Abstract
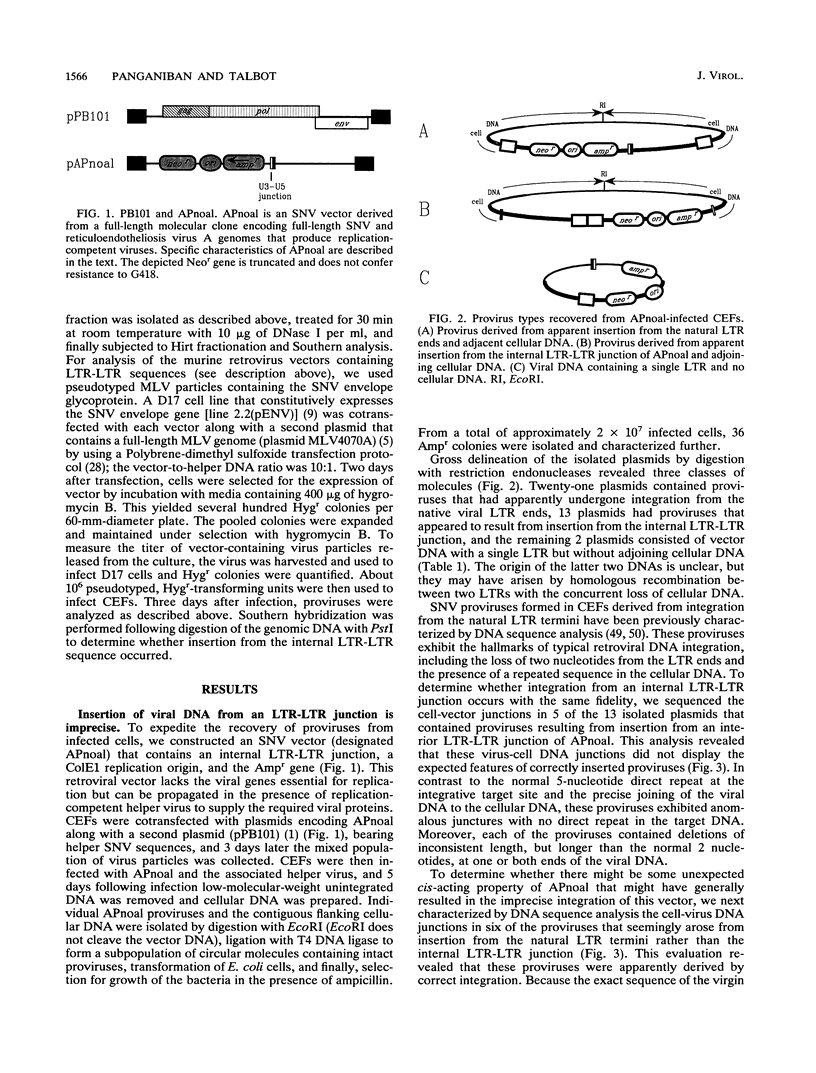
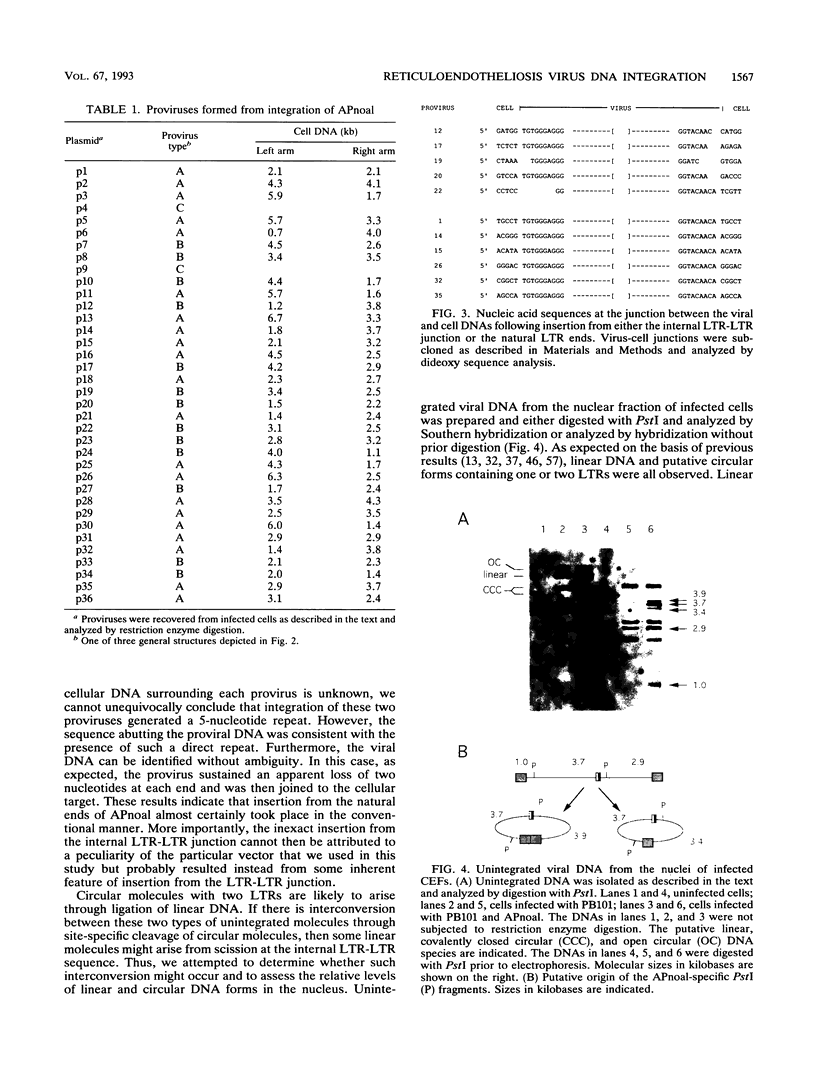
To examine the fidelity and efficiency of integration from a covalently closed long terminal repeat (LTR)-LTR sequence in vivo, we isolated individual spleen necrosis virus proviruses that arose following infection of chicken embryo fibroblasts (CEFs) and sequenced the provirus-cell DNA junctions. Some but not all CEF preparations allowed efficient insertion from the internal sequence. Moreover, in contrast to integration from the normal ends of the viral DNA, which occurs with precision with respect to the viral DNA, insertion from the internal sequence was not precise. In particular, there were short deletions of variable size from the viral DNA and these proviruses were not flanked by short direct repeats. Although this imprecise insertion can be efficient in CEFs, such integration is very inefficient in two other cell types (D17 and QT47) that support the replication of reticuloendotheliosis viruses. Thus, it is possible that there is a cell-specific factor(s) in CEFs required for efficient but imprecise insertion or, alternatively, D17 and QT47 cells contain a factor that abrogates integration from an internal LTR-LTR junction. Virus particles released from CEFs do not efficiently use the LTR-LTR junction following infection of D17 cells. Therefore, if there is a CEF-specific factor required for insertion, it does not appear to be transferred through particles.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bandyopadhyay P. K., Temin H. M. Expression from an internal AUG codon of herpes simplex thymidine kinase gene inserted in a retrovirus vector. Mol Cell Biol. 1984 Apr;4(4):743–748. doi: 10.1128/mcb.4.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacid M., Hunter E., Aaronson S. A. Avian reticuloendotheliosis viruses: evolutionary linkage with mammalian type C retroviruses. J Virol. 1979 May;30(2):508–514. doi: 10.1128/jvi.30.2.508-514.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P. O., Bowerman B., Varmus H. E., Bishop J. M. Correct integration of retroviral DNA in vitro. Cell. 1987 May 8;49(3):347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- Brown P. O., Bowerman B., Varmus H. E., Bishop J. M. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S. K., Oliff A. I., Linemeyer D. L., Lander M. R., Lowy D. R. Genomes of murine leukemia viruses isolated from wild mice. J Virol. 1981 Sep;39(3):777–791. doi: 10.1128/jvi.39.3.777-791.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobrinik D., Katz R., Terry R., Skalka A. M., Leis J. Avian sarcoma and leukosis virus pol-endonuclease recognition of the tandem long terminal repeat junction: minimum site required for cleavage is also required for viral growth. J Virol. 1987 Jun;61(6):1999–2008. doi: 10.1128/jvi.61.6.1999-2008.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colicelli J., Goff S. P. Mutants and pseudorevertants of Moloney murine leukemia virus with alterations at the integration site. Cell. 1985 Sep;42(2):573–580. doi: 10.1016/0092-8674(85)90114-x. [DOI] [PubMed] [Google Scholar]
- Colicelli J., Goff S. P. Sequence and spacing requirements of a retrovirus integration site. J Mol Biol. 1988 Jan 5;199(1):47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- Delwart E. L., Panganiban A. T. Role of reticuloendotheliosis virus envelope glycoprotein in superinfection interference. J Virol. 1989 Jan;63(1):273–280. doi: 10.1128/jvi.63.1.273-280.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar R., McClements W. L., Enquist L. W., Vande Woude G. F. Nucleotide sequences of integrated Moloney sarcoma provirus long terminal repeats and their host and viral junctions. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3937–3941. doi: 10.1073/pnas.77.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower L. A., Varmus H. E. A mutant murine leukemia virus with a single missense codon in pol is defective in a function affecting integration. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6461–6465. doi: 10.1073/pnas.81.20.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyk G., Longiaru M., Cobrinik D., Kowal R., deHaseth P., Skalka A. M., Leis J. Circles with two tandem long terminal repeats are specifically cleaved by pol gene-associated endonuclease from avian sarcoma and leukosis viruses: nucleotide sequences required for site-specific cleavage. J Virol. 1985 Nov;56(2):589–599. doi: 10.1128/jvi.56.2.589-599.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch E., Temin H. M. Formation and structure of infectious DNA of spleen necrosis virus. J Virol. 1977 Jan;21(1):119–130. doi: 10.1128/jvi.21.1.119-130.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988 Aug 12;54(4):497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- Golomb M., Grandgenett D. P. Endonuclease activity of purified RNA-directed DNA polymerase from avian myeloblastosis virus. J Biol Chem. 1979 Mar 10;254(5):1606–1613. [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Grandgenett D. P., Golomb M., Vora A. C. Activation of an Mg2+-dependent DNA endonuclease of avian myeloblastosis virus alpha beta DNA polymerase by in vitro proteolytic cleavage. J Virol. 1980 Jan;33(1):264–271. doi: 10.1128/jvi.33.1.264-271.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandgenett D. P., Vora A. C., Schiff R. D. A 32,000-dalton nucleic acid-binding protein from avian retravirus cores possesses DNA endonuclease activity. Virology. 1978 Aug;89(1):119–132. doi: 10.1016/0042-6822(78)90046-6. [DOI] [PubMed] [Google Scholar]
- Grandgenett D. P., Vora A. C., Swanstrom R., Olsen J. C. Nuclease mechanism of the avian retrovirus pp32 endonuclease. J Virol. 1986 Jun;58(3):970–974. doi: 10.1128/jvi.58.3.970-974.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntaka R. V., Richards O. C., Shank P. R., Kung H. J., Davidson N. Covalently closed circular DNA of avian sarcoma virus: purification from nuclei of infected quail tumor cells and measurement by electron microscopy and gel electrophoresis. J Mol Biol. 1976 Sep 15;106(2):337–357. doi: 10.1016/0022-2836(76)90090-5. [DOI] [PubMed] [Google Scholar]
- Herr W. Nucleotide sequence of AKV murine leukemia virus. J Virol. 1984 Feb;49(2):471–478. doi: 10.1128/jvi.49.2.471-478.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hishinuma F., DeBona P. J., Astrin S., Skalka A. M. Nucleotide sequence of acceptor site and termini of integrated avian endogenous provirus ev1: integration creates a 6 bp repeat of host DNA. Cell. 1981 Jan;23(1):155–164. doi: 10.1016/0092-8674(81)90280-4. [DOI] [PubMed] [Google Scholar]
- Hughes S. H., Mutschler A., Bishop J. M., Varmus H. E. A Rous sarcoma virus provirus is flanked by short direct repeats of a cellular DNA sequence present in only one copy prior to integration. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4299–4303. doi: 10.1073/pnas.78.7.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter E., Bhown A. S., Bennett J. C. Amino-terminal amino acid sequence of the major structural polypeptides of avian retroviruses: sequence homology between reticuloendotheliosis virus p30 and p30s of mammalian retroviruses. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2708–2712. doi: 10.1073/pnas.75.6.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju G., Skalka A. M. Nucleotide sequence analysis of the long terminal repeat (LTR) of avian retroviruses: structural similarities with transposable elements. Cell. 1980 Nov;22(2 Pt 2):379–386. doi: 10.1016/0092-8674(80)90348-7. [DOI] [PubMed] [Google Scholar]
- Katzman M., Katz R. A., Skalka A. M., Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989 Dec;63(12):5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai S., Nishizawa M. New procedure for DNA transfection with polycation and dimethyl sulfoxide. Mol Cell Biol. 1984 Jun;4(6):1172–1174. doi: 10.1128/mcb.4.6.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFemina R. L., Callahan P. L., Cordingley M. G. Substrate specificity of recombinant human immunodeficiency virus integrase protein. J Virol. 1991 Oct;65(10):5624–5630. doi: 10.1128/jvi.65.10.5624-5630.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leis J., Duyk G., Johnson S., Longiaru M., Skalka A. Mechanism of action of the endonuclease associated with the alpha beta and beta beta forms of avian RNA tumor virus reverse transcriptase. J Virol. 1983 Feb;45(2):727–739. doi: 10.1128/jvi.45.2.727-739.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobel L. I., Murphy J. E., Goff S. P. The palindromic LTR-LTR junction of Moloney murine leukemia virus is not an efficient substrate for proviral integration. J Virol. 1989 Jun;63(6):2629–2637. doi: 10.1128/jvi.63.6.2629-2637.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequences at host-proviral junctions for mouse mammary tumour virus. Nature. 1981 Jan 22;289(5795):253–258. doi: 10.1038/289253a0. [DOI] [PubMed] [Google Scholar]
- Misra T. K., Grandgenett D. P., Parsons J. T. Avian retrovirus pp32 DNA-binding protein. I. Recognition of specific sequences on retrovirus DNA terminal repeats. J Virol. 1982 Oct;44(1):330–343. doi: 10.1128/jvi.44.1.330-343.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panet A., Baltimore D. Characterization of endonuclease activities in Moloney murine leukemia virus and its replication-defective mutants. J Virol. 1987 May;61(5):1756–1760. doi: 10.1128/jvi.61.5.1756-1760.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban A. T., Fiore D. Ordered interstrand and intrastrand DNA transfer during reverse transcription. Science. 1988 Aug 26;241(4869):1064–1069. doi: 10.1126/science.2457948. [DOI] [PubMed] [Google Scholar]
- Panganiban A. T., Temin H. M. Circles with two tandem LTRs are precursors to integrated retrovirus DNA. Cell. 1984 Mar;36(3):673–679. doi: 10.1016/0092-8674(84)90347-7. [DOI] [PubMed] [Google Scholar]
- Panganiban A. T., Temin H. M. The retrovirus pol gene encodes a product required for DNA integration: identification of a retrovirus int locus. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7885–7889. doi: 10.1073/pnas.81.24.7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban A. T., Temin H. M. The terminal nucleotides of retrovirus DNA are required for integration but not virus production. Nature. 1983 Nov 10;306(5939):155–160. doi: 10.1038/306155a0. [DOI] [PubMed] [Google Scholar]
- Pauza C. D. Two bases are deleted from the termini of HIV-1 linear DNA during integrative recombination. Virology. 1990 Dec;179(2):886–889. doi: 10.1016/0042-6822(90)90161-j. [DOI] [PubMed] [Google Scholar]
- Quinn T. P., Grandgenett D. P. Genetic evidence that the avian retrovirus DNA endonuclease domain of pol is necessary for viral integration. J Virol. 1988 Jul;62(7):2307–2312. doi: 10.1128/jvi.62.7.2307-2312.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M. J., Schwartzberg P. L., Goff S. P. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989 Jul 14;58(1):47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- Schwartzberg P., Colicelli J., Goff S. P. Construction and analysis of deletion mutations in the pol gene of Moloney murine leukemia virus: a new viral function required for productive infection. Cell. 1984 Jul;37(3):1043–1052. doi: 10.1016/0092-8674(84)90439-2. [DOI] [PubMed] [Google Scholar]
- Scott M. L., McKereghan K., Kaplan H. S., Fry K. E. Molecular cloning and partial characterization of unintegrated linear DNA from gibbon ape leukemia virus. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4213–4217. doi: 10.1073/pnas.78.7.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank P. R., Hughes S. H., Kung H. J., Majors J. E., Quintrell N., Guntaka R. V., Bishop J. M., Varmus H. E. Mapping unintegrated avian sarcoma virus DNA: termini of linear DNA bear 300 nucleotides present once or twice in two species of circular DNA. Cell. 1978 Dec;15(4):1383–1395. doi: 10.1016/0092-8674(78)90063-6. [DOI] [PubMed] [Google Scholar]
- Shank P. R., Varmus H. E. Virus-specific DNA in the cytoplasm of avian sarcoma virus-infected cells is a precursor to covalently closed circular viral DNA in the nucleus. J Virol. 1978 Jan;25(1):104–104. doi: 10.1128/jvi.25.1.104-104.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman P. A., Dickson M. L., Fyfe J. A. Human immunodeficiency virus type 1 integration protein: DNA sequence requirements for cleaving and joining reactions. J Virol. 1992 Jun;66(6):3593–3601. doi: 10.1128/jvi.66.6.3593-3601.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih C. C., Stoye J. P., Coffin J. M. Highly preferred targets for retrovirus integration. Cell. 1988 May 20;53(4):531–537. doi: 10.1016/0092-8674(88)90569-7. [DOI] [PubMed] [Google Scholar]
- Shimotohno K., Mizutani S., Temin H. M. Sequence of retrovirus provirus resembles that of bacterial transposable elements. Nature. 1980 Jun 19;285(5766):550–554. doi: 10.1038/285550a0. [DOI] [PubMed] [Google Scholar]
- Shimotohno K., Temin H. M. No apparent nucleotide sequence specificity in cellular DNA juxtaposed to retrovirus proviruses. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7357–7361. doi: 10.1073/pnas.77.12.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G., Shinnick T. M., Verma I. M., Lerner R. A. Nucleotide sequence of Moloney leukemia virus: 3' end reveals details of replications, analogy to bacterial transposons, and an unexpected gene. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3302–3306. doi: 10.1073/pnas.77.6.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beveren C., Goddard J. G., Berns A., Verma I. M. Structure of Moloney murine leukemia viral DNA: nucleotide sequence of the 5' long terminal repeat and adjacent cellular sequences. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3307–3311. doi: 10.1073/pnas.77.6.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink C., van Gent D. C., Elgersma Y., Plasterk R. H. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J Virol. 1991 Sep;65(9):4636–4644. doi: 10.1128/jvi.65.9.4636-4644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Temin H. M. Construction of a helper cell line for avian reticuloendotheliosis virus cloning vectors. Mol Cell Biol. 1983 Dec;3(12):2241–2249. doi: 10.1128/mcb.3.12.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver T. A., Talbot K. J., Panganiban A. T. Spleen necrosis virus gag polyprotein is necessary for particle assembly and release but not for proteolytic processing. J Virol. 1990 Jun;64(6):2642–2652. doi: 10.1128/jvi.64.6.2642-2652.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yoshimura F. K., Weinberg R. A. Restriction endonuclease cleavage of linear and closed circular murine leukemia viral DNAs: discovery of a smaller circular form. Cell. 1979 Feb;16(2):323–332. doi: 10.1016/0092-8674(79)90009-6. [DOI] [PubMed] [Google Scholar]