Abstract
Co-assembly of KCNQ1 α-subunits with KCNE1 β-subunits results in the channel complex underlying the cardiac IKs current in vivo. Like other voltage-gated K+ channels, KCNQ1 has a tetrameric configuration. The S6 segment of each subunit lines the ion channel pore with the lower part forming the activation gate. To determine residues involved in protein–protein interactions in the C-terminal part of S6 (S6T), alanine and tryptophan perturbation scans were performed from residue 348–362 in the KCNQ1 channel. Several residues were identified to be relevant in channel gating, as substitutions affected the activation and/or deactivation process. Some mutations (F351A and V355W) drastically altered the gating characteristics of the resultant KCNQ1 channel, to the point of mimicking the IKs current. Furthermore, mutagenesis of residue L353 to an alanine or a charged residue impaired normal channel closure upon hyperpolarization, generating a constitutively open phenotype. This indicates that the L353 residue is essential for stabilizing the closed conformation of the channel gate. These findings together with the identification of several LQT1 mutations in the S6 C-terminus of KCNQ1 underscore the relevance of this region in KCNQ1 and IKs channel gating.
The IKs channel is formed by co-assembly of KCNQ1 (KvLQT1) α-subunits with KCNE1 (minK) β-subunits (Sanguinetti et al. 1996; Barhanin et al. 1996). Co-assembly of the KCNQ1 subunits with KCNE1 results in an increase in current amplitude and an approximately 100-fold slower rate of activation compared with KCNQ1 alone. The IKs current contributes to the ‘repolarizing reserve’ in the ventricular repolarization (Jost et al. 2005; Roden, 2006). Mutations in KCNQ1 can cause a reduction in this ventricular repolarization reserve, leading to the long QT syndrome (LQT1) (Chiang & Roden, 2000; Keating & Sanguinetti, 2001; Kass & Moss, 2003; Moss, 2003).
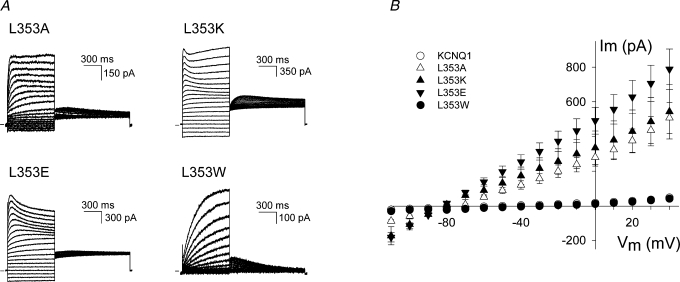
KCNQ1 α-subunits tetramerize to form a voltage-gated potassium channel that responds to changes in membrane potential by opening or closing of its potassium-selective pore. Like other voltage-gated K+ channels, each KCNQ1 α-subunit contains six transmembrane segments (S1–S6) with a voltage-sensing domain formed by S1–S4, and a pore domain (S5, pore loop and S6). The 3D crystal structures of voltage-independent and voltage-dependent bacterial and mammalian K+ channels (Doyle et al. 1998; Jiang et al. 2003; Kuo et al. 2003; Long et al. 2005a) unravelled the molecular mechanism of K+ selectivity and ion permeation through the channel pore. Substituted cysteine accessibility studies showed that in Shaker-type Kv channels, the gate that actively opens and closes the channel pore is formed by the lower part of the S6 segment (S6 tail or S6T) (Liu et al. 1997; del Camino & Yellen, 2001). The exact molecular mechanism underlying the opening and closing of the channel pore is still unknown, although most evidence indicates that kinking or swivelling of the S6 helix at a flexible gating ‘hinge’ results in the reorientation of residues around the S6 bundle crossing (Bright & Sansom, 2004; Grottesi et al. 2005). In most Kv channels, this flexibility is created by a highly conserved glycine residue near the middle of S6 (Ding et al. 2005) or by a PXP motif further downstream in the S6 segment (del Camino et al. 2000; Labro et al. 2003). The crystal structure of Kv1.2 shows that the S6 helix of Shaker-type Kv channels is indeed bent around the PXP motif, thereby orienting the cytoplasmic end of S6 towards the S4–S5 linker (Long et al. 2005a). As a consequence, specific interactions between the S6T and the S4–S5 linker might provide a coupling mechanism between S4 movement and opening or closing of the channel gate (Long et al. 2005b).
Thus, the S6T region plays a central role in channel gating, and the elucidation of the Kv1.2 crystal structure has provided new insights in the structure and function of this region. However, the structure of the S6 segment of KCNQ1 may be different from other Kv channels as KCNQ1 channels lack the glycine in the middle of S6 and contain a PAG sequence instead of PXP (Fig. 1). The PAG motif has been shown to form the ‘hinge’ region in KCNQ1 gating (Seebohm et al. 2006). To determine which residues in the S6T region are important for the gating process of KCNQ1, alanine and tryptophan perturbation scanning of residues 348–362 was performed.
Figure 1. Sequence alignment of the C-terminal part of S6 of several Kv channels.
Sequence alignment of the C-terminal part of S6 shows the conservation of the region between several members of Kv channels. Boxed residues in hERG and HCN2 have been shown to face the S4–S5 linker (Tristani-Firouzi et al. 2002; Decher et al. 2004; Ferrer et al. 2006). In bold, the conserved glycine and the PXP motif. This PXP motif is lacking in KcsA and KvAP. KCNQ1 has a PAG sequence instead of the PXP motif, and it does not possess the conserved glycine.
Methods
Molecular biology
hKCNQ1 was expressed using a pIRES2-EGFP expression vector resulting in separate expression of both genes (BD Biosciences, San Jose, CA, USA). Mutations were introduced with a PCR reaction using mutant primers and the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). After PCR based mutagenesis, a PvuI–EcoRI fragment containing the mutation was cut out of the PCR-amplified vector and ligated in hKCNQ1/pIRES2-EGFP to replace the wild-type sequence. Double-stranded sequencing of the exchanged fragment and the adjacent sequence confirmed the presence of the desired modification and the absence of unwanted mutations. Plasmid DNA for mammalian expression was obtained by amplification in XL2 Bluescript cells (Stratagene), and then isolated from the bacterial cells with the endotoxin-free Maxiprep kit (Sigma, St Louis, MO, USA). The cDNA concentration was determined with UV absorption.
Electrophysiology
CHO-K1 cells were cultured in Ham's F12 medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA). The cells were transfected with 6 μg cDNA of wild-type or mutant KCNQ1 following the fugene transfection method (Roche Diagnostics, Basel, Switzerland). Sixteen hours after transfection the cells were trypsinized and GFP fluorescent cells were used for analysis within 12 h.
Current recordings were made with a Multiclamp-700B amplifier (Axon Instruments, Union City, CA, USA) in the whole-cell configuration of the patch clamp technique. Experiments were performed at room temperature (20–23°C); current recordings were low-pass filtered and sampled at 1–10 kHz with a Digidata 1322A data acquisition system (Axon Instruments). Command voltages and data storage were controlled with pCLAMP8 software (Axon Instruments). Patch pipettes were pulled from 1.2 mm borosilicate glass capillaries (World Precision Instruments, inc., Sarasota, FL, USA) with a P-2000 puller (Sutter Instruments, Novato, CA, USA). After heat polishing, the resistance of the patch pipettes was < 3 MΩ in the standard extracellular solution.
The cells were perfused continuously with a bath solution containing (mm): 145 NaCl, 4 KCl, 1.8 CaCl2, 1 MgCl2, 10 Hepes, 10 glucose, and adjusted to pH 7.35 with NaOH. The pipette solution contained (mm): 110 KCl, 5 K4BAPTA, 5 K2ATP, 1 MgCl2, 10 Hepes and was adjusted to pH 7.2 using KOH. To determine the reversal potentials of the instantaneous current component of mutants L353A, L353E and L353K both a normal external solution, composed as described above, and a high external potassium [K+]o (in mm: 133 NaCl, 16 KCl, 1.8 CaCl2, 1 MgCl2, 10 Hepes, 10 glucose, and adjusted to pH 7.35 with NaOH) solution were used. Junction potentials were zeroed with the filled pipette in the bath solution. The remaining liquid junction potential was estimated to be 1.7 mV and was not corrected. After achieving a gigaohm seal, the whole-cell configuration was obtained by suction. Capacitive transients were elicited to determine the capacitive surface area and access resistance. The access resistance varied from 3 to 9 MΩ without compensation and was below 3 MΩ after whole-cell compensation. Experiments were excluded from analysis if the voltage errors originating from the series resistance were greater than 5 mV.
Data analysis
The holding potential was −80 mV and the interpulse interval was at least 15 s. The voltage protocols were adjusted to adequately determine the biophysical properties of the WT and mutant channels (see figure legends). The time constants for activation and deactivation were determined by fitting the current recordings with a single or double exponential function. The voltage dependence of channel activation was fitted with a Boltzmann equation: y = 1/1 + exp(−(E −V1/2)/k), in which k represents the slope factor, E the applied voltage, and V1/2 the voltage at which 50% of the channels are activated referred to as the midpoint potential. Both the voltage dependence of activation and the slope factor of the Boltzmann equation were used to calculate the Gibbs free energy of activation at 0 mV (ΔG0): ΔG0 = 0.0002389zFV1/2, with the factor 0.0002389 to express the values in kcal mol−1 (Li-Smerin et al. 2000a). ΔΔG0 was calculated as (ΔG0mutant − ΔG0WT). Standard errors of ΔG0 and ΔΔG0 were calculated by linear error propagation according to Yifrach et al. (Yifrach & MacKinnon, 2002).
For mutants L353A, L353K and L353E, current recordings from the voltage ramp protocol were corrected for capacitive currents that were evoked by the voltage ramp. This capacitive current was calculated using the equation: Ic = Cm (dV/dt), where Cm is the cell capacity, and subtracted from the current recordings.
Results are expressed as mean ± s.e.m. with n the number of cells analysed. For the statistical analysis, a Student's unpaired t test was performed.
Molecular modelling
The homology model of the S4–S6 domain of KCNQ1 was based on the Kv1.2 crystal structure (Long et al. 2005a). After aligning the sequence of the S4–S6 sequence of both channels (for the S6 sequence, the alignment shown in Fig. 1 was used), the KCNQ1 homology model was generated using Swiss-Model (http://swissmodel.expasy.org//SWISS-MODEL.html).
Results
Substitution scan of the S6T region in KCNQ1: kinetic analyses of WT and mutant KCNQ1 channels
Due to the sequence differences between KCNQ1 and other Kv channels, there may be subtle yet relevant differences in the intracellular gate region although the overall structure of the S6 segment of KCNQ1 and Kv channels is probably similar (Fig. 1). To evaluate the relevance of each residue in the S6T region, an alanine substitution scan of residues G348 to K362 was performed with the aim to reduce the side chain volume, while preserving a potential α-helical structure (Fig. 1). In addition, the native alanine residue at position 352 and residues with small side chains (G348, G350 and V355) were substituted by a tryptophan to induce a significant increase in side chain volume. Alanine and tryptophan perturbation mutagenesis has been used previously to explore secondary structures and protein-facing residues of both water-exposed and membrane-embedded domains in several types of voltage-gated potassium channels (Hong & Miller, 2000; Li-Smerin et al. 2000a,b; Rocheleau et al. 2006). If the native residue is directed towards the protein interface, both substitutions would result in a sterical disruption of the native structure, presumably leading to altered gating properties. The tryptophan substitution would cause a perturbation by inducing a bulky side chain, while the alanine substitution would lead to a loss of protein–protein contacts by introducing a small side chain. Except for mutations G348W and G350W, all mutant channels generated time-dependent K+ currents in the voltage range between −110 and +70 mV (Table 1). WT KCNQ1 channels elicited currents with an exponential time course and an apparent threshold of activation of approximately −40 mV. Upon membrane repolarization, a slow deactivation process was preceded by a transient increase in current, a ‘hooked tail’, resulting from a rapid recovery of channel inactivation (Fig. 2A) (Barhanin et al. 1996; Sanguinetti et al. 1996). The mutants K354A, Q357A, K358A, R360A, Q361A and K362A generated channels with biophysical properties similar to WT KCNQ1 (Table 1), which indicated that these residues are not located at a crucial protein interface.
Table 1.
Biophysical characteristics for WT KCNQ1 and mutant channels
| Activation | Deactivation | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| V1/2 (mV) (mV) | k (mV) (mV) | n | τ(V1/2 +60) (ms) | n | τ(V1/2 −100) (ms) | τ(V1/2 −50) (ms) | n | ΔG0 | ΔΔG0 | |
| WT KCNQ1 | −1.8 ± 2.5 | 16 ± 0.8 | 8 | 21 ± 2.0 | 15 | 100 ± 11 | 410 ± 56 | 13 | −0.07 ± 0.09 | |
| G348A | −25 ± 0.5 | 10 ± 1.0 | 5 | 63 ± 7.0 | 8 | 120 ± 13 | 320 ± 36 | 5 | −1.47 ± 0.15 | −1.39 ± 0.17 |
| G348W | NC | NC | — | NC | — | NC | NC | — | — | — |
| S349A | 24 ± 2.0 | 15 ± 2.0 | 5 | 390 ± 57 | 8 | 120 ± 28* | 680 ± 81 | 4 | 0.94 ± 0.15 | 1.01 ± 0.17 |
| G350A | 26 ± 3.4 | 16 ± 1.0 | 5 | 16 ± 1.8 | 9 | 130 ± 44 | 290 ± 27 | 5 | 0.95 ± 0.14 | 1.02 ± 0.17 |
| G350W | NC | NC | — | NC | — | NC | NC | — | — | — |
| G348AG350A | 9.2 ± 2.7 | 15 ± 1.0 | 5 | 120 ± 7.7 | 5 | 130 ± 17 | 530 ± 36 | 5 | 0.36 ± 0.11 | 0.43 ± 0.14 |
| F351A | 46 ± 1.8 | 8.4 ± 0.6 | 5 | 640 ± 81** | 9 | 170 ± 21 | 1000 ± 130*** | 5 | 3.21 ± 0.26 | 3.28 ± 0.28 |
| A352W | 22 ± 3.5 | 14 ± 1.2 | 5 | 240 ± 30 | 10 | 185 ± 46* | 470 ± 43 | 6 | 0.92 ± 0.16 | 0.99 ± 0.19 |
| L353A | −1.0 ± 2.1 | 18 ± 2.0 | 5 | 28 ± 2.1 | 10 | 150 ± 12 | 570 ± 34 | 9 | −0.03 ± 0.07 | 0.04 ± 0.11 |
| L353W | 15 ± 3.0 | 16 ± 0.8 | 6 | 210 ± 25 | 8 | 230 ± 30* | 1500 ± 110 | 6 | 0.55 ± 0.11 | 0.62 ± 0.15 |
| L353K | 2.7 ± 3.8 | 19 ± 2.1 | 5 | 10 ± 2.1 | 5 | 320 ± 50 | 1800 ± 170 | 7 | 0.08 ± 0.12 | 0.16 ± 0.15 |
| L353E | −22 ± 3.0 | 17 ± 1.1 | 3 | 28 ± 6.6 | 5 | 170 ± 40 | 280 ± 78 | 4 | −0.76 ± 0.11 | −0.69 ± 0.15 |
| L353P | NC | NC | — | NC | — | NC | NC | — | — | — |
| K354A | −1.7 ± 2.1 | 14 ± 0.9 | 5 | 29 ± 1.7 | 10 | 96 ± 23 | 390 ± 77 | 7 | −0.07 ± 0.09 | 0.00 ± 0.13 |
| V355A | −8.0 ± 2.5 | 12 ± 0.9 | 5 | 53 ± 4.8 | 8 | 73 ± 4.1 | 170 ± 53 | 5 | −0.39 ± 0.12 | −0.32 ± 0.16 |
| V355W | 29 ± 2.9 | 12 ± 1.0 | 6 | 380 ± 60** | 8 | 58 ± 11 | 260 ± 38 | 6 | 1.42 ± 0.18 | 1.49 ± 0.20 |
| Q356A | 8.4 ± 2.2 | 12 ± 1.1 | 5 | 24 ± 1.7 | 5 | 300 ± 96 | 2100 ± 260 | 6 | 0.41 ± 0.11 | 0.48 ± 0.15 |
| Q357A | −0.2 ± 2.7 | 16 ± 1.7 | 5 | 29 ± 1.7 | 10 | 93 ± 5.7 | 300 ± 31 | 5 | −0.01 ± 0.10 | 0.06 ± 0.14 |
| K358A | 4.1 ± 2.4 | 13 ± 0.8 | 6 | 30 ± 5.5 | 6 | 120 ± 15 | 510 ± 74 | 5 | 0.18 ± 0.11 | 0.26 ± 0.14 |
| Q359A | 8.5 ± 2.1 | 18 ± 1.5 | 6 | 28 ± 2.3 | 9 | 220 ± 33 | 1400 ± 120 | 6 | 0.28 ± 0.07 | 0.35 ± 0.12 |
| R360A | 6.8 ± 2.5 | 15 ± 1.2 | 5 | 21 ± 1.5 | 12 | 120 ± 11 | 390 ± 47 | 5 | 0.27 ± 0.10 | 0.34 ± 0.14 |
| Q361A | 0.8 ± 1.8 | 16 ± 0.9 | 6 | 24 ± 3.3 | 9 | 120 ± 13 | 580 ± 82 | 4 | 0.03 ± 0.07 | 0.10 ± 0.11 |
| K362A | 6.8 ± 0.7 | 18 ± 1.7 | 5 | 36 ± 5.2 | 8 | 78 ± 14 | 450 ± 130 | 4 | 0.22 ± 0.03 | 0.29 ± 0.10 |
Values are means ± s.e.m. The voltage dependence of activation of the channels is represented by V1/2 and k which are the midpoint of activation and the inverted factor of the slope of the used Boltzmann equation, respectively. Time constants were derived from mono-exponential fits to activating or deactivating currents. τ(V1/2 +60mV) is the time constant of activation at the potential V1/2 +60 mV. τ(V1/2 −50mV) and τ(V1/2 −100mV) are the time constants for deactivation at the potentials V1/2 −50 mV and V1/2 −100 mV, respectively. n represents the number of cells analysed. ΔG0 represents the Gibbs free energy of activation at 0 mV. ΔΔG0 was calculated as (ΔG0mutant − ΔG0WT). ΔG0 and ΔΔG0 are expressed in kcal mol−1. ΔΔG0 values in bold were above or approached the threshold (see text). Standard errors of ΔG0 and ΔΔG0 were calculated according to Yifrach et al. (Yifrach & MacKinnon, 2002). L353P is a mutant involved in LQT1 (Splawski et al. 1998). For the activation of the mutant Q356A, a double exponential fit was used; the time constant of the fast component is shown.
Determined at −90 mV
determined at +80 mV
determined at −20 mV. NC, no current observed between −110 mV and +70 mV.
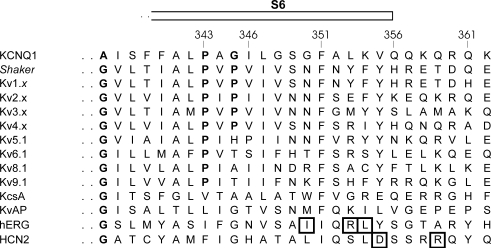
Figure 2. Biophysical properties of WT KCNQ1 and mutant channels in region 348–350.
A, representative current traces for the WT KCNQ1 channel and several mutants expressed in CHO-K1 cells. Cells were clamped at a holding potential of −80 mV, and 800–1500 ms pulses to voltages between −60 to +70 mV were imposed in steps of 10 mV. Tail currents were recorded by stepping to −40 mV (note the differences in scale bars). B, voltage dependence of activation. Activation curves were obtained by plotting the normalized tail currents as a function of the step potential. The continuous lines represent the average Boltzmann fit and were compared with WT (dotted line). Parameters are shown in Table 1. C, activation and deactivation time constants derived from mono-exponential fits to the raw current traces were plotted as a function of applied potential. Mutation S349A slowed the activation kinetics without affecting the channel deactivation rate. The mutant channel G350A showed an accelerated deactivation between potentials −20 to −70 mV.
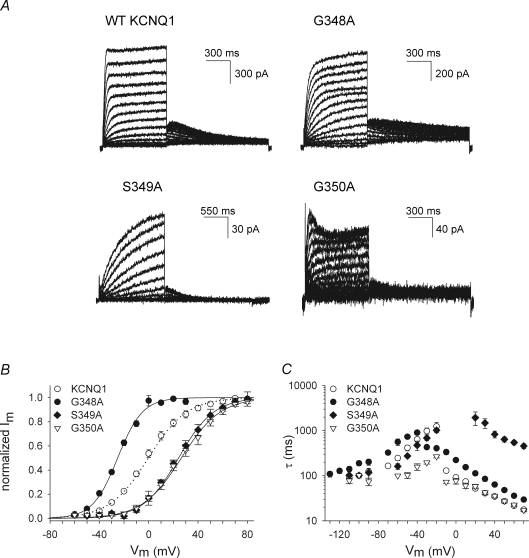
Some mutations (G348A, S349A, G350A, F351A, A352W, V355W) affected the gating properties of the mutant channel profoundly (Figs 2 and 3, Table 1). The mutant G348A shifted the voltage dependence of activation 25 mV towards negative potentials compared with WT, while the S349A and G350A mutations resulted in a significant positive shift (Fig. 2B). In addition to the shifts in voltage dependence of activation, the gating kinetics of the mutant S349A were strongly altered compared with WT (Fig. 2C). To account for the shift in the voltage dependence of activation, the time constants of activation were calculated at the midpoint (V1/2) + 60 mV, and the deactivation time constants were calculated at (V1/2) −50 mV and (V1/2) −100 mV (Table 1). Mutating residues G348 and G350 to a tryptophan was not tolerated since mutant channels G348W and G350W failed to display potassium selective currents within the voltage range of −110 to +70 mV. These mutants were retained in the endoplasmic reticulum (ER) as was shown by confocal imaging (Supplemental Fig. S1). At position 355, an alanine substitution caused a pronounced increase in inactivation compared with WT (P < 0.001; at +70 mV: V355A 38 ± 4%, n = 8; WT KCNQ1 11 ± 3%, n = 8). This was associated with a 2- to 3-fold slowing of the activation time constants (Fig. 3A and C, Table 1). The mutants F351A, A352W and V355W displayed a pronounced shift of the voltage dependence towards positive potentials and a marked slowing of the activation kinetics (Fig. 3B and C). The V355W mutant further demonstrated an acceleration of the deactivation kinetics (Fig. 3C, Table 1).
Figure 3. Biophysical properties of mutants in region 351–355.
A, representative current traces for four mutants in KCNQ1 obtained with a protocol as in Fig. 2A. For the F351A mutant, cells were clamped at a holding potential of −80 mV and 4000 ms pulses between −10 to +70 mV were imposed. Tail currents were recorded by stepping to −40 mV. B, voltage dependence of activation obtained as in Fig. 2B. The continuous lines indicate the average Boltzmann fits with the parameters shown in Table 1. C, activation and deactivation time constants obtained from a mono-exponential fit to the raw current traces. The F351A and A352W mutations caused a slowing of the activation kinetics without affecting deactivation. The V355W mutation slowed the activation and accelerated the deactivation kinetics.
At positions 356 and 359, the mutants Q356A and Q359A slowed the deactivation kinetics by 2- to 3-fold, without affecting the voltage dependence of activation (Table 1). In addition to the slower deactivation, the Q356A mutant displayed a bi-exponential activation time course. The fast component was comparable to the WT activation kinetics, while the time constants of the second component were approximately 8-fold slower (τ at +70 mV: ∼200 ms) (Supplemental Fig. 2S).
In order to quantify the observed effects on the voltage dependence of activation, ΔΔG0 was calculated for the several mutations (Table 1). Using a threshold of 1 kcal mol−1 for a relevant perturbation, five mutants scored as ‘high impact’ (G348A, S349A, G350A, F351A, V355W), and one mutant approached the threshold value (A352W) (Hong & Miller, 2000; Li-Smerin et al. 2000a,b; Rocheleau et al. 2006).
In summary, alanine or tryptophan substitutions at several positions in the S6T region of KCNQ1 resulted in channels with altered biophysical properties, which indicated that these residues contribute to a critical protein–protein interface within the KCNQ1 protein.
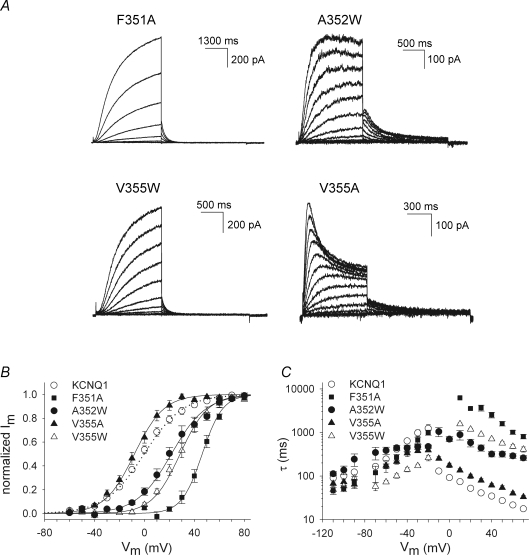
Constitutively conducting channel L353A
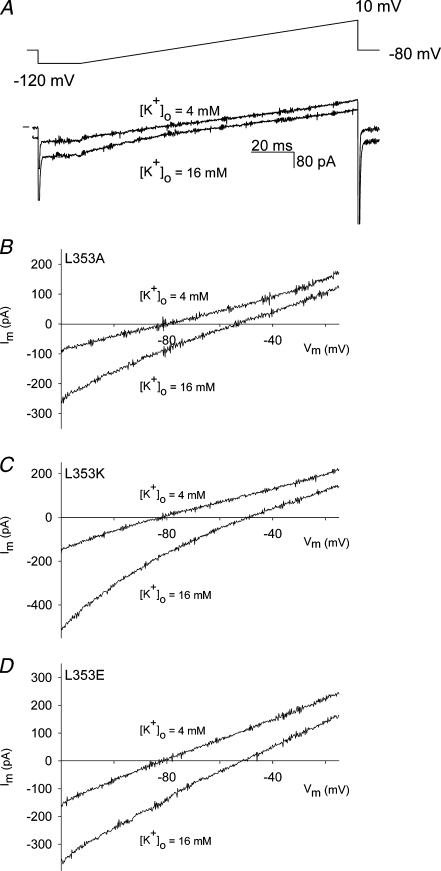
A remarkable behaviour was observed when the L353 residue in KCNQ1 was mutated to an alanine. The gating properties of the time-dependent component of the L353A current were not significantly different from WT KCNQ1 (Table 1). However, the L353A mutant displayed a constitutive conductance at hyperpolarized potentials (Fig. 4). To further examine the potential relevance of L353 in the gating process of KCNQ1, this residue was substituted by a tryptophan, a lysine and a glutamate introducing a bulky hydrophobic and a positively and negatively charged amino acid, respectively. Figure 4 shows the raw current traces for these L353 substitutions. The activation kinetics for the L353A, L353K and L353E mutants were comparable to WT KCNQ1, but were slowed for the L353W mutant. For the L353K and L353E mutants, a large fraction of the current was instantaneous, similar to the results for the L353A mutant. At +70 mV the ratio of voltage-independent to total current (voltage-independent + peak voltage-dependent current) was: for L353A, 54 ± 2% (n = 16); L353K, 70 ± 5% (n = 9); L353E, 81 ± 2% (n = 11) (Fig. 4). In contrast, the L353W mutant did not show an instantaneous current component. Furthermore, the L353K mutant showed a modest inward rectification behaviour that was not observed for the WT KCNQ1 channel or the L353A, L353E and L353W mutants which all showed a linear I–V relationship (Supplemental Fig. 3S). The inward rectification of the L353K mutant was not due to increased inactivation in comparison with WT KCNQ1, as was shown by tail current analysis in high Rb+ and high K+ using the approach of previous studies (Pusch et al. 2000; Seebohm et al. 2003) (Supplemental Fig. 4S).
Figure 4. L353A, L353K and L353E display a constitutively conducting phenotype.
A, representative current traces for the mutants L353A, L353K, L353E and L353W. The cells were clamped at a holding potential of −80 mV, and 800 ms pulses to voltages between −110 to +70 mV were imposed in steps of 10 mV. Tail currents were recorded by stepping to −40 mV. The horizontal bar on the left indicates the zero current level. B, instantaneous current amplitudes (determined after the capacitive transient) as a function of pulse potential for the WT KCNQ1 and mutants L353A (n = 16), L353K (n = 9), L353E (n = 8) and L353W (n = 14). For the L353A, L353K and L353E channels a clear instantaneous current was observed at voltages below −20 mV with a reversal potential of −80 mV, while the L353W mutant was able to close normally.
To further investigate whether the instantaneous current component observed for mutants L353A, L353K and L353E originated from a potassium-selective conductance, we determined the reversal potential both in normal extracellular solution ([K+]o = 4 mm) and in an external solution with an elevated concentration of potassium ([K+]o = 16 mm), using a voltage ramp protocol (Fig. 5). During the time frame of this ramp protocol, no significant activation of the voltage-dependent current component occurred, as no obvious deactivating current was observed upon repolarization to −80 mV in the high [K+]o solution (Fig. 5A). In the normal [K+]o solution, the reversal potential for the instantaneous component was around −80 mV (L353A: −81 ± 1 mV, n = 17; L353K: −81 ± 1 mV, n = 13; L353E: −82 ± 1 mV, n = 5), while after wash-in of the elevated [K+]o solution the reversal potential shifted in a positive direction (L353A: −52 ± 1 mV, n = 9; L353K: −47 ± 1 mV, n = 6; L353E: −51 ± 1 mV, n = 5), as would be expected from the shift in the Nernst potential for a potassium-selective current (Fig. 5B, C and D).
Figure 5. Reversal potential of the instantaneous current component for mutants L353A, L353K and L353E obtained with a voltage ramp.
A, representative current traces for the mutant L353A obtained with a voltage ramp shown on top in normal external K+ solution ([K+]o = 4 mm) (upper current trace) and after wash-in with high external K+ solution ([K+]o = 16 mm) (lower current trace). Horizontal bar on the left indicates the zero current level. During the time frame of the ramp protocol, no significant activation of the voltage-dependent current occurred. B, C and D, current–voltage relationships from the ramp protocol for L353A, L353K, L353E after correction for the capacitive current. In the normal external K+ solution (4 mm), the reversal potential was around −80 mV, while in the high K+ solution (16 mm) the reversal potential was approximately −54 mV as would be expected for a K+-selective current.
In summary, mutating residue L353 to an alanine, a lysine and a glutamate resulted in a constitutively conducting phenotype, a behaviour that was not observed when the L353 residue was mutated to a tryptophan.
Discussion
Perturbation mutagenesis of the S6T in KCNQ1 indicates marked effects on channel gating
Mutagenesis studies in several Kv channels have indicated the involvement of the S6T region in channel gating (Ding & Horn, 2002, 2003; Hackos et al. 2002; Rich et al. 2002).
During gating, the S6 segment is thought to undergo rotational and angular movements (Perozo et al. 1999; Fedida & Hesketh, 2001; Liu et al. 2001). Therefore, the S6T region is expected to be highly dynamic and it may display different state-dependent interactions during gating, including transient ones. To evaluate the relevance of each residue for channel gating in the S6T region of KCNQ1, we performed a perturbation scan with the intention of disrupting protein–protein interactions. Because the protein interface of the S6T within the channel protein may differ in the open and closed state, perturbation mutagenesis in this dynamic region is expected to affect both states and the transitional state between them.
Mutations of several residues did not affect channel gating (K354, Q357, K358, R360, Q361, K362) and therefore it is unlikely that these residues would be directed towards the protein interface. These residues are probably facing the lipid or an aqueous environment in the KCNQ1 protein and were classified as ‘no impact’. In contrast, other mutations strongly affected the gating characteristics of the mutant channels. Based on the ΔΔG0 values, several mutations were classified as ‘high impact’ (G348A, S349A, G350A, F351A, A352W, V355W). The mutant G348A displayed a negative ΔΔG0 value signifying that the mutation caused a relative stabilization of the open state over the closed state. The S349A, G350A, F351A, A352W and V355W mutations resulted in a positive value of ΔΔG0 indicating that these mutations caused a relative stabilization of the closed state over the open state (Li-Smerin et al. 2000a).
Between positions 348 and 350, alanine substitutions markedly altered the gating characteristics with |ΔΔG0| values above the threshold of 1 kcal mol−1. In addition, the S349A mutation caused a strong slowing of the activation kinetics. Thus, even relatively small alterations in side chain volume at these positions resulted in a severe disruption of the gating properties of the resultant channels (residue volumes for glycine: 60.1 Å3; serine: 89.0 Å3; alanine: 88.6 Å3). Tryptophan substitutions (residue volume: 227.8 Å3) of residues G348 and G350 disrupted the trafficking of the mutant channel proteins, as was shown by confocal microscopy (Supplemental Fig. S1), and no current was observed between −110 mV and +70 mV.
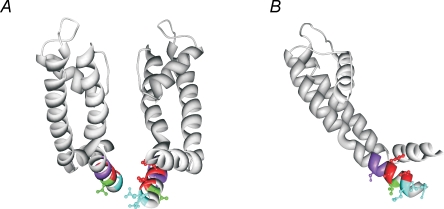
Due to their small size, glycine residues have been shown to be important in protein packing or folding (Russ & Engelman, 2000); the lack of a side chain provides an interaction surface for the side chains of other residues. As mutation G348A caused a relative stabilization of the open state (negative ΔΔG0 value) and mutations S349A and G350A resulted in a relative stabilization of the closed state (positive ΔΔG0 value), these residues are probably directed towards a different protein environment (Li-Smerin et al. 2000a). To identify the potential protein interfaces of these residues, a homology model for KCNQ1 was constructed based on the Kv1.2 crystal structure, similar to the approach used by Seebohm et al. (Long et al. 2005a; Seebohm et al. 2006). In this homology model, the GSG residues are intercalated between the bottom of the S5 helix and the S4–S5 linker (Fig. 6). Therefore, substitution of these residues by an alanine or tryptophan would disturb the putative protein interactions at these interfaces.
Figure 6. Homology model of the S6T region of KCNQ1 based on the Kv1.2 crystal structure.
A and B, homology model of KCNQ1 based on the Kv1.2 crystal structure, which is representative for the open state. In this model, ‘high impact’ residues G348, S349 and G350 are indicated in purple. These residues are intercalated between the S4–S5 linker and the S5 segment. Starting at residue F351, the side chains of the ‘high impact’ residues F351, A352 and V355 are indicated in red. Residues F351 and V355 are directed towards the S4–S5 linker. The residue L353 (indicated in green) is orientated downwards in the channel pore. Residues Q356 and Q359 (indicated in blue), both affecting the deactivation rate upon substitution, are directed towards the channel pore.
Because of the helix-destabilizing effect of glycine residues, the G348 and G350 residues could also result in a flexible region or a kink in the helix (O'Neil & DeGrado, 1990; Blaber et al. 1993). However, the double mutant channel G348A–G350A generated robust voltage- and time-dependent K+ current (Table 1), arguing against a requirement for flexibility around residues G348 and G350.
Mutations F351A and V355W generate an IKs-like phenotype
Between positions 351 and 362, alanine or tryptophan substitution of residues F351, A352 and V355 resulted in a strong shift of the voltage dependence of activation towards positive potentials (ΔΔG0 ≥ 1 kcal mol−1) and a slowing of the activation kinetics. In addition, mutant V355W accelerated the deactivation kinetics. These gating alterations reflect a relative destabilization of the open state versus the closed state or a stabilization of the closed state of the channel over the open state. Combined with the absence of obvious inactivation or ‘hooked’ tail currents, mutations F351A and V355W caused an ‘IKs-like’ phenotype during channel opening (Fig. 3).
As these ‘high impact’ mutations had a drastic effect on channel gating, it is likely that these substitutions disrupt protein–protein interactions in the KCNQ1 channel (Hong & Miller, 2000; Li-Smerin et al. 2000a,b; Rocheleau et al. 2006). Although the exact mechanism for electromechanical coupling is not unravelled yet, several studies in different types of Kv channels demonstrated the importance of specific interactions between the activation gate region and the S4–S5 linker (Lu et al. 2002; Tristani-Firouzi et al. 2002; Decher et al. 2004; Ferrer et al. 2006). The Kv1.2 crystal structure further showed that the S4–S5 linker crosses over the S6T region of the same subunit, forming several amino acid contacts with it (Long et al. 2005a,b).
Based on these studies, the S4–S5 linker could form the protein interaction site of the ‘high impact’ residues F351, A352 and V355 in the KCNQ1 channel. However, we did not determine the interacting protein interface, and therefore can not exclude other channel regions.
In the homology model of KCNQ1 based on the Kv1.2 structure, which is representative for the open state, the residues F351 and V355 might indeed be directed towards the S4–S5 linker (Fig. 6).
As mutants F351A and V355W mimic the cardiac IKs current during channel opening, it is tempting to speculate that these residues are relevant in the IKs channel. The exact binding site and location of KCNE1 in the IKs channel has been controversial. Most evidence indicates that the KCNE1 subunit is located near the S6 helices of KCNQ1 and specific interactions between the S6 segment and KCNE1 have been reported (Tai & Goldstein, 1998; Tapper & George., 2001; Melman et al. 2001, 2002, 2004; Chen et al. 2003a; Panaghie et al. 2006). However, a recent study suggested the proximity of the KCNE1 subunit with the S4 segment of KCNQ1 (Nakajo & Kubo, 2007). In addition, the mechanism of gating modulation of the KCNQ1 channel by the KCNE1 subunit has been a subject of debate. Association of KCNE1 with the KCNQ1 channel might induce a local conformational change in the KCNQ1 protein (Lerche et al. 2000) or modulate a transition state of the KCNQ1 channel during channel opening or closing (Melman et al. 2001). Recently, it has been proposed that KCNE1 impacts on the S4 movement, directly by interacting with S4 or indirectly by binding to the pore domain (Nakajo & Kubo, 2007). The F351A and V355W mutations could simulate the effect of KCNE1 on the KCNQ1 protein by inducing a local conformational change in the KCNQ1 protein similar to the one possibly caused by the KCNE1 subunit or by modulating the transitional states which could be affected by KCNE1 during the channel gating process.
Mutations Q356A and Q359A slow the deactivation kinetics
Although activation of the Q356A mutant showed a bi-exponential time course, alanine substitutions of both Q356 and Q359 did not induce a significant alteration of the voltage dependence of activation, and the ΔΔG0 value did not reach the cutoff. However, the deactivation kinetics were slowed by 2- to 3-fold. Deactivation kinetics have been used previously as an additional parameter for perturbation mutagenesis, but a specific cutoff value has not been defined (Monks et al. 1999; Hong & Miller, 2000). Because of their more subtle effects, we classified these mutants as ‘low impact’ (Table 1). The slower deactivation for these mutants suggests that these residues might contribute to a protein interface in the closed state or during the closing transition. In the KCNQ1 homology model, these residues are directed towards the channel pore (Fig. 6). To orientate residue Q356 and Q359 towards the protein interface, a slight rotation of the S6T region would be needed. This could indicate that the S6T region of KCNQ1 is swivelled in comparison with the situation in Kv1.2, although no experimentalevidence arguing against or in favour of this hypothesis is present at the moment. However, as the S6T region forms a dynamic region during channel gating and the Kv1.2 crystal structure is representative for the open state, it is conceivable that this swivel motion occurs during the gating process, thereby reorienting residues Q356 and Q359 towards the protein interface upon channel closure.
The L353 residue is crucial for normal KCNQ1 channel closure
When residue L353 was replaced by an alanine, the resultant channel was unable to close completely. A large fraction of the conductance was voltage independent, while the remaining fraction was voltage dependent with gating properties similar to WT (Fig. 4). Both the voltage-independent and voltage-dependent current component were potassium selective, which indicated that they originated from a potassium-selective flux through the channel pore (Fig. 5). According to the Nernst equation, the reversal potential in the 4 mm [K+]o solution would be −89 mV, in the 16 mm [K+]o solution the reversal potential would be −54 mV. The slight difference between the experimental and the calculated values of the reversal potential can be attributed partly to the remaining junction potential which was not corrected and partly to the small but finite relative sodium conductance (e.g. for Kv 1.5: PNa/PK = 0.007; for KCNQ2/Q3: PNa/PK < 0.005) (Snyders et al. 1993; Prole & Marrion, 2004). Thus, although this mutant did not reach the cutoff value for ΔΔG0 and thus could not be classified as ‘high impact’ in terms of closed–open equilibrium, it is essential for channel closure because of the drastic disturbance of the closed state.
To further investigate the role of the L353 residue in the KCNQ1 channel, the residue was mutated to a lysine (residue volume: 168.6 Å3), a glutamate (residue volume: 138.4 Å3) and a tryptophan (residue volume: 227.8 Å3). The lysine and glutamate resulted in a constitutively conducting phenotype, though the L353W mutant was able to close normally (Fig. 4).
Several mutations which resulted in a constitutively conducting channel have been described for the KCNQ1 channel. Mutations S140G and V141M located in the S1 domain of KCNQ1 generated constitutively open IKs channels (Chen et al. 2003b; Hong et al. 2005). Furthermore, the KCNQ1 channel becomes constitutively open by association with KCNE2 and KCNE3 to generate potassium ‘leak’ current in gastrointestinal epithelia. This phenomenon was coupled to a limited amount of net positive charge in the S4 segment of KCNQ1 facilitating the conversion of KCNQ1 in a voltage-independent channel (Panaghie & Abbott, 2007).
Several mechanisms might explain the constitutively conducting phenotype of the L353A, L353E and L353K mutants. For the Shaker channel, it has been argued that a constitutively conducting phenotype could be the result of a disturbed closed–open equilibrium in favour of the open state, enabling opening of the activation gate without voltage sensor activation (Sukhareva et al. 2003). Indeed, a less stringent interaction between the activation gate and the voltage sensor could destabilize the closed conformation of the channel as was also shown in the hERG channel (Ferrer et al. 2006). In the hERG channel, L666 was implicated in restoring normal channel closure of a constitutively open HBS6 chimera. This L666 residue is located adjacent to the residue homologous to residue L353 in KCNQ1. In addition, a specific interaction was identified between residue L666 in the S6T region and residue D540 in the S4–S5 linker (Ferrer et al. 2006).
The observed disruption of channel closure of the mutants L353A, L353E and L353K could be attributed to a loss of stabilizing protein–protein contacts with the channel core. Seebohm et al. suggested the presence of hydrophobic protein contacts in KCNQ1 between residues L273 (S5), V310 (pore helix) and F340 (S6) which would stabilize the closed state of the channel and substitutions at these positions generated a constitutively conducting phenotype (Seebohm et al. 2005). As a leucine residue has a large hydrophobic side chain, the constitutively open phenotype could be due to a loss of hydrophobic interaction with the channel core. Thus, it is conceivable that residue L353 contributes to a crucial protein interaction within the channel protein in the closed state although our experiments did not elucidate the interacting protein interface. Substitution by an alanine or a charged residue would destabilize the closed conformation of the activation gate leading to voltage-independent opening of the channel.
Homology modelling based on the Kv1.2 crystal structure suggested that the L353 residue is directed downwards in the channel pore (Fig. 6). However, as this structure is representative for the open state, a reorientation of the L353 residue towards the channel protein could occur during channel closure.
Another possible mechanism for constitutive conductance is a direct disruption of the seal of the channel pore in the closed state. This would imply that the L353 residue forms the activation ‘gate residue’ of the channel. At position 353, the bulky hydrophobic tryptophan did not display constitutive conduction, which suggests that it allowed the channel to close normally. In contrast, the charged lysine and glutamate did not and the channels were constitutively open possibly by electrostatic repulsion between the four lysine and glutamate residues in the tetrameric configuration (Fig. 4). The altered rectification of the L353K mutant could indicate that the L353 residue faces the permeation pathway (Supplemental Fig. 3S) as the local electrical field induced by the four positive charges is expected to produce inward rectification compared with WT (Perez-Cornejo & Begenisich, 1994). However, the inward rectification behaviour of the L353K mutant could be simulated by increasing the inner barrier by only 0.5 RT (RT = 583 cal/mol) and introduction of a negative charge at position 353 (L353E) did not result in an opposite rectifying behaviour. Thus, these results do not provide conclusive evidence that the L353 residue would face the centre of the pore, forming the activation ‘gate residue’ of KCNQ1.
LQT1 mutants in the S6T region underscore its relevance in the IKs channel
The observation that disease-causing LQT1 mutations (S349W, S349P, G350R, F351S, L353P) have been identified at several positions in the S6T region that we showed to be of ‘high impact’ for gating or crucial for channel closure, further emphasizes the relevance of this region in KCNQ1 and IKs channel functionality (Splawski et al. 1998, 2000; Napolitano et al. 2005). The LQT1 mutation L353P was further examined. Our results show that the L353P mutant channel failed to generate a K+ current in the voltage range of −110 mV to +70 mV (Table 1) possibly by disruption of the channel structure as the mutant showed impaired trafficking, which was confirmed by confocal microscopy (Supplementary Fig. S1).
In summary, alanine/tryptophan mutagenesis in the S6T region of KCNQ1 identified several residues relevant in channel gating. Most mutations affected the voltage dependence and kinetics of activation. Some ‘high impact’ residues mimicked the IKs current. Furthermore, introduction of an alanine or a charged amino acid at position 353 resulted in a constitutively conducting phenotype arguing for a crucial role of the L353 residue in the closed conformation of the activation gate region of KCNQ1.
Acknowledgments
We would like to thank Tessa de Block, Tine Bruyns and Evy Mayeur for their excellent technical assistance and Jean-Pierre Timmermans for the use of the confocal microscope. I.R.B. and A.J.L. are doctoral and post-doctoral fellows with the ‘Fonds voor Wetenschappelijk Onderzoek Vlaanderen’ (FWO). This work was supported by grants of the ‘Fonds voor Wetenschappelijk Onderzoek Vlaanderen’ FWO-0085.04, FWO-0152.06, FWO-1.5.047.05, FWO-1.5.044.07.
Supplemental material
Online supplemental material for this paper can be accessed at:
http://jp.physoc.org/cgi/content/full/jphysiol.2007.145813/DC1 and http://www.blackwell-synergy.com/doi/suppl/10.1113/jphysiol.2007.145813
References
- Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, Romey G. K(v)LQT1 and IsK (minK) proteins associate to form the IKs cardiac potassium current. Nature. 1996;384:78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- Blaber M, Zhang XJ, Matthews BW. Structural basis of amino acid alpha helix propensity. Science. 1993;260:1637–1640. doi: 10.1126/science.8503008. [DOI] [PubMed] [Google Scholar]
- Bright JN, Sansom MSP. Kv channel S6 helix as a molecular switch: simulation studies. IEE Proc – Nanobiotechnology. 2004;151:17–27. doi: 10.1049/ip-nbt:20040101. [DOI] [PubMed] [Google Scholar]
- Chen H, Sesti F, Goldstein SA. Pore- and state-dependent cadmium block of Iks channels formed with Mink-55C and wild-type KCNQ1 subunits. Biophys J. 2003a;84:3679–3689. doi: 10.1016/S0006-3495(03)75097-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Xu SJ, Bendahhou S, Wang XL, Wang Y, Xu WY, Jin HW, Sun H, Su XY, Zhuang QN, Yang YQ, Li YB, Liu Y, Xu HJ, Li XF, Ma N, Mou CP, Chen Z, Barhanin J, Huang W. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003b;299:251–254. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- Chiang CE, Roden DM. The long QT syndromes: genetic basis and clinical implications. J Am Coll Cardiol. 2000;36:1–12. doi: 10.1016/s0735-1097(00)00716-6. [DOI] [PubMed] [Google Scholar]
- Decher N, Chen J, Sanguinetti MC. Voltage-dependent gating of hyperpolarization-activated, cyclic nucleotide-gated pacemaker channels: molecular coupling between the S4–S5 and C-linkers. J Biol Chem. 2004;279:13859–13865. doi: 10.1074/jbc.M313704200. [DOI] [PubMed] [Google Scholar]
- del Camino D, Holmgren M, Liu Y, Yellen G. Blocker protection in the pore of a voltage-gated K+ channel and its structural implications. Nature. 2000;403:321–325. doi: 10.1038/35002099. [DOI] [PubMed] [Google Scholar]
- del Camino D, Yellen G. Tight steric closure at the intracellular activation gate of a voltage- gated K+ channel. Neuron. 2001;32:649–656. doi: 10.1016/s0896-6273(01)00487-1. [DOI] [PubMed] [Google Scholar]
- Ding S, Horn R. Tail end of the s6 segment: role in permeation in shaker potassium channels. J Gen Physiol. 2002;120:87–97. doi: 10.1085/jgp.20028611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S, Horn R. Effect of S6 tail mutations on charge movement in Shaker potassium channels. Biophys J. 2003;84:295–305. doi: 10.1016/S0006-3495(03)74850-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S, Ingleby L, Ahern CA, Horn R. Investigating the putative glycine hinge in shaker potassium channel. J Gen Physiol. 2005;126:213–226. doi: 10.1085/jgp.200509287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle DA, Cabral JM, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- Fedida D, Hesketh JC. Gating of voltage-dependent potassium channels. Prog Biophys Mol Biol. 2001;75:165–199. doi: 10.1016/s0079-6107(01)00006-2. [DOI] [PubMed] [Google Scholar]
- Ferrer T, Rupp J, Piper DR, Tristani-Firouzi M. The S4–S5 linker directly couples voltage sensor movement to the activation gate in the human ether-a-go-go-related gene (hERG) K+ channel. J Biol Chem. 2006;281:12858–12864. doi: 10.1074/jbc.M513518200. [DOI] [PubMed] [Google Scholar]
- Grottesi A, Domene C, Hall B, Sansom MS. Conformational dynamics of M2 helices in KirBac channels: helix flexibility in relation to gating via molecular dynamics simulations. Biochem. 2005;44:14586–14594. doi: 10.1021/bi0510429. [DOI] [PubMed] [Google Scholar]
- Hackos DH, Chang TH, Swartz KJ. Scanning the intracellular s6 activation gate in the shaker K+ channel. J Gen Physiol. 2002;119:521–532. doi: 10.1085/jgp.20028569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K, Piper DR, Diaz-Valdecantos A, Brugada J, Oliva A, Burashnikov E, Santos-de-Soto J, Grueso-Montero J, Diaz-Enfante E, Brugada P, Sachse F, Sanguinetti MC, Brugada R. De novo KCNQ1 mutation responsible for atrial fibrillation and short QT syndrome in utero. Cardiovasc Res. 2005;68:433–440. doi: 10.1016/j.cardiores.2005.06.023. [DOI] [PubMed] [Google Scholar]
- Hong KH, Miller C. The lipid–protein interface of a Shaker K+ channel. J Gen Physiol. 2000;115:51–58. doi: 10.1085/jgp.115.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- Jost N, Virag L, Bitay M, Takacs J, Lengyel C, Biliczki P, Nagy Z, Bogats G, Lathrop DA, Papp JG, Varro A. Restricting excessive cardiac action potential and QT prolongation: a vital role for IKs in human ventricular muscle. Circulation. 2005;112:1392–1399. doi: 10.1161/CIRCULATIONAHA.105.550111. [DOI] [PubMed] [Google Scholar]
- Kass RS, Moss AJ. Long QT syndrome: novel insights into the mechanisms of cardiac arrhythmias. J Clin Invest. 2003;112:810–815. doi: 10.1172/JCI19844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating MT, Sanguinetti MC. Molecular and cellular mechanisms of cardiac arrhythmias. Cell. 2001;104:569–580. doi: 10.1016/s0092-8674(01)00243-4. [DOI] [PubMed] [Google Scholar]
- Kuo A, Gulbis JM, Antcliff JF, Rahman T, Lowe ED, Zimmer J, Cuthbertson J, Ashcroft FM, Ezaki T, Doyle DA. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 2003;300:1922–1926. doi: 10.1126/science.1085028. [DOI] [PubMed] [Google Scholar]
- Labro AJ, Raes AL, Bellens I, Ottschytsch N, Snyders DJ. Gating of Shaker-type channels requires the flexibility of S6 caused by prolines. J Biol Chem. 2003;278:50724–50731. doi: 10.1074/jbc.M306097200. [DOI] [PubMed] [Google Scholar]
- Lerche C, Seebohm G, Wagner CI, Scherer CR, Dehmelt L, Abitbol I, Gerlach U, Brendel J, Attali B, Busch AE. Molecular impact of MinK on the enatiospecific block of Iks by chromanols. Br J Pharmacol. 2000;131:1503–1506. doi: 10.1038/sj.bjp.0703734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Smerin Y, Hackos DH, Swartz KJ. A localized interaction surface for voltage-sensing domains on the pore domain of a K+ channel. Neuron. 2000a;25:411–423. doi: 10.1016/s0896-6273(00)80904-6. [DOI] [PubMed] [Google Scholar]
- Li-Smerin Y, Hackos DH, Swartz KJ. alpha-helical structural elements within the voltage-sensing domains of a K+ channel. J Gen Physiol. 2000b;115:33–50. doi: 10.1085/jgp.115.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Holmgren M, Jurman ME, Yellen G. Gated access to the pore of a voltage-dependent K+ channel. Neuron. 1997;19:175–184. doi: 10.1016/s0896-6273(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Liu YS, Sompornpisut P, Perozo E. Structure of the KcsA channel intracellular gate in the open state. Nat Struct Biol. 2001;8:883–887. doi: 10.1038/nsb1001-883. [DOI] [PubMed] [Google Scholar]
- Long SB, Campbell EB, MacKinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005a;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- Long SB, Campbell EB, MacKinnon R. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science. 2005b;309:903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- Lu Z, Klem AM, Ramu Y. Coupling between voltage sensors and activation gate in voltage-gated K+ channels. J Gen Physiol. 2002;120:663–676. doi: 10.1085/jgp.20028696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melman YF, Domenech A, de la LS, McDonald TV. Structural determinants of KvLQT1 control by the KCNE family of proteins. J Biol Chem. 2001;276:6439–6444. doi: 10.1074/jbc.M010713200. [DOI] [PubMed] [Google Scholar]
- Melman YF, Krumerman A, McDonald TV. A single transmembrane site in the KCNE-encoded proteins controls the specificity of KvLQT1 channel gating. J Biol Chem. 2002;277:25187–25194. doi: 10.1074/jbc.M200564200. [DOI] [PubMed] [Google Scholar]
- Melman YF, Um SY, Krumerman A, Kagan A, McDonald TV. KCNE1 Binds to the KCNQ1 Pore to Regulate Potassium Channel Activity. Neuron. 2004;42:927–937. doi: 10.1016/j.neuron.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Monks SA, Needleman DJ, Miller C. Helical structure and packing orientation of the S2 segment in the Shaker K+ channel. J Gen Physiol. 1999;113:415–423. doi: 10.1085/jgp.113.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss AJ. Long QT syndrome. JAMA. 2003;289:2041–2044. doi: 10.1001/jama.289.16.2041. [DOI] [PubMed] [Google Scholar]
- Nakajo K, Kubo Y. KCNE1 and KCNE3 stabilize and/or slow voltage sensing S4 segment of KCNQ1 channel. J Gen Physiol. 2007;130:269–281. doi: 10.1085/jgp.200709805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano C, Priori SG, Schwartz PJ, Bloise R, Ronchetti E, Nastoli J, Bottelli G, Cerrone M, Leonardi S. Genetic testing in the long QT syndrome: development and validation of an efficient approach to genotyping in clinical practice. JAMA. 2005;294:2975–2980. doi: 10.1001/jama.294.23.2975. [DOI] [PubMed] [Google Scholar]
- O'Neil KT, DeGrado WF. A thermodynamic scale for the helix-forming tendencies of the commonly occurring amino acids. Science. 1990;250:646–651. doi: 10.1126/science.2237415. [DOI] [PubMed] [Google Scholar]
- Panaghie G, Abbott GW. The Role of S4 Charges in Voltage-dependent and Voltage-independent KCNQ1 Potassium Channel Complexes. J Gen Physiol. 2007;129:121–133. doi: 10.1085/jgp.200609612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaghie G, Tai KK, Abbott GW. Interaction of KCNE subunits with the KCNQ1 K+ channel pore. J Physiol. 2006;570:455–467. doi: 10.1113/jphysiol.2005.100644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Cornejo P, Begenisich T. The multi-ion nature of the pore in Shaker K+ channels. Biophys J. 1994;66:1929–1938. doi: 10.1016/S0006-3495(94)80986-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perozo E, Cortes DM, Cuello LG. Structural rearrangements underlying K+-channel activation gating. Science. 1999;285:73–78. doi: 10.1126/science.285.5424.73. [DOI] [PubMed] [Google Scholar]
- Prole DL, Marrion NV. Ionic Permeation and Conduction Properties of Neuronal KCNQ2/KCNQ3 Potassium Channels. Biophys J. 2004;86:1454–1469. doi: 10.1016/S0006-3495(04)74214-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusch M, Bertorello L, Conti F. Gating and flickery block differentially affected by rubidium in homomeric KCNQ1 and heteromeric KCNQ1/KCNE1 potassium channels. Biophys J. 2000;78:211–226. doi: 10.1016/S0006-3495(00)76586-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich TC, Yeola SW, Tamkun MM, Snyders DJ. Mutations throughout the S6 region of the hKv1.5 channel alter the stability of the activation gate. Am J Physiol Cell Physiol. 2002;282:C161–C171. doi: 10.1152/ajpcell.00232.2001. [DOI] [PubMed] [Google Scholar]
- Rocheleau JM, Gage SD, Kobertz WR. Secondary structure of a KCNE cytoplasmic domain. J Gen Physiol. 2006;128:721–729. doi: 10.1085/jgp.200609657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden DM. Long QT syndrome: reduced repolarization reserve and the genetic link. J Intern Med. 2006;259:59–69. doi: 10.1111/j.1365-2796.2005.01589.x. [DOI] [PubMed] [Google Scholar]
- Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix–helix association. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- Sanguinetti MC, Curran ME, Zou A, Shen J, Spector PS, Atkinson DL, Keating MT. Coassembly of K(v)LQT1 and minK (IsK) proteins to form cardiac IKs potassium channel. Nature. 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- Seebohm G, Sanguinetti MC, Pusch M. Tight coupling of rubidium conductance and inactivation in human KCNQ1 potassium channels. J Physiol. 2003;552:369–378. doi: 10.1113/jphysiol.2003.046490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebohm G, Strutz-Seebohm N, Ureche ON, Baltaev R, Lampert A, Kornichuk G, Kamiya K, Wuttke TV, Lerche H, Sanguinetti MC, Lang F. Differential roles of S6 domain hinges in the gating of KCNQ potassium channels. Biophys J. 2006;90:2235–2244. doi: 10.1529/biophysj.105.067165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebohm G, Westenskow P, Lang F, Sanguinetti MC. Mutation of colocalized residues of the pore helix and transmembrane segments S5 and S6 disrupt deactivation and modify inactivation of KCNQ1 K+ channels. J Physiol. 2005;563:359–368. doi: 10.1113/jphysiol.2004.080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyders DJ, Tamkun MM, Bennett PB. A rapidly activating and slowly inactivating potassium channel cloned from human heart. Functional analysis after stable mammalian cell culture expression. J Gen Physiol. 1993;101:513–543. doi: 10.1085/jgp.101.4.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splawski I, Shen J, Timothy KW, Lehmann MH, Priori S, Robinson JL, Moss AJ, Schwartz PJ, Towbin JA, Vincent GM, Keating MT. Spectrum of mutations in long-QT syndrome genes. KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 2000;102:1178–1185. doi: 10.1161/01.cir.102.10.1178. [DOI] [PubMed] [Google Scholar]
- Splawski I, Shen J, Timothy KW, Vincent GM, Lehmann MH, Keating MT. Genomic structure of three long QT syndrome genes: KVLQT1, HERG, and KCNE1. Genomics. 1998;51:86–97. doi: 10.1006/geno.1998.5361. [DOI] [PubMed] [Google Scholar]
- Sukhareva M, Hackos DH, Swartz KJ. Constitutive activation of the Shaker Kv channel. J Gen Physiol. 2003;122:541–556. doi: 10.1085/jgp.200308905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai KK, Goldstein SAN. The conduction pore of a cardiac potassium channel. Nature. 1998;391:605–608. doi: 10.1038/35416. [DOI] [PubMed] [Google Scholar]
- Tapper AR, George AL., Jr Location and orientation of minK within the Iks potassium. 2001. [DOI] [PubMed]
- Tristani-Firouzi M, Chen J, Sanguinetti MC. Interactions between S4–S5 linker and S6 transmembrane domain modulate gating of HERG K+ channels. J Biol Chem. 2002;277:18994–19000. doi: 10.1074/jbc.M200410200. [DOI] [PubMed] [Google Scholar]
- Yifrach O, MacKinnon R. Energetics of pore opening in a voltage-gated K+ channel. Cell. 2002;111:231–239. doi: 10.1016/s0092-8674(02)01013-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.