Abstract
Mutations in the PAX6 gene have been implicated in aniridia, a congenital malformation of the eye with severe hypoplasia of the iris. However, not all aniridia cases can be explained by mutations in the PAX6 gene. The purpose of this study was to enhance the molecular diagnosis of aniridia using multiplex ligation-dependent probe amplification (MLPA). Total genomic DNA was isolated from peripheral blood of 70 unrelated probands affected with aniridia. Polymerase chain reaction (PCR) was performed followed by automated bidirectional sequencing. Additionally, MLPA was performed. We identified 24 different point mutations in the PAX6 gene in 34 patients after sequencing. In eight additional patients, we identified a deletion of one or more exons of the PAX6 gene or in the 3′ regulatory region of the PAX6 gene using MLPA. This work demonstrates the necessity to screen for larger deletions in the region of the PAX6 gene in addition to the sequencing of exons in the PAX6 gene. The mutation detection rate will increase from 49% to 60%. This shows that MLPA substantially enhances the molecular diagnosis of aniridia.
Introduction
Aniridia (AN2; OMIM 106210) is a genetic disorder, which is characterized by iris hypoplasia as well as several other ocular features such as cataract, glaucoma, corneal opacity, and nystagmus. The aniridia gene was mapped to chromosome 11p13, and no other locus besides chromosome 11p13 was convincingly implicated in aniridia [1]. Heterozygous mutations in the paired box gene 6 (PAX6; OMIM 607108), located on human chromosome 11p13, were previously implicated in familial and sporadic aniridia. The functional relationship between the gene and the disease was further enhanced by the finding that PAX6 mutations cause the small eye (sey) phenotype in mice [2]. PAX6 is clearly the major if not only gene responsible for the disease. A frequently updated summary of PAX6 mutations, which are associated with aniridia and related disorders, is documented at the PAX6 allelic variation database [3]. PAX6 is highly conserved, and missense mutations can also cause dysfunction of the protein [4]. In isolated aniridia cases, published PAX6 alterations comprise mainly point mutations [1]. The majority of these mutations result in premature termination of the PAX6 protein. A proportion of aniridia cases are associated with the WAGR (Wilms tumor, aniridia, genitourinary abnormalities, mental retardation) syndrome. WAGR is a contiguous gene syndrome arising from deletions of chromosome 11p13, which encompass at least both the PAX6 and WT1 genes. In contrast, only a few isolated aniridia patients with a gross deletion or a cytogenetic translocation in the PAX6 region have been reported in the literature [5-10]. Gross deletions may be underreported due to the fact that these are not always detected by FISH (fluorescent in situ hybridization) and that these are not detected by the common mutation detection methods such as SSCP, DHPLC, or sequencing.
We performed a mutation analysis for PAX6 in 70 unrelated patients with aniridia. By scanning the 14 exons of PAX6, 24 different point mutations were identified in 34 patients. In addition, we detected gross deletions in another eight patients using multiplex ligation-dependent probe amplification (MLPA).
Methods
Patients
The patient group consisted of 70 patients who were referred to our laboratory with the clinical diagnosis of isolated aniridia. The study was performed according to a protocol approved by the local ethics committee.
DNA analysis
Genomic DNA was isolated from peripheral blood. All 14 exons of the PAX6 gene were amplified and sequenced bidirectionally on an ABI PRISM 377 sequencer (Applied Biosystems, Foster City, CA) using standard technology and the primers described in Table 1. The mutation nomenclature is according to the recommendations of the Human Genome Variation Society [11]. cDNA nucleotide numbering (reference sequence NM_001604.3 from GenBank) starts with +1 at the A of the ATG translation initiation codon. Probes for MLPA analysis (SALSA MLPA Kits P097 and P219) were purchased from MRC Holland (Amsterdam, the Netherlands). MLPA was performed according to the manufacturer’s instructions. In summary, 100 ng DNA was denatured and hybridized overnight at 60 °C with SALSA probe mix. After treating the samples with ligase 65 for 15 min at 54 °C, polymerase chain reaction (PCR) amplification was performed with the specific SALSA FAM PCR primers. Electrophoresis of PCR products was performed using an ABI PRISM 310 (Applied Biosystems). Data analysis was performed by exporting the peak areas to a Microsoft Excel file. Sample-related and peak-related differences in PCR and electrophoresis efficiency were corrected by first calculating the peak area relative to the sum of peak areas per sample and subsequently expressing each normalized peak area relative to the mean of that peak across samples. To detect deviating peaks, each normalized peak was divided by the mean of that peak in the control samples. Peak areas outside the range 0.7–1.3 times the control peak area were considered abnormal with those below 0.7 representing deletions and those above 1.3 representing duplications. All normal samples run in one assay showed standard deviations of less than 10% of the mean. Several control samples were included in each MLPA experiment. Each result was confirmed by two independent tests.
Table 1. List of primers used to perform amplification and sequencing of the 14 PAX6 exons.
|
Exon |
Forward primer |
Reverse primer |
| 1 |
AGGGAACCGTGGCTCGGC |
GGGTGAGGGAAGTGGCTGC |
| 2 |
TTATCTCTCACTCTCCAGCC |
GGAGACCTGTCTGAATATTGC |
| 3 |
TCAGAGAGCCCATCGACGTAT |
CTGTTTGTGGGTTTTGAGCC |
| 4 |
AGTTCAGGCCTACCTGATGC |
GTCGCGAGTCCCTGTGTC |
| 5 |
CTCCCTCATCTTCCTCTTCC |
GGGGTCCATAATTAGCATCG |
| 6–7 |
GGGCTACAAATGTAATTTTAAGAAA |
AGAGAGGGTGGGAGGAGGTA |
| 8 |
GAGCTGAGATGGGTGACTG |
GAGAGTAGGGGACAGGCAAA |
| 9 |
AGACTACACCAGGCCCCTTT |
TGAAGATGTGGCATTTACTTTGA |
| 10 |
GGAACCAGTTTGATGCACAG |
ACTCTGTACAAGCACCTCTGTCTC |
| 11 |
GGGCTCGACGTAGACACAGT |
GGAAACTGAGGGCAAGAGAA |
| 12 |
CGGGCTCTGACTCTCACTCT |
GCCACTCCTCACTTCTCTGG |
| 13 |
GCTGTGGCTGTGTGATGTGT |
AGGAGATTCTGTTTGGGTA |
| 14 | TCCATGTCTGTTTCTCAAAGG | TCAACTGTTGTGTCCCCATAG |
Sequences of primers used to perform amplification and sequencing of the 14 PAX6 exons.
Long range polymerase chain reaction
For confirmation of the PAX6 exon 9 deletion, PCR was performed with the following primer combination: 5′-GGA CTT CGG TGC CAG GGC AAC CTA-3′ (forward) located in exon 8 and 5′-TGG CTG CTA GTC TTT CTC GGG CAA ACA-3′ (reverse) located in exon 10.
Fluorescent in situ hybridazation
FISH analysis was performed according a standard protocol using the FISH probes, FAT5 (PAX6), p60 (D11S324), and B2.1 (WT1) as described by Crolla et al. [7].
Results
We performed a mutation analysis for PAX6 in 70 unrelated patients with isolated aniridia. By scanning the 14 exons of the PAX6 gene, 24 different point mutations were identified in 34 patients (Table 2). We identified the following types of mutations: a frameshift mutation in nine patients; a splice-site mutation in seven patients; a nonsense mutation in 11 patients; in three patients a no-stop change; and in four patients a missense mutation.
Table 2. Point mutations in the PAX6 gene.
|
Patient |
DNA change |
Consequence |
Reference |
| 1 and 53 |
c.-52+1G>A |
Splice defect |
[15] |
| 6 and 74 |
c.301delG |
p.Glu101LysfsX22 |
Database* |
| 9 |
c.489T>G |
p.Tyr163X |
New |
| 11, 12 |
c.399+5G>A |
Splice defect |
New |
| 16 |
c.181_189delinsCA |
p.Tyr61GlnfsX15 |
New |
| 19 |
c.34G>C |
p.Gly12Arg |
New |
| 20 |
c.406C>T |
p.Gln136X |
[16] |
| 21 |
c.-52+5G>C |
Splice defect |
New |
| 22, 48, 75 |
c.1268A>T |
p.X423Leu |
[17] |
| 23, 29, 79 |
c.718C>T |
p.Arg240X |
[18] |
| 26 |
c.640A>G |
p.Arg214Gly |
New |
| 30, 49 |
c.114_121del8 |
p.Pro39HisfsX13 |
New |
| 35 |
c.844_845delCC |
p.Pro282X |
New |
| 36 |
c.112delC |
p.Arg38GlyfsX15 |
New |
| 37 |
c.117insGGCC |
p.Cys40GlyfsX15 |
New |
| 41, 51, 68 |
c.949C>T |
p.Arg317X |
[16] |
| 42 |
c.299G>A |
p.Trp100X |
[19] |
| 44 |
c.77G>C |
p.Arg26Pro |
New |
| 58 |
c.365C>A |
p.Ser122X |
New |
| 61 |
c.400–1G>C |
Splice defect |
New |
| 63 |
c.143T>A |
p.Val48Glu |
New |
| 64 |
c.1074+1G>A |
Splice defect |
[15] |
| 66 |
c.265_266insC |
p.Gln89ProfsX2 |
New |
| 76 | c.781C>T | p.Arg261X | [15] |
The asterisk indicates the Human PAX6 Allelic Variant Database.
It is known that PAX6 missense mutations may give rise to variant phenotypes [4]. Of the four patients with missense mutations in our study, three have phenotypes within the classical aniridia spectrum. The fourth case, patient 44 with the p.Arg26Pro mutation, is a familial aniridia case with an affected mother and daughter. The daughter has ectopic pupils and marked atrophy of the iris stroma as well as congenital nystagmus and cataract. The mother has nystagmus, microcornea, coloboma of the iris with iridocorneal adhesions, some stromal atrophy, and peripheral coloboma of the choroid of one eye.
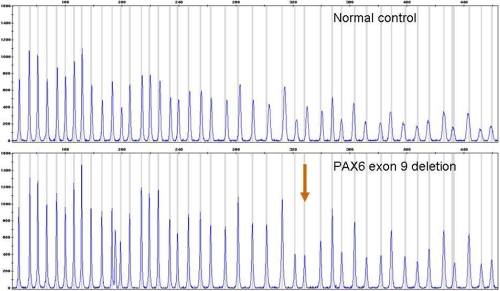
We examined the remaining 36 patients in which no mutation was yet identified by means of the multiplex ligation-dependent probe amplification (MLPA) method. An example MLPA result is shown in Figure 1. Using the P219 MLPA kit, a deletion was detected in eight isolated aniridia patients (Table 3). In six cases, a deletion of several contiguous exons was found; in one case, a deletion of a single exon was found; and in another case, a deletion outside the coding region of PAX6 was identified. These first six deletions were confirmed with the MLPA P097 kit. This MLPA kit is an early version of the PAX6 kit with probes for a limited number of PAX6 exons. The deletion of exon 9 was confirmed by long range PCR. The last deletion is a deletion of three contiguous probes of ELP4 and DCDC1 (Figure 2).
Figure 1.
Detection of PAX6 exon 9 deletion by MLPA. Electropherograms are from a normal control and from the patient with an exon 9 deletion. The deletion is apparent by a ~50% reduction in peak area of the PAX6 exon 9 specific probe (red arrow).
Table 3. Gross deletions identified by MLPA kit P219 in eight isolated aniridia patients and in two WAGR patients as well as the results of the FISH analysis on these patients.
|
Patient |
Deleted PAX6 exons |
Other deleted probes* |
Confirmation |
Deleted FISH probes |
| 4 |
exon 8–14 |
- |
MLPA kit P097 |
- |
| 5 |
exon 8–14 |
- |
MLPA kit P097 |
- |
| 17 |
exon 9 |
- |
Long range PCR |
- |
| 45 |
exon 9–14 |
- |
MLPA kit P097 |
- |
| 50 |
exon 1–14 |
DCDC1 exon 4 - LOC646008 exon 4 |
MLPA kit P097 |
ND** |
| 57 |
exon 1–14 |
ELP4 - DKFZ exon 3 |
MLPA kit P097 |
ND** |
| 69 |
exon 1–14 |
DCDC1 exon 4- DKFZ exon 1 |
MLPA kit P097 |
- |
| 71 |
- |
DCDC1 exon 4 - ELP4 |
contiguous probes |
- |
| WAGR1 |
exon 1–14 |
DCDC1 exon 4 – HIPK3 |
MLPA kit P097 |
FAT5, p60, B2.1 |
| WAGR2 | exon 1–14 | BDNF exon 2 - CD44 | MLPA kit P097 | FAT5, p60, B2.1 |
The asterisk indicates that the first and the last deleted probes are mentioned. The double asterisk indicates that no lymphocytes were available for FISH analysis.
Figure 2.
Example of normalized MLPA results. A and B: The height of the columns represents the dosage of the respective segments in the genomic DNA with two alleles (value of about one corresponds to two alleles). The light blue columns represent the 11p13 specific probes from telomere to centromere. The brown columns represent the deleted probes. The allele dosage of the deleted probes was found in the range of about 0.5, which corresponds to one allele. The dark blue columns represent the control probes. C: Long range PCR for PAX6 exons 8–10 confirmed the deletion. The deleted allele is more strongly amplified than the normal allele due to preferential amplification of smaller fragments. Lane 1 is a 100 bp ladder size marker; lane 2 is the DNA of the patient; and lane 3 is a control DNA.
For validation of the MLPA method, we also analyzed the DNA of two WAGR patients with deletions that were previously detected by FISH (Table 3). One WAGR patient showed a deletion of all 11p13 probes while the other WAGR patient had a deletion of DCDC1 exon 1 up to HIPK3 including PAX6 and WT1.
Discussion
In this study, we showed that by using MLPA, it is possible to extend the number of confirmed aniridia cases at the molecular level. In our series of 70 isolated aniridia cases, we detected PAX6 pathogenic sequence changes in 42 patients; 34 of whom had single nucleotide mutations (49%) and eight had deletions detected by MLPA (11%). In three of these eight cases, we found a deletion of all PAX6 exons including the surrounding genes. In four of the eight cases, there is a partial deletion of PAX6. In one case, a deletion of ELP4 and DCDC1 was found, leaving PAX6 itself intact. ELP4 and DCDC1 are located downstream of PAX6. Deletions in this region were described previously in aniridia patients [10,12]. This region contains 3′ regulatory elements for PAX6 [13,14].
In spite of the multiplicity and high sensitivity of methods used, we could not give a molecular diagnosis to 40% of the patients investigated. This may be partly the result of misdiagnosis; in other cases, it might be the result of the methods used. Point mutations or deletions might occur in regions of PAX6 that are not currently investigated such as intronic or remote promoter regions or in the 3′ regulatory region. Balanced rearrangements are not excluded either. There is no evidence to suspect that other genes besides PAX6 are responsible for aniridia [1].
Until now, most large deletions in or near PAX6 have been identified by FISH or Southern blot analysis. However, these techniques lack the resolution of MLPA in detecting smaller deletions or are not generally applicable to detect all kind of deletions. For our MLPA analysis, we used the P219 MLPA kit, which is an improved version of the P097 kit. Six of the eight deletions are detected by both kits. The P097 kit failed to detect two deletions as this kit lacks the relevant probes.
Young individuals with aniridia but no family history of aniridia are usually tested by high resolution cytogenetic or FISH analysis. This is done to detect a contiguous gene deletion involving 11p13 and consequently to identify individuals with an increased risk for Wilms tumor. It is now also possible to detect this type of deletion by using MLPA, which has the advantage of a higher resolution than FISH analysis and strongly reduces the time and cost of analysis.
References
- 1.Prosser J, van Heyningen V. PAX6 Mutations Reviewed. Hum Mutat. 1998;11:93–108. doi: 10.1002/(SICI)1098-1004(1998)11:2<93::AID-HUMU1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 2.van Heyningen V, Williamson KA. PAX6 in sensory development. Hum Mol Genet. 2002;11:1161–7. doi: 10.1093/hmg/11.10.1161. [DOI] [PubMed] [Google Scholar]
- 3.Brown A, McKie M, van Heyningen V, Prosser J. The human PAX6 mutation database. Nucleic Acids Res. 1998;26:259–64. doi: 10.1093/nar/26.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanson I, Churchill A, Love J, Axton R, Moore T, Clarke M, Meire F, van Heyningen V. Missense mutations in the most ancient residues of the PAX6 paired domain underlie a spectrum of human congenital eye malformations. Hum Mol Genet. 1999;8:165–72. doi: 10.1093/hmg/8.2.165. [DOI] [PubMed] [Google Scholar]
- 5.Fantes J, Redeker B, Breen M, Boyle S, Brown J, Fletcher J, Jones S, Bickmore W, Fukushima Y, Mannens M, Danes S, van Heyningen V, Hanson I. Aniridia-associated cytogenetic rearrangements suggest that a position effect may cause the mutant phenotype. Hum Mol Genet. 1995;4:415–22. doi: 10.1093/hmg/4.3.415. [DOI] [PubMed] [Google Scholar]
- 6.Drechsler M, Royer-Pokora BA. LINE element is present at the site of a 300-kb deletion starting in intron 10 of the PAX6 gene in a case of familial aniridia. Hum Genet. 1996;98:297–303. doi: 10.1007/s004390050210. [DOI] [PubMed] [Google Scholar]
- 7.Crolla JA, Cawdery JE, Oley CA, Young ID, Gray J, Fantes J, van Heyningen VA. FISH approach to defining the extent and possible clinical significance of deletions at the WAGR locus. J Med Genet. 1997;34:207–12. doi: 10.1136/jmg.34.3.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao LY, Huff V, Strong LC, Saunders GF. Mutation in the PAX6 gene in twenty patients with aniridia. Hum Mutat. 2000;15:332–9. doi: 10.1002/(SICI)1098-1004(200004)15:4<332::AID-HUMU5>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 9.Vincent MC, Pujo AL, Olivier D, Calvas P. Screening for PAX6 gene mutations is consistent with haploinsufficiency as the main mechanism leading to various ocular defects. Eur J Hum Genet. 2003;11:163–9. doi: 10.1038/sj.ejhg.5200940. [DOI] [PubMed] [Google Scholar]
- 10.D’Elia AV, Pellizzari L, Fabbro D, Pianta A, Divizia MT, Rinaldi R, Grammatico P, Arduino C, Damante G. A deletion 3′ to the PAX6 gene in familial aniridia cases. Mol Vis. 2007;13:1245–50. [PubMed] [Google Scholar]
- 11.Den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15:7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 12.Lauderdale JD, Wilensky JS, Oliver ER, Watson DS, Glaser T. 3′ deletions cause aniridia by preventing PAX6 gene expression. Proc Natl Acad Sci USA. 2000;97:13755–9. doi: 10.1073/pnas.240398797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tyas DA, Simpson TI, Carr CB, Kleinjan DA, van Heyningen V, Mason JO, Price DJ. Functional conservation of Pax6 regulatory elements in humans and mice demonstrated with a novel transgenic reporter mouse. BMC Dev Biol. 2006;6:21. doi: 10.1186/1471-213X-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kikuta H, Laplante M, Navratilova P, Komisarczuk AZ, Engström PG, Fredman D, Akalin A, Caccamo M, Sealy I, Howe K, Ghislain J, Pezeron G, Mourrain P, Ellingsen S, Oates AC, Thisse C, Thisse B, Foucher I, Adolf B, Geling A, Lenhard B, Becker TS. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 2007;17:545–55. doi: 10.1101/gr.6086307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gronskov K, Rosenberg T, Sand A, Brondum-Nielsen K. Mutational analysis of PAX 6: 16 novel mutations including 5 missense mutations with a mild aniridia phenotype. Eur J Hum Genet. 1999;7:274–86. doi: 10.1038/sj.ejhg.5200308. [DOI] [PubMed] [Google Scholar]
- 16.Davis A, Cowell JK. Mutations in the PAX 6 gene in patients with hereditary aniridia. Hum Mol Genet. 1993;2:2093–7. doi: 10.1093/hmg/2.12.2093. [DOI] [PubMed] [Google Scholar]
- 17.Singh S, Chao LY, Mishra R, Davies J, Saunders GF. Missense mutation at the C-terminus of PAX6 negatively modulates homeodomain function. Hum Mol Genet. 2001;10:911–8. doi: 10.1093/hmg/10.9.911. [DOI] [PubMed] [Google Scholar]
- 18.Glaser T, Walton DS, Maas RL. Genomic structure, evolutionary conservation and aniridia mutations in the human PAX6 gene. Nat Genet. 1992;2:232–9. doi: 10.1038/ng1192-232. [DOI] [PubMed] [Google Scholar]
- 19.Martha A, Ferrel RE, Mintz-Hittner H, Lyons LA, Saunders GF. Paired box mutations in familial and sporadic aniridia predicts truncated aniridia proteins. Am J Hum Genet. 1994;54:801–11. [PMC free article] [PubMed] [Google Scholar]