottom upwards. FT=normally spliced transcript (702 bp). AbnT=abnormal transcripts.
Figure 2.
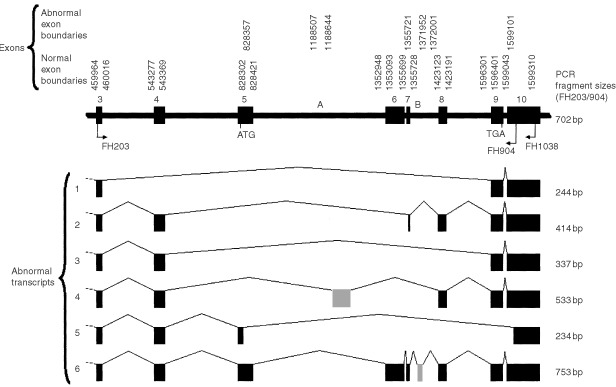
Abnormal FHIT transcripts found in this study and their generation by alternative splice site usage. Only the region amplified by the FH203S/FH1038 primers is shown. The FHIT coding sequence start and stop codons are indicated and the origins of two abnormal exons found (grey) are labelled A and B. Introns are shown on a 500 times smaller scale than exons. Numbering of the first and last bases of exons is from Genbank accession number NT_005607.3.
Table 2. Distribution of HR HPV, abnormal FHIT transcripts (A) and abnormal FHIT protein expression (B) in histologically normal and abnormal cervical epithelium.
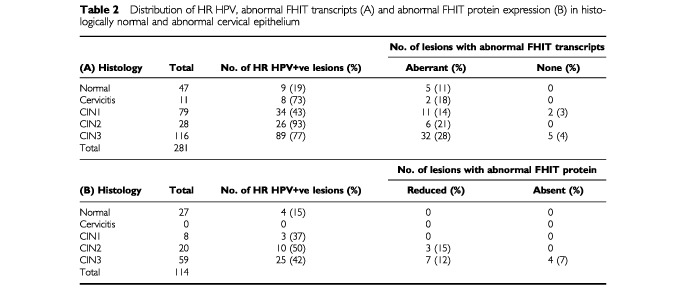
Figure 3 shows the FHIT protein staining criteria used in grading individual lesions. FHIT protein expression was readily detectable in all normal epithelium and CIN1 lesions and in most CIN2/3 lesions but was totally absent in four CIN3 lesions (Table 2B). One of these was associated with HPV18 but the remaining three contained no detectable HR HPV. After adjusting for HR HPV, abnormal FHIT protein expression (reduced or absent) was also found to be associated with increasing CIN grade (Ptrend=0.004).
Figure 3.
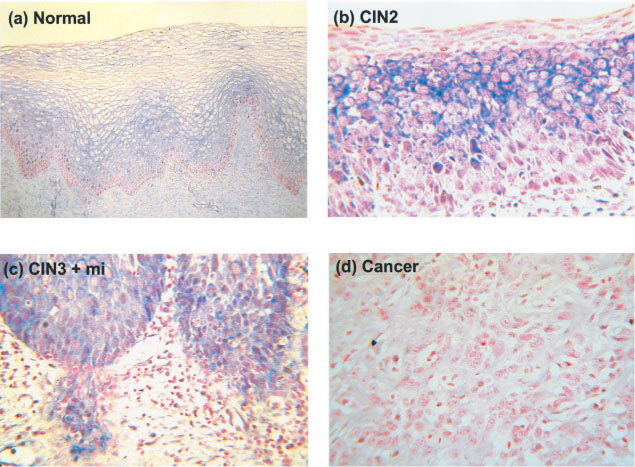
Immunohistochemical staining with FHIT antibody. (A) Normal epithelium (positive control) ×100, (B) a positive CIN2 lesion×350, (C) a positive CIN3 lesion showing microinvasion×350 and (D) a negative squamous cell carcinoma×350. Stromal cells present in all sections were used as an internal negative control.
DISCUSSION
A full length FHIT transcript was detected in all normal cervical tissue samples and in all but 3% (7 out of 223) of CIN lesions (Figure 1, Table 2A). The precise significance of its detection in almost all CIN lesions is unclear since it could have been derived from abnormal and/or adjacent normal epithelium. The presence of abnormal FHIT transcripts could be clearly identified and their nature defined by nucleic acid sequencing. They were found to be generated by alternative splice site usage and cryptic alternative splice sites were identified in exons 5, 7 and 10 and in introns 5 and 7 (Figure 2). Our results show that abnormal transcripts can be found in both histologically normal and abnormal tissues (Table 2A).
When compared against stromal cells within the same section, expression of FHIT protein in epithelial cells was found in all normal tissues and low grade CIN1 as well as in most high grade CIN2/3 lesions (Table 2B, Figure 3). This is to be expected if full length FHIT transcripts are expressed in most CIN lesions (Figure 1, Table 2A). However, abnormal FHIT protein expression was found in a proportion (17%) of exclusively CIN2 and CIN3 lesions and interestingly, three of the four CIN3 lesions which showed no FHIT protein expression also had no detectable HR HPV. Similar abnormal FHIT protein expression has previously been found in 52% of high grade lesions associated with cancer but only in 8% of those not associated with cancer (Connolly et al, 2000). This suggests that these CIN2/3 lesions could carry an increased risk of malignancy.
Recent evidence suggests that FHIT instability may play a synergistic role with HR HPV in the pathogenesis of high grade cervical lesions (Butler et al, 2000). HPV16 genomes have been found integrated in the vicinity of the FHIT coding sequence and the presence of HPV16 DNA was associated with LOH in the FHIT region (Muller et al, 1998; Butler et al, 2000; Yoshino et al, 2000). Detailed typing of HR HPV in our study did not show any associations between abnormal FHIT transcript expression and presence of specific HR HPV types or all HR HPV taken together. This is consistent with a negative correlation found between the presence of HPV E6/E7 transcripts and abnormal FHIT transcripts in cervical cancers (Segawa et al, 1999).
Our results indicate that, after adjusting for HR HPV, abnormal FHIT transcript expression (Ptrend<0.001) or abnormal FHIT protein expression (Ptrend<0.004) represents a risk factor for the development of high grade CIN lesions. This suggests that genomic instability at the FHIT locus induced by other risk factors (e.g. cigarette smoking, alcohol consumption or concurrent chronic inflammation) could account for the development of those high grade cervical lesions which are HR HPV negative and which may have a lower risk of cancer and tend to regress in the absence of persistent HR-HPV infection. For clinical or epidemiological purposes, testing for FHIT protein, rather than FHIT transcripts, would have obvious advantages since it can be carried out using low cost and simple procedures on routine paraffin sections.
Acknowledgments
We would like to thank the Frances and Augustus Newman Foundation for financial support.
References
- BirrerMJHendricksDFarleyJSundborgMJBonomeTWaltsMJGeradtsJ1999Abnormal Fhit expression in malignant and premalignant lesions of the cervix Cancer Res 5952705274 [PubMed] [Google Scholar]
- BoschFXManosMMMunozNShermanMJansenAMPetoJSchiffmanMHMorenoVKurmanRShahKV1995Prevalence of human papillomavirus in cervical cancer: a worldwide perspective JNCI 8778847707393 [Google Scholar]
- ButlerDCollinsCMabrukMWalshCBLeaderMBKayEW2000Deletion of the FHIT gene in neoplastic and invasive cervical lesions is related to high-risk HPV infection but is independent of histopathological features J Pathol 192502510 [DOI] [PubMed] [Google Scholar]
- ChellyJKaplanJCMairePGautronSKahnA1988Transcription of the dystrophin gene in human muscle and non-muscle tissues Nature 333858860 [DOI] [PubMed] [Google Scholar]
- ConnollyDCGreenspanDLWuRRenXDunnRLShahKVJonesRWBoschFXMunozNChoKR2000Loss of fhit expression in invasive cervical carcinomas and intraepithelial lesions associated with invasive disease Clin Cancer Res 635053510 [PubMed] [Google Scholar]
- ChuTYShenCYChiouYSLuJJPerngCLYuMSLiuHS1998HPV-associated cervical cancers show frequent allelic loss at 3p14 but no apparent aberration of FHIT mRNA IJC 75199204 [DOI] [PubMed] [Google Scholar]
- CuzickJSasieniPSingerA1996Risk factors for invasive cervix cancer in young women Eur J Cancer 32A836841 [DOI] [PubMed] [Google Scholar]
- FongKMBiesterveldEJVirmaniAWistubaISekidoYBaderSAAhmadianMOngSTRassoolFVZimmermanPVGiacconeGGazdarAFMinnaJD1997FHIT and FRA3B 3p14.2 allele loss are common in lung cancer and preneoplastic bronchial lesions and are associated with cancer-related FHIT cDNA splicing variations Cancer Res 5722562267 [PubMed] [Google Scholar]
- GreenspanDLConnollyDCWuRLeiRYVogelsteinJTKimYTMokJEMunozNBoschFXShahKChoKR1997Loss of FHIT expression in cervical carcinoma cell lines and primary tumors Cancer Res 5746924698 [PubMed] [Google Scholar]
- HoLTerryGMansellBButlerBSingerA1994Detection of DNA and E7 transcripts of human papillomavirus types 16, 18, 31 and 33, TGFβ and GM-CSF transcripts in cervical cancers and precancers Arch Virol 1397985 [DOI] [PubMed] [Google Scholar]
- LinWMMichalopulosEADhuranderNChengPCRobinsonWAshfaqRColemanRLMullerCY2000Allelic loss and microsatellite alternations of chromosome 3p14.2 are more frequent in recurrent cervical dysplasias Clinical Cancer Res 614101414 [PubMed] [Google Scholar]
- LorinczATReidRJensonABGreenbergMDLancasterWKurmanRJ1992Human papillomavirus infection of the cervix: relative risk associations of 15 common anogenital types Obs Gynaecol 79328337 [DOI] [PubMed] [Google Scholar]
- MansellBHoLTerryGSingerACuzickJ1994Semi-quantitative human papillomavirus DNA detection in the management of women with minor cytological abnormality B J Obs Gynaecol 101807809 [DOI] [PubMed] [Google Scholar]
- MullerCYO'BoyleJDFongKMWistubaIIBiesterveldEAhmadianMMillerDSGazdarAFMinnaJD1998Abnormalities of fragile histidine triad genomic and complementary DNAs in cervical cancer: association with human papillomavirus type JNCI 90433439 [DOI] [PubMed] [Google Scholar]
- NakagawaSYoshikawaHKimuraMKawanaKMatsumotoKOndaTKinoNYamadaMYasugiTTaketaniY1999A possible involvement of aberrant expression of the FHIT gene in the carcinogenesis of squamous cell carcinoma of the uterine cervix BJC 79589594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SambrookJFritschEFManiatisT1989Molecular Cloning. A laboratory manual2nd edition,Cold Spring Harbor Laboratory Press [Google Scholar]
- SozziGVeroneseMLNegriniMBaffaRCotticelliMGInoueHTornielliSPilottiSGregorioLDPastorinoUPierottiMAOhtaMHuebnerKCroceCM1996The FHIT gene at 3p14.2 is abnormal in lung cancer Cell 851726 [DOI] [PubMed] [Google Scholar]
- SchantzSPHuangQShahKinnerMurtyVSHsuTCYuGAndersenPEHuvosAGChagantiSK2000Mutagen sensitivity and environmental exposures as contributing causes of chromosome 3p losses in head and neck cancers Carcinogenesis 2112391246 [PubMed] [Google Scholar]
- SegawaTSasagawaTYamazakiHSakaikeJIshikawaHInoueM1999Fragile histidine triad transcription abnormalities and human papillomavirus E6-E7 mRNA expression in the development of cervical carcinoma Cancer 8520012010 [PubMed] [Google Scholar]
- SuTHWangJCTsengHHChangCPChangTAWeiHJChangJG1998Analysis of FHIT transcripts in cervical and endometrial cancers IJC 76216222 [DOI] [PubMed] [Google Scholar]
- SzarewskiAJarvisMJSasieniPAndersonMEdwardsRSteeleSJGuillebaudJCuzickJ1996Effect of smoking cessation on cervical lesion size Lancet 347941943 [DOI] [PubMed] [Google Scholar]
- TerryGHoLLondesboroughPCuzickJMielzynska-LohnasILorinczA2001Detection of high-risk HPV types by the Hybrid Capture 2 test J Med Virol 65155162 [PubMed] [Google Scholar]
- WagatsumaMHashimotoKMatsukuraT1990Analysis of integrated human papillomavirus type 16 DNA in cervical cancers: amplification of viral sequences together with cellular flanking sequences J Virol 642813821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WalboomersJMJacobsMVManosMMBoschFXKummerJAShahKVSnijdersPJPetoJMeijerCJMunozN1999Human papillomavirus is a necessary cause of invasive cervical cancer worldwide J Pathol 1891219 [DOI] [PubMed] [Google Scholar]
- WilkeCMHallBKMillerDHogeAParadeeWSmithDIGloverTW1995Identification of a spontaneous HPV16 integration site within the FRA3B region: direct evidence for the coincidence of fragile sites and viral integration sites Am J Hum Genet 57A18A18 [Google Scholar]
- WistubaIIMontellanoFDMilchgroubSVirmaniAKBehrensCChenHAhmadianMNowakJAMullerCMinnaJGazdarAF1997Deletions of chromosome 3p are frequent and early events in the pathogenesis of uterine cervical carcinoma Cancer Res 5731543158 [PubMed] [Google Scholar]
- YoshinoKEnomotoTNakamuraTNakashimaRWadaHSaitohJNodaKMurataY1998Aberrant FHIT transcripts in squamous cell carcinoma of the uterine cervix IJC 76176181 [DOI] [PubMed] [Google Scholar]
- YoshinoKEnomotoTNakamuraTSunHOzakiKNakashimaRWadaHSaitohJWatanabeYNodaKMurataY2000FHIT alterations in cancerous and non-cancerous cervical epithelium Int J Cancer 85613 [DOI] [PubMed] [Google Scholar]
- zur HausenH1999Immortalization of human cells and their malignant conversion by high risk human papillomavirus genotypes Semin Cancer Biol 9405411 [DOI] [PubMed] [Google Scholar]


