Abstract
Nitric oxide is a potential regulator of mitochondrial biogenesis. Therefore, we investigated if mice deficient in endothelial nitric oxide synthase (eNOS−/−) or neuronal NOS (nNOS−/−) have attenuated activation of skeletal muscle mitochondrial biogenesis in response to exercise. eNOS−/−, nNOS−/− and C57Bl/6 (CON) mice (16.3 ± 0.2 weeks old) either remained in their cages (basal) or ran on a treadmill (16 m min−1, 5% grade) for 60 min (n = 8 per group) and were killed 6 h after exercise. Other eNOS−/−, nNOS−/− and CON mice exercise trained for 9 days (60 min per day) and were killed 24 h after the last bout of exercise training. eNOS−/− mice had significantly higher nNOS protein and nNOS−/− mice had significantly higher eNOS protein in the EDL, but not the soleus. The basal mitochondrial biogenesis markers NRF1, NRF2α and mtTFA mRNA were significantly (P< 0.05) higher in the soleus and EDL of nNOS−/− mice whilst basal citrate synthase activity was higher in the soleus and basal PGC-1α mRNA higher in the EDL. Also, eNOS−/− mice had significantly higher basal citrate synthase activity in the soleus but not the EDL. Acute exercise increased (P< 0.05) PGC-1α mRNA in soleus and EDL and NRF2α mRNA in the EDL to a similar extent in all genotypes. In addition, short-term exercise training significantly increased cytochrome c protein in all genotypes (P< 0.05) in the EDL. In conclusion, eNOS and nNOS are differentially involved in the basal regulation of mitochondrial biogenesis in skeletal muscle but are not critical for exercise-induced increases in mitochondrial biogenesis in skeletal muscle.
In skeletal muscle the regulation of mitochondrial biogenesis is particularly important as mitochondria are highly abundant and play a vital role in skeletal muscle metabolism (for reviews see Schrauwen & Hesselink, 2004; Lowell & Shulman, 2005; Hood et al. 2006).
There is emerging evidence that nitric oxide (NO) may be an important regulator of the mitochondrial biogenesis pathway (Nisoli et al. 2003; Nisoli et al. 2004). Although the process is still unclear in skeletal muscle, NO may be playing a role in the regulation of skeletal muscle mitochondrial biogenesis since skeletal muscle expresses several isoforms of nitric oxide synthase (NOS) (Hussain et al. 1997; Lau et al. 2000) and NOS (from all isoforms) is constitutively active at rest in rodent skeletal muscle (Balon & Nadler, 1994; Roy et al. 1998; Roberts et al. 1999). Both endothelial (eNOS) and neuronal (nNOS) isoforms are expressed within skeletal muscle fibres, with eNOS more abundant in oxidative, and nNOS more abundant in glycolytic rodent skeletal muscle (Kobzik et al. 1995; Hussain et al. 1997; Kapur et al. 1997; Lau et al. 2000). NO donors increase mitochondrial biogenesis in L6 muscle cells (Nisoli et al. 2004) and our recent short-term l-NAME study revealed some aspects of the mitochondrial respiratory chain are decreased in rodent skeletal muscle under basal (non-exercise) conditions (Wadley & McConell, 2007). There is also evidence of isoform specific alteration of basal mitochondrial biogenesis in skeletal muscle from mice deficient in eNOS (eNOS−/−) and nNOS (nNOS−/−) (Momken et al. 2002, 2004; Nisoli et al. 2004; Schild et al. 2006). eNOS−/− mice have decreased PGC-1α mRNA levels and reduced mitochondrial density (Nisoli et al. 2004). In contrast, nNOS−/− mice display characteristics consistent with increased skeletal muscle mitochondrial biogenesis, such as increased citrate synthase activity (Schild et al. 2006), although the levels of the mitochondrial biogenesis markers (i.e. PGC-1α, NRF1, NRF2 and mtTFA) were not measured.
NO production, total NOS activity and the downstream messenger of NO, cGMP, increase during contraction in rodent skeletal muscle (Balon & Nadler, 1994; Roberts et al. 1999; Lau et al. 2000) and may therefore be involved in the up-regulation of mitochondrial biogenesis by exercise. However, the role(s) of the eNOS and nNOS isoforms in the regulation of exercise-induced mitochondrial biogenesis are limited and equivocal. eNOS−/− mice have decreased cytochrome c oxidase (COX) activity following voluntary wheel running, suggestive of impaired mitochondrial biogenesis (Momken et al. 2004). However it is unclear if the effect of eNOS knockout is due solely to impairments in COX following wheel running, or extends to the mitochondrial biogenesis pathway per se, since other mitochondrial enzymes such as citrate synthase activity were not impaired (Momken et al. 2004). Thus it is important to determine if mitochondrial biogenesis markers (i.e. PGC-1α, NRF1, NRF2 and mtTFA) are attenuated in eNOS−/− mice following either acute exercise or exercise training. Also, a confounding factor with eNOS−/− mice is that voluntary physical activity levels are much lower than control mice, so the exercise stimulus from voluntary running is not equal (Momken et al. 2004). We have recently shown that 2 days of total NOS inhibition via the non-specific NOS inhibitor NG-nitro-l-arginine methyl ester (l-NAME) does not attenuate the exercise-induced increase in PGC-1α mRNA in rodent skeletal muscle subjected to the same acute exercise stimulus (Wadley & McConell, 2007). Therefore, it is unclear if eNOS or nNOS are involved in the regulation of exercise-induced mitochondrial biogenesis in skeletal muscle. Since several days of pharmacological NOS inhibition results in a number of adverse effects in rodents (i.e. reduced body and muscle mass and development of hypertension) (Scrogin et al. 1998; Wang et al. 2001) it is not possible to use the l-NAME treatment model for short-term training studies. Furthermore, it is not known if nNOS−/− mice have normal increases in exercise-induced mitochondrial biogenesis. Thus, the present study investigated the activation of exercise-induced mitochondrial biogenesis following acute exercise and short-term training in the skeletal muscle of eNOS−/− and nNOS−/− mice.
We tested the hypothesis that NO derived from either eNOS or nNOS or both plays a role in skeletal mitochondrial biogenesis. Based on our previous findings in rats following acute exercise and NOS inhibition (Wadley & McConell, 2007), we hypothesized that eNOS−/− and nNOS−/− mice would have normal increases in mitochondrial biogenesis markers in skeletal muscle following acute exercise and short-term exercise training. We also tested the hypotheses that eNOS−/− mice would have lower markers and nNOS−/− mice would have higher markers of basal mitochondrial biogenesis in skeletal muscle.
Methods
Animal care and dietary treatment
Mice homozygous for targeted disruption of the nNOS gene (B6,129-NOS1tm1plh, nNOS−/−; Huang et al. 1993) were purchased from Jackson Laboratories (Bar Harbor, ME, USA, stock no. 002633) and a colony was established by backcrossing the nNOS−/− onto a C57BL/6 background for at least six generations and genotyped from tail clippings taken at day 20 (Choate et al. 2001). eNOS knock-out mice as developed in the C57BL/6 strain (Huang et al. 1995) were also purchased from Jackson Laboratories (eNOS−/−; stock no. 002684). Following establishment of the nNOS−/− and eNOS−/− colonies, male laboratory bred homozygous (from sibling matings) and C57BL/6 mice (CON) were used in all experiments (16.3 ± 0.2 weeks old). Animals were housed in an environmentally controlled laboratory (temperature 22°C) with a 12 : 12 h light–dark cycle. Animals were familiarized to treadmill running 2 weeks prior to being killed, on two separate days for 20 min each day. The University of Melbourne Animal Experimentation Ethics Committee approved all experimental procedures.
Animals were assigned to one of three groups on the basis of whether they (1) remained in their cage (basal; n = 8), (2) ran on a motor driven treadmill at 16 m min−1 on a 5% incline for 60 min and were killed 6 h later (acute ex; n = 8), or (3) exercise trained for 9 days and were killed 24 h after the last training session (trained; n = 8). The exercise training protocol consisted of running on a treadmill for 60 min per day on a 5% incline for 9 days, with 2 days rest between training day five and seven. On the first 3 days of training mice ran at 16 m min−1 and the speed was then progressively increased so that by the final day of training mice ran at 19 m min−1.
For all mice, food was withdrawn for 1 h prior to their being killed. Mice were then anaesthetized with an intraperitoneal injection of pentobarbital sodium (Sigma, St Louis, MO, USA; 170 mg kg−1) followed by cervical dislocation. Soleus and EDL muscles were rapidly excised, frozen and stored in liquid N2.
Preparation of mouse tissue
RNA was isolated from frozen mouse soleus and EDL using Trizol (Invitrogen, Melbourne, Australia). For immunoblotting and enzyme activity, frozen muscle (30 μl of buffer per mg of muscle) was homogenized in freshly prepared ice-cold buffer (50 mm Tris at pH 7.5 containing 1 mm EDTA, 10% v/v glycerol, 1% v/v Triton X-100, 50 mm NaF, 5 mm Na4P2O7, 1 mm DTT, 1 mm PMSF and 5 μl ml−1 Protease Inhibitor Cocktail (P8340, Sigma)). Tissue lysates were incubated on ice for 20 min and then spun at 16 000 g for 20 min at 4°C. Protein concentration was determined using a bicinchoninic acid (BCA) protein assay (Pierce, Rockford, IL, USA) with BSA as the standard.
Gene expression
RNA integrity was verified and the concentration determined on the Experion Automated Electrophoresis System (Bio-Rad Laboratories, NSW, Australia). First strand cDNA was generated from 0.5 μg RNA using AMV Reverse Transcriptase (Promega, Madison, WI, USA) as previously described (Wadley et al. 2001). Primers and FAM-labelled probes were designed using the Roche Universal ProbeLibrary (Roche, http://www.roche-applied-science.com/sis/rtpcr/upl/index.jsp) from gene sequences obtained from GenBank (PGC-1α: BC066868.1; PGC-1β: NM_133249.1; NRF1: NM_010938.2; NRF2α: M74515; mtTFA: NM_009360.2; COX III: AK131579). With the exception of the mitochondria-encoded COX III, all primer sequences were intron-spanning. Also, primer sequences were validated using BLAST (Altschul et al. 1990) to ensure each primer was homologous with the desired mRNA of mouse skeletal muscle. The primer sequences are shown in Table 1. Real-time PCR was performed in triplicate using the Rotor-gene 3000 system (Corbett Research, Sydney, Australia). β2-Microglobulin (cat. no. Mm00437762_m1) was assessed using predesigned/prevalidated FAM-labelled Assays-on-Demand from Applied Biosystems (Foster City, CA, USA). All genes were normalized to β2-microglobulin using the 2− Δ ΔCt method. β2-Microglobulin mRNA levels have been reported not to change in response to an acute bout of endurance exercise in skeletal muscle (Mahoney et al. 2004; Wadley & McConell, 2007). Furthermore, there was no difference in β2-microglobulin mRNA levels measured from the 2-Ct values (2 way ANOVA genotype × exercise intervention) for either the soleus or EDL muscles (data not shown).
Table 1.
Gene primer sequences
| Gene | Sense primer (5′–3′) | Antisense primer (5′–3′) | UPL probe# |
|---|---|---|---|
| PGC-1α | GAGTCTGAAAGGGCCAAGC | GTAAATCACACGGCGCTCTT | 29 |
| PGC-1β | CTGACGGTGGAGCTTTGC | TTGGTGTCTGGCTTGAAGG | 25 |
| NRF1 | GCTGCTTTCAGTCCTTCTGG | GTGTTCAGTTTGGGTCACTCC | 25 |
| NRF2α | CTCGGAGCAGGTGACGAG | TGGACCAGCGTATAGGATCA | 66 |
| mtTFA | CAAAGGATGATTCGGCTCAG | AAGCTGAATATATGCCTGCTTTTC | 94 |
| COX III | TAGCCTCGTACCAACACATGA | AGTGGTGAAATTCCTGTTGGA | 66 |
UPL: Universal ProbeLibrary (Roche, http://www.roche-applied-science.com/sis/rtpcr/upl/index.jsp).
Immunoblotting
Total lysates for determination of PGC-1α, COX IV, cytochrome c, p38 MAPK, AMP-activated protein kinase (AMPKα), eNOS and nNOS were solubilized in Laemmli sample buffer. Equal amounts of proteins were separated by SDS-PAGE and electrotransfer of proteins from the gel to PVDF membranes (25 mmol l−1 Tris, pH 8.3, 192 mmol l−1 glycine, and 20% v/v methanol) was performed for 90 min at 95 V (constant). Blots were probed with anti-PGC-1α rabbit polyclonal (Chemicon, Temecula, CA, USA), anti-COX IV mouse monoclonal (Molecular Probes, Eugene, OR, USA), anti-cytochrome c mouse monoclonal (BD Biosciences Pharmingen, San Diego, CA, USA), anti-p38 rabbit polyclonal MAPK (Cell Signalling, Hartsfordshire, England), anti-AMPKα rabbit polyclonal (Cell Signalling Technology), anti-eNOS mouse monoclonal (BD Biosciences) and anti-nNOS mouse monoclonal (BD Biosciences) antibodies. Binding was detected with IRDye™ 800-conjugated antirabbit IgG (Rockland, Gilbertsville, PA, USA) or IRDye™ 680-conjugated anti-mouse IgG (Molecular Probes) secondary antibodies. As a loading control, blots were then reprobed with anti-α-tubulin mouse monoclonal antibody (Sigma). Data were normalized to the level of α-tubulin and expressed as the ratio of integrated intensity following infrared detection (Odyssey Imaging system, LI-COR Biosciences, Lincoln, NE, USA). For p38 MAPK and AMPKα signalling, membranes were then stripped (2% SDS (w/v) in 25 mm glycine, pH 2.0) and re-probed with anti-phospho-p38 MAPK rabbit polyclonal antibody (Cell Signalling) or anti-phospho-AMPK Thr172 antibody (Upstate Biotechnology, New York) and phosphorylation was expressed relative to p38 MAPK or AMPKα protein abundance.
To test if there was compensatory expression of iNOS in the eNOS−/− and nNOS−/− mice, we performed immunoblotting as described above using 90 μg of total lysates from soleus and EDL of all genotypes with anti-iNOS rabbit polyclonal antibody (BD Biosciences), with mouse macrophage lysate used as a positive control (BD Biosciences).
Citrate synthase activity
Citrate synthase activity was measured spectrophotometrically on a 96-well plate at room temperature by following the increase in 5,5′-dithiobis-2-nitrobenzoate (DTNB) at 412 nm using the muscle homogenates and expressed in μmol min−1 g−1 of total protein (Srere, 1969).
Statistical analyses
Results were analysed using either one-factor analysis of variance (basal levels between genotypes) or two-factor ANOVA (genotype × exercise intervention) where appropriate. If these analyses revealed a significant interaction, specific differences between mean values were located using Fisher's least significance difference test. All data are presented as means ± s.e.m. The level of significance was set at P < 0.05.
Results
Anatomical data
The mass of the soleus and EDL muscles were significantly (P< 0.05) lower in nNOS−/− mice compared to CON and eNOS−/− mice (Table 2). Also, the soleus mass of eNOS−/− mice was significantly higher compared to CON and nNOS−/− mice (Table 2). Body mass was not significantly different between the genotypes (Table 2).
Table 2.
Anatomical data
| CON | eNOS−/− | nNOS−/− | |
|---|---|---|---|
| Body mass (g) | 26.8 ± 0.2 | 26.6 ± 0.4 | 26.3 ± 0.4 |
| Soleus mass (mg) | 11.6 ± 5.8 | 13.4 ± 6.7†# | 7.7 ± 2.0† |
| EDL mass (mg) | 18.4 ± 0.9 | 16.9 ± 1.7 | 11.9 ± 0.8† |
Values are means ± s.e.m.n = 16 for CON, n = 11 for eNOS−/− and n = 14 for nNOS−/−.
P <0.05 versus CON
P < 0.05 versus nNOS−/− (one-way ANOVA).
Compensatory effects of eNOS or nNOS knockout on skeletal muscle
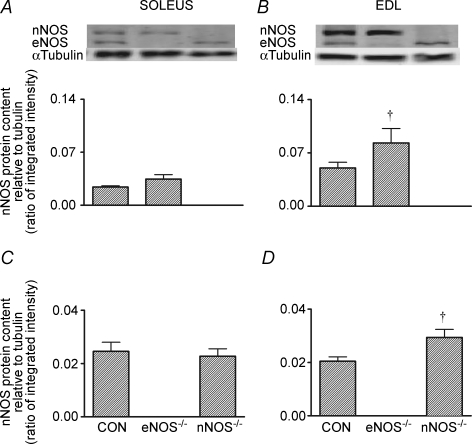
Compared to CON mice, soleus eNOS expression was similar in nNOS−/− mice and nNOS expression was similar in eNOS−/− mice (i.e. no compensation evident; Fig. 1A and C, respectively). However in the EDL, nNOS expression was significantly higher in eNOS−/− mice and eNOS expression was significantly higher in nNOS−/− mice (P< 0.05; Fig. 1B and D, respectively). We were not able to detect the expression of the iNOS isoform in the soleus or EDL muscles of any genotype (data not shown).
Figure 1.
Basal levels of nNOS (A and B) and eNOS (C and D) protein abundance in the soleus and EDL of CON and NOS knockout mice Values are means ± s.e.m. Western blots are representative from one mouse from each genotype. n = 8 for all groups except n = 7 for the EDL of eNOS−/− mice due to lack of available sample. †P <0.05 versus CON mice (one-way ANOVA).
Effect of eNOS or nNOS knockout on basal levels of mitochondrial biogenesis markers and phosphorylation of p38 MAPK and AMPKα
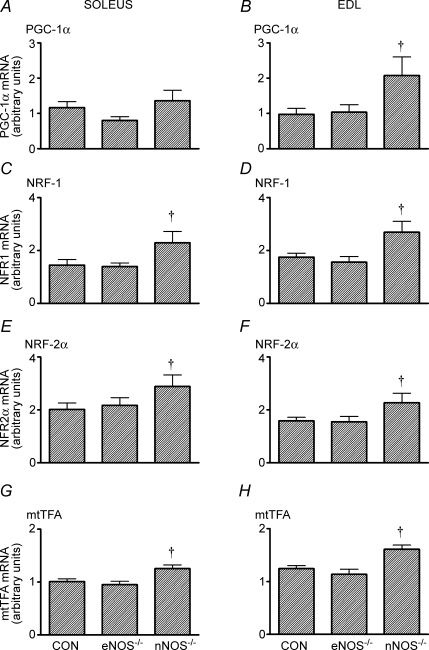
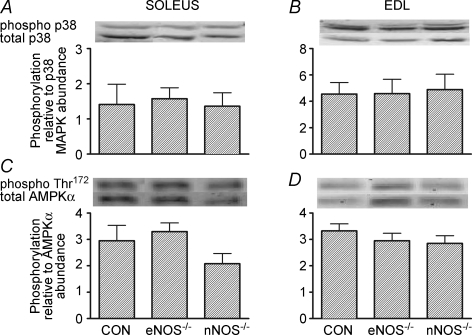
In the EDL, basal levels of PGC-1α mRNA were higher in the nNOS−/− mice than the two other groups (P< 0.05; Fig. 2B). The basal mRNA levels of NRF-1, NRF-2 and mtTFA were significantly higher in the soleus and EDL of nNOS−/− mice compared to CON and eNOS−/− mice (Fig. 2, P < 0.05). Although PGC-1β and COX III mRNA appeared to be higher in the EDL of nNOS−/− mice, these variables were not significantly different compared to CON and eNOS−/− mice (Table 3, P = 0.20 and 0.12, one-way ANOVA for basal PGC-1β and COX III mRNA, respectively). Basal phosphorylation of p38 MAPK and AMPKα Thr172 was not different among genotypes in the soleus or EDL (Fig. 3). Basal citrate synthase activity was significantly higher in the soleus of nNOS−/− mice compared to CON and eNOS−/− mice (Table 3, P < 0.05). Also, eNOS−/− mice had significantly elevated basal citrate synthase activity compared to CON mice (Table 3, P < 0.05). In the EDL, there were no differences in the citrate synthase activity between the genotypes (Table 3).
Figure 2.
Basal levels of PGC-1α (A and B), NRF1 (C and D), NRF2α (E and F) and mtTFA (G and H) mRNA in the soleus and EDL of CON and NOS knockout mice Values are means ± s.e.m.n = 8 for all groups. †P <0.05 versus CON mice (one-way ANOVA).
Table 3.
Basal levels of mitochondrial biogenesis markers and citrate synthase activity in the soleus and EDL of CON and NOS knockout mice
| CON | eNOS−/− | nNOS−/− | |
|---|---|---|---|
| SOLEUS | |||
| PGC-1β mRNA (arbitrary units) | 1.24 ± 0.10 | 1.54 ± 0.12 | 1.53 ± 0.17 |
| COX III mRNA (arbitrary units) | 1.07 ± 0.10 | 0.92 ± 0.07 | 1.08 ± 0.13 |
| Citrate synthase activity (μmol min−1 (g protein)−1) | 778 ± 16 | 924 ± 62† | 1180 ± 41†# |
| EDL | |||
| PGC-1β mRNA (arbitrary units) | 0.75 ± 0.11 | 0.85 ± 0.15 | 1.17 ± 0.21 |
| COX III mRNA (arbitrary units) | 1.18 ± 0.14 | 1.07 ± 0.14 | 1.64 ± 0.29 |
| Citrate synthase activity (μmol min−1 (g protein)−1) | 772 ± 14 | 781 ± 16 | 818 ± 23 |
Values are means ± s.e.m.n = 8 for all groups.
P <0.05 versus CON mice;
P <0.05 versus eNOS−/− mice (one-way ANOVA).
Figure 3.
Basal levels of phosphorylated p38 MAPK (A and B) and phosphorylated AMPKα Thr172 (C and D) in the soleus and EDL of CON and NOS knockout mice Values are means ± s.e.m.n = 8 for all groups. Western blots are representative from one mouse from each genotype.
Effect of eNOS or nNOS knockout on mitochondrial biogenesis following acute exercise
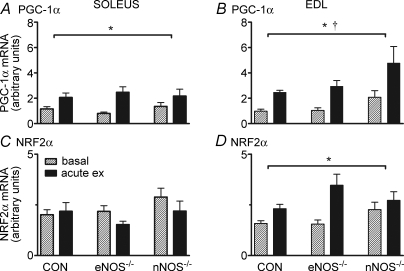
The acute exercise bout resulted in significantly higher PGC-1α mRNA levels 6 h following exercise in the soleus and EDL for all genotypes (P< 0.05; main effect for exercise, Fig. 4A and B). Six hours following acute exercise, NRF2α mRNA levels were also significantly increased in all genotypes in the EDL (P< 0.05; main effect for exercise, Fig. 4C), but there was no change in any genotype in the soleus (P> 0.05; Fig. 4D). NRF-1 mRNA was not changed 6 h following acute exercise in any genotype (data not shown).
Figure 4.
PGC-1α (A and B) and NRF2α (C and D) mRNA in the soleus and EDL of CON and NOS knockout mice under basal conditions and 6 h after 60 min of treadmill running (acute ex) Values are means ± s.e.m.n = 8 for all groups. *P <0.05 main effect for exercise, †P <0.05 main effect for genotype (two-way ANOVA).
Effect of eNOS or nNOS knockout on mitochondrial biogenesis following short-term exercise training
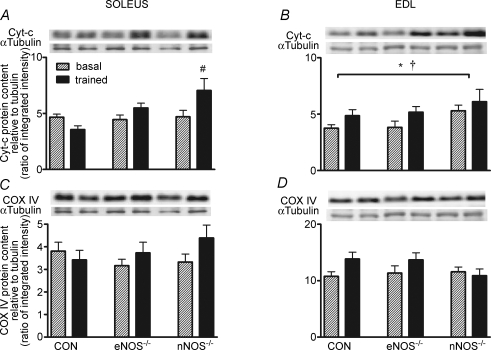
Short-term endurance training significantly increased cytochrome c protein levels in the soleus of nNOS−/− mice and in the EDL of CON, eNOS−/− and nNOS−/− mice (P< 0.05; main effect for exercise, Fig. 5A and B, respectively). Howver, short-term training did not increase COX IV protein levels in the soleus or EDL of CON, eNOS−/− and nNOS−/− mice (Fig. 5C and D). Furthermore, PGC-1α protein levels were not significantly altered by short-term training in the soleus or EDL in any genotype and neither were the levels of α-tubulin (data not shown).
Figure 5.
Cytochrome c (A and B) and COX IV (C and D) protein abundance in the soleus and EDL of CON and NOS knockout mice under basal conditions and after short-term training (trained) Values are means ± s.e.m.n = 8 for all groups. *P <0.05 main effect for exercise training, †P <0.05 main effect for genotype, #P < 0.05 versus basal (two-way ANOVA). Western blots are representative from one mouse from each genotype.
Discussion
A major finding of this study was that acute exercise increased mitochondrial biogenesis markers to a similar extent in CON, eNOS−/− and nNOS−/− mice. In addition, a modest but similar increase in cytochrome c protein abundance in CON, eNOS−/− and nNOS−/− mice was observed following short-term exercise training. These findings, when combined with our previous results using l-NAME ingestion in rats (Wadley & McConell, 2007), suggest that NO is not critical for increases in skeletal muscle mitochondrial biogenesis that are associated with exercise. Another important finding of the present study was that basal levels of mitochondrial biogenesis markers are elevated in the skeletal muscle of nNOS−/− mice. This finding would suggest a negative regulatory role for the nNOS isoform on basal mitochondrial biogenesis.
In agreement with our hypothesis, we found that ablation of either eNOS or nNOS in mice does not attenuate the exercise-induced increase in the mitochondrial biogenesis markers following acute exercise. These findings are in line with our previous study that found that pharmacological NOS inhibition did not alter the increases in PGC-1α mRNA following acute exercise in rat skeletal muscle (Wadley & McConell, 2007). Thus, eNOS and nNOS do not appear to be involved in the regulation of acute exercise-induced mitochondrial biogenesis in skeletal muscle.
The present study also found that eNOS or nNOS knockout in mice did not attenuate the small but significant increase in skeletal muscle cytochrome c protein content following short-term endurance training (Fig. 5A and B). Somewhat surprisingly, COX IV protein (Fig. 5C and D) and PGC-1α protein (data not shown) did not increase following short-term training. However, examination of the literature reveals that, unlike rats that have a robust increase in skeletal muscle mitochondrial biogenesis markers following a few weeks of endurance training (Murakami et al. 1998; Taylor et al. 2005), the endurance training response of mice is modest (Ikeda et al. 2006; Jorgensen et al. 2007) and varies greatly between strains (Massett & Berk, 2005). In particular, C57/Bl6 mice (i.e. same genetic background as the mice used in the present study) display only modest increases in cytochrome c protein content following 4 weeks of treadmill running at a similar intensity and duration as the protocol employed in the present study (Massett & Berk, 2005). Others have also demonstrated that increases in skeletal muscle mitochondrial cytochrome c protein can occur following short-term exercise training in mice without increases in PGC-1α protein (Ikeda et al. 2006). Therefore, the normal increase in cytochrome c protein following short-term training, taken with the normal increases in PGC-1α and NRF2α mRNA following acute exercise in eNOS−/− and nNOS−/− mice, suggests that neither eNOS nor nNOS are involved in exercise-induced mitochondrial biogenesis.
An interesting finding of the present study was that in the EDL, gene knockout of eNOS significantly increased the level of nNOS and vice versa (Fig. 1B and D) when compared to CON mice. This could suggest some type of compensatory mechanism; indeed nNOS protein is also elevated in the aorta of eNOS−/− mice (Biecker et al. 2004), although the effects observed in the present study appear specific to the muscle examined, since there were no differences in the soleus muscle. Our finding of higher alternative NOS isoforms in the EDL of eNOS−/− and nNOS−/− mice is in contrast to no differences found in the EDL by Lau et al. (2000) despite age and genetic background being similar for both studies. Thus, the differences between the studies are difficult to reconcile. However, the data for eNOS and nNOS protein in the EDL reported by Lau et al. (2000) are quite variable, with the standard errors ranging between 10% and 50% of the mean. Furthermore, the statistical power reported by Lau et al. (2000) is unclear as sample size was not reported. Nevertheless, an important finding from the present study is the need to always examine the expression of the alternative isoforms following gene knockout and not make too many assumptions based on the findings of previous studies.
Our finding of increased basal citrate synthase activity in nNOS−/− mice is in agreement with the findings of Schild et al. (2006) and we extend these findings to show numerous mitochondrial biogenesis markers are up-regulated in the soleus and EDL muscles of these mice. The mechanism for the increased basal mitochondrial biogenesis in nNOS−/− mice is unclear. It is unlikely that the increased basal mitochondrial biogenesis in nNOS−/− mice could be due to the compensatory increase in eNOS protein that was observed in the EDL (Fig. 1D), since basal mitochondrial biogenesis markers were also up-regulated in the soleus muscle of nNOS−/− mice (Fig. 2) despite eNOS protein being unaltered in the soleus (Fig. 1C). It is also unlikely that the increased basal mitochondrial biogenesis in nNOS−/− mice is due to elevated phosphorylation of p38 MAPK or AMPKα Thr172. Elevated basal phosphorylation of p38 MAPK, a key upstream mitochondrial biogenesis signalling kinase (Akimoto et al. 2005), has been observed in the hearts of nNOS−/− mice (Kinugawa et al. 2005), but in the current study no elevation of basal phosphorylation of p38 MAPK was observed in the skeletal muscle of nNOS−/− mice (Fig. 3). AMPK is a known stimulator of mitochondrial biogenesis in skeletal muscle (Suwa et al. 2003), although basal levels of AMPKα Thr172 were not different for any genotype in the soleus and EDL (Fig. 3). Therefore, the mechanism whereby nNOS knockout increases basal levels of skeletal muscle mitochondrial biogenesis is an important question that remains to be answered.
A somewhat surprising finding in the present study was that skeletal muscle mitochondrial biogenesis markers were not significantly reduced in eNOS−/− mice compared with CON mice. It is possible that previous findings of reduced mitochondrial biogenesis in skeletal muscle of eNOS−/− mice (Momken et al. 2002, 2004; Nisoli et al. 2003, 2004) may be explained by the confounding effects of obesity. Indeed, Valerio et al. (2006) have observed reduced mitochondrial biogenesis in adipose tissue and skeletal muscle in several rodent models of obesity. Previous studies by Momken et al. (2002, 2004) have shown reduced oxidative capacity in the soleus of eNOS−/− mice, consistent with impaired mitochondrial biogenesis, although these were much older and obese mice. In addition, Nisoli et al. reported reduced mitochondrial biogenesis in several tissues of eNOS−/− mice (Nisoli et al. 2003), including skeletal muscle (Nisoli et al. 2004). Although the eNOS−/− mice used by Nisoli et al. (2003, 2004) were derived from the same line as ours (Huang et al. 1995), for reasons we cannot explain they were also obese at a similar age to our eNOS−/− mice (i.e. male mice ∼12 weeks old weigh ∼40 g) (Nisoli et al. 2003), whereas others report similar body mass at a similar age to ours (i.e. male eNOS−/− mice ∼12 weeks old weigh ∼24 g) (Kubis et al. 2002; Morishita et al. 2005). In the present study no differences in body mass were observed between the three mice genotypes (∼26 g; Table 1) at ∼16 weeks of age. However, the role of eNOS in skeletal muscle mitochondrial biogenesis should not be discounted since a recent study reported reduced mitochondrial content and impaired β-oxidation in skeletal muscle from eNOS−/− mice that have normal body and fat mass (Le Gouill et al. 2007), suggestive of impaired mitochondrial biogenesis. However the eNOS−/− mice used by Le Gouill et al. (2007) were generated using a different method of eNOS gene disruption (Shesely et al. 1996) compared to the present study (Huang et al. 1995), making comparisons difficult.
In conclusion, knocking out either the eNOS or the nNOS isoform does not attenuate the exercise-induced increase in mitochondrial biogenesis markers following acute exercise or short-term training in mouse skeletal muscle. These results indicate that neither nNOS nor eNOS is critical for normal increases in skeletal muscle mitochondrial biogenesis in response to exercise. In contrast the present study observed increased basal levels of several mitochondrial biogenesis markers in the skeletal muscle of nNOS−/− mice, suggesting an inhibitory role for nNOS in the basal regulation of mitochondrial biogenesis.
Acknowledgments
This work was supported by grants from the National Health and Medical Research Council (GKM 350483) and The University of Melbourne–CSIRO Collaborative Research Program.
References
- Akimoto T, Pohnert SC, Li P, Zhang M, Gumbs C, Rosenberg PB, Williams RS, Yan Z. Exercise stimulates Pgc-1α transcription in skeletal muscle through activation of the p38 MAPK pathway. J Biol Chem. 2005;280:19587–19593. doi: 10.1074/jbc.M408862200. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Balon TW, Nadler JL. Nitric oxide release is present from incubated skeletal muscle preparations. J Appl Physiol. 1994;77:2519–2521. doi: 10.1152/jappl.1994.77.6.2519. [DOI] [PubMed] [Google Scholar]
- Biecker E, Neef M, Sagesser H, Shaw S, Koshy A, Reichen J. Nitric oxide synthase 1 is partly compensating for nitric oxide synthase 3 deficiency in nitric oxide synthase 3 knock-out mice and is elevated in murine and human cirrhosis. Liver Int. 2004;24:345–353. doi: 10.1111/j.1478-3231.2004.0933.x. [DOI] [PubMed] [Google Scholar]
- Choate JK, Danson EJ, Morris JF, Paterson DJ. Peripheral vagal control of heart rate is impaired in neuronal NOS knockout mice. Am J Physiol Heart Circ Physiol. 2001;281:H2310–H2317. doi: 10.1152/ajpheart.2001.281.6.H2310. [DOI] [PubMed] [Google Scholar]
- Hood DA, Irrcher I, Ljubicic V, Joseph AM. Coordination of metabolic plasticity in skeletal muscle. J Exp Biol. 2006;209:2265–2275. doi: 10.1242/jeb.02182. [DOI] [PubMed] [Google Scholar]
- Huang PL, Dawson TM, Bredt DS, Snyder SH, Fishman MC. Targeted disruption of the neuronal nitric oxide synthase gene. Cell. 1993;75:1273–1286. doi: 10.1016/0092-8674(93)90615-w. [DOI] [PubMed] [Google Scholar]
- Huang PL, Huang Z, Mashimo H, Bloch KD, Moskowitz MA, Bevan JA, Fishman MC. Hypertension in mice lacking the gene for endothelial nitric oxide synthase. Nature. 1995;377:239–242. doi: 10.1038/377239a0. [DOI] [PubMed] [Google Scholar]
- Hussain SN, El-Dwairi Q, Abdul-Hussain MN, Sakkal D. Expression of nitric oxide synthase isoforms in normal ventilatory and limb muscles. J Appl Physiol. 1997;83:348–353. doi: 10.1152/jappl.1997.83.2.348. [DOI] [PubMed] [Google Scholar]
- Ikeda S, Kawamoto H, Kasaoka K, Hitomi Y, Kizaki T, Sankai Y, Ohno H, Haga S, Takemasa T. Muscle type-specific response of PGC-1α and oxidative enzymes during voluntary wheel running in mouse skeletal muscle. Acta Physiol (Oxf) 2006;188:217–223. doi: 10.1111/j.1748-1716.2006.01623.x. [DOI] [PubMed] [Google Scholar]
- Jorgensen SB, Treebak JT, Viollet B, Schjerling P, Vaulont S, Wojtaszewski JF, Richter EA. Role of AMPKα2 in basal, training-, and AICAR-induced GLUT4, hexokinase II, and mitochondrial protein expression in mouse muscle. Am J Physiol Endocrinol Metab. 2007;292:E331–E339. doi: 10.1152/ajpendo.00243.2006. [DOI] [PubMed] [Google Scholar]
- Kapur S, Bedard S, Marcotte B, Cote CH, Marette A. Expression of nitric oxide synthase in skeletal muscle: a novel role for nitric oxide as a modulator of insulin action. Diabetes. 1997;46:1691–1700. doi: 10.2337/diab.46.11.1691. [DOI] [PubMed] [Google Scholar]
- Kinugawa S, Huang H, Wang Z, Kaminski PM, Wolin MS, Hintze TH. A defect of neuronal nitric oxide synthase increases xanthine oxidase-derived superoxide anion and attenuates the control of myocardial oxygen consumption by nitric oxide derived from endothelial nitric oxide synthase. Circ Res. 2005;96:355–362. doi: 10.1161/01.RES.0000155331.09458.A7. [DOI] [PubMed] [Google Scholar]
- Kobzik L, Stringer B, Balligand JL, Reid MB, Stamler JS. Endothelial type nitric oxide synthase in skeletal muscle fibers: mitochondrial relationships. Biochem Biophys Res Commun. 1995;211:375–381. doi: 10.1006/bbrc.1995.1824. [DOI] [PubMed] [Google Scholar]
- Kubis N, Besnard S, Silvestre JS, Feletou M, Huang PL, Levy BI, Tedgui A. Decreased arteriolar density in endothelial nitric oxide synthase knockout mice is due to hypertension, not to the constitutive defect in endothelial nitric oxide synthase enzyme. J Hypertens. 2002;20:273–280. doi: 10.1097/00004872-200202000-00017. [DOI] [PubMed] [Google Scholar]
- Lau KS, Grange RW, Isotani E, Sarelius IH, Kamm KE, Huang PL, Stull JT. nNOS and eNOS modulate cGMP formation and vascular response in contracting fast-twitch skeletal muscle. Physiol Genomics. 2000;2:21–27. doi: 10.1152/physiolgenomics.2000.2.1.21. [DOI] [PubMed] [Google Scholar]
- Le Gouill E, Jimenez M, Binnert C, Pierre-Yves J, Thalmann S, Nicod P, Scherrer U, Vollenweider P. Diabetes. 2007. eNOS knock-out mice have defective mitochondrial β-oxidation. in press. [DOI] [PubMed] [Google Scholar]
- Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384–387. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- Mahoney DJ, Carey K, Fu MH, Snow R, Cameron-Smith D, Parise G, Tarnopolsky MA. Real-time RT-PCR analysis of housekeeping genes in human skeletal muscle following acute exercise. Physiol Genomics. 2004;18:226–231. doi: 10.1152/physiolgenomics.00067.2004. [DOI] [PubMed] [Google Scholar]
- Massett MP, Berk BC. Strain-dependent differences in responses to exercise training in inbred and hybrid mice. Am J Physiol Regul Integr Comp Physiol. 2005;288:R1006–R1013. doi: 10.1152/ajpregu.00476.2004. [DOI] [PubMed] [Google Scholar]
- Momken I, Fortin D, Serrurier B, Bigard X, Ventura-Clapier R, Veksler V. Endothelial nitric oxide synthase (NOS) deficiency affects energy metabolism pattern in murine oxidative skeletal muscle. Biochem J. 2002;368:341–347. doi: 10.1042/BJ20020591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momken I, Lechene P, Ventura-Clapier R, Veksler V. Voluntary physical activity alterations in endothelial nitric oxide synthase knockout mice. Am J Physiol Heart Circ Physiol. 2004;287:H914–H920. doi: 10.1152/ajpheart.00651.2003. [DOI] [PubMed] [Google Scholar]
- Morishita T, Tsutsui M, Shimokawa H, Sabanai K, Tasaki H, Suda O, Nakata S, Tanimoto A, Wang KY, Ueta Y, Sasaguri Y, Nakashima Y, Yanagihara N. Nephrogenic diabetes insipidus in mice lacking all nitric oxide synthase isoforms. Proc Natl Acad Sci U S A. 2005;102:10616–10621. doi: 10.1073/pnas.0502236102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Shimomura Y, Yoshimura A, Sokabe M, Fujitsuka N. Induction of nuclear respiratory factor-1 expression by an acute bout of exercise in rat muscle. Biochim Biophys Acta. 1998;1381:113–122. doi: 10.1016/s0304-4165(98)00018-x. [DOI] [PubMed] [Google Scholar]
- Nisoli E, Clementi E, Paolucci C, Cozzi V, Tonello C, Sciorati C, Bracale R, Valerio A, Francolini M, Moncada S, Carruba MO. Mitochondrial biogenesis in mammals: the role of endogenous nitric oxide. Science. 2003;299:896–899. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- Nisoli E, Falcone S, Tonello C, Cozzi V, Palomba L, Fiorani M, Pisconti A, Brunelli S, Cardile A, Francolini M, Cantoni O, Carruba MO, Moncada S, Clementi E. Mitochondrial biogenesis by NO yields functionally active mitochondria in mammals. Proc Natl Acad Sci U S A. 2004;101:16507–16512. doi: 10.1073/pnas.0405432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CK, Barnard RJ, Jasman A, Balon TW. Acute exercise increases nitric oxide synthase activity in skeletal muscle. Am J Physiol Endocrinol Metab. 1999;277:E390–E394. doi: 10.1152/ajpendo.1999.277.2.E390. [DOI] [PubMed] [Google Scholar]
- Roy D, Perreault M, Marette A. Insulin stimulation of glucose uptake in skeletal muscles and adipose tissues in vivo is NO dependent. Am J Physiol Endocrinol Metab. 1998;274:E692–E699. doi: 10.1152/ajpendo.1998.274.4.E692. [DOI] [PubMed] [Google Scholar]
- Schild L, Jaroscakova I, Lendeckel U, Wolf G, Keilhoff G. Neuronal nitric oxide synthase controls enzyme activity pattern of mitochondria and lipid metabolism. FASEB J. 2006;20:145–147. doi: 10.1096/fj.05-3898fje. [DOI] [PubMed] [Google Scholar]
- Schrauwen P, Hesselink MK. Oxidative capacity, lipotoxicity, and mitochondrial damage in type 2 diabetes. Diabetes. 2004;53:1412–1417. doi: 10.2337/diabetes.53.6.1412. [DOI] [PubMed] [Google Scholar]
- Scrogin KE, Hatton DC, Chi Y, Luft FC. Chronic nitric oxide inhibition with L-NAME: effects on autonomic control of the cardiovascular system. Am J Physiol Regul Integr Comp Physiol. 1998;274:R367–R374. doi: 10.1152/ajpregu.1998.274.2.R367. [DOI] [PubMed] [Google Scholar]
- Shesely EG, Maeda N, Kim HS, Desai KM, Krege JH, Laubach VE, Sherman PA, Sessa WC, Smithies O. Elevated blood pressures in mice lacking endothelial nitric oxide synthase. Proc Natl Acad Sci U S A. 1996;93:13176–13181. doi: 10.1073/pnas.93.23.13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srere P. Citrate synthase. In: Lowenstein JM, editor. Methods in Enzymology. vol. 13. New York: Academic Press; 1969. pp. 3–11. [Google Scholar]
- Suwa M, Nakano H, Kumagai S. Effects of chronic AICAR treatment on fiber composition, enzyme activity, UCP3, and PGC-1 in rat muscles. J Appl Physiol. 2003;95:960–968. doi: 10.1152/japplphysiol.00349.2003. [DOI] [PubMed] [Google Scholar]
- Taylor EB, Lamb JD, Hurst RW, Chesser DG, Ellingson WJ, Greenwood LJ, Porter BB, Herway ST, Winder WW. Endurance training increases skeletal muscle LKB1 and PGC-1α protein abundance: effects of time and intensity. Am J Physiol Endocrinol Metab. 2005;289:E960–E968. doi: 10.1152/ajpendo.00237.2005. [DOI] [PubMed] [Google Scholar]
- Valerio A, Cardile A, Cozzi V, Bracale R, Tedesco L, Pisconti A, Palomba L, Cantoni O, Clementi E, Moncada S, Carruba MO, Nisoli E. TNF-α downregulates eNOS expression and mitochondrial biogenesis in fat and muscle of obese rodents. J Clin Invest. 2006;116:2791–2798. doi: 10.1172/JCI28570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadley GD, McConell GK. Effect of nitric oxide synthase inhibition on mitochondrial biogenesis in rat skeletal muscle. J Appl Physiol. 2007;102:314–320. doi: 10.1152/japplphysiol.00549.2006. [DOI] [PubMed] [Google Scholar]
- Wadley GD, Tunstall RJ, Sanigorski A, Collier GR, Hargreaves M, Cameron-Smith D. Differential effects of exercise on insulin-signaling gene expression in human skeletal muscle. J Appl Physiol. 2001;90:436–440. doi: 10.1152/jappl.2001.90.2.436. [DOI] [PubMed] [Google Scholar]
- Wang MX, Murrell DF, Szabo C, Warren RF, Sarris M, Murrell GA. Nitric oxide in skeletal muscle: inhibition of nitric oxide synthase inhibits walking speed in rats. Nitric Oxide. 2001;5:219–232. doi: 10.1006/niox.2001.0348. [DOI] [PubMed] [Google Scholar]