Abstract
Mitochondrial Ca2+ uptake and poly(ADP-ribose) polymerase-1 (PARP-1) activation are both required for glutamate-induced excitotoxic neuronal death. Since activation of the glutamate receptors can induce increased levels of reactive oxygen species (ROS), we investigated the relationship of mitochondrial Ca2+ uptake and ROS generation, and the possibility that ROS increase is a required signal for PARP-1 activation in cultured striatal neurons. Based on the spatial profile of NMDA-induced ROS generation, we found that only mitochondria showed a significant ROS increase within 30 min after NMDA receptor activation. This ROS increase was inhibited by the mitochondrial complex inhibitors rotenone and oligomycin, but not by the cytosolic phospholipase A2 or xanthine oxidase inhibitors. Mitochondrial ROS generation was also inhibited by both removal of Ca2+ from extracellular medium and blockage of mitochondrial Ca2+ uptake by either a mitochondrial uncoupler or a Ca2+ uniporter inhibitor. Furthermore, both DNA damage and PARP-1 activation induced by NMDA treatment was inhibited by blocking mitochondrial Ca2+ uptake or by antioxidants. Our results demonstrate that ROS production during the early stage of acute excitotoxicity derives primarily from mitochondria and is Ca2+-dependent. More importantly, the increase of mitochondrial ROS serves as a signal for PARP-1 activation, suggesting that concomitant mitochondrial Ca2+ uptake and PARP-1 activation constitute a unified mechanism for excitotoxic neuronal death.
Glutamate excitotoxicity is the central mechanism underlying delayed neuronal death in either acute injury such as stroke and trauma, or neurodegenerative diseases such as Alzheimer's disease (Choi & Rothman, 1990; Hynd et al. 2004; Nicholls, 2004). Under pathological conditions, glutamate undergoes an uncontrolled release, which leads to a massive influx of extracellular Ca2+ mainly through N-methyl-d-aspartate (NMDA) receptors. The overload of cytosolic Ca2+ initiates a series of cytotoxic events including mitochondrial Ca2+ overload (Peng et al. 1998), mitochondrial depolarization (White & Reynolds, 1996), ATP depletion (Gunter & Gunter, 1994), generation of ROS (Reynolds & Hastings, 1995) and nitric oxide (NO) (Urushitani et al. 1998), delayed Ca2+ deregulation (DCD) (Vesce et al. 2004), opening of the mitochondrial permeability transition pore (PTP) (Alano et al. 2002), activation of PARP-1 (Mandir et al. 2000), and release of cytochrome c and apoptosis-inducing factor (AIF) (Wang et al. 2004).
In this cascade, two events are critical for excitotoxic neuronal death: mitochondrial Ca2+ uptake and PARP-1 activation, which is caused by DNA damage. Studies have shown that either eliminating the driving force for mitochondrial Ca2+ uptake by depolarizing mitochondria (Stout et al. 1998), or genetically knocking out PARP-1 (Yu et al. 2002) protects neurons from excitotoxic death. However, a unified scheme linking these two components of excitotoxicity is still lacking. NO has been implicated as a signal for PARP-1 activation (Zhang et al. 1994). Interestingly, mitochondrial uncoupler did not change NO production while it prevented neuronal death (Stout et al. 1998). This suggests that other signals, which are inhibited by blocking mitochondrial Ca2+ uptake, also participate in activating PARP-1. Evidence suggests that mitochondrial Ca2+ increase leads to ROS production (Urushitani et al. 2001). Because superoxide (O2·−) can react with NO to form peroxynitrite (ONOO−), which is more deleterious for DNA damage (Kennedy et al. 1997; Szabo & Ohshima, 1997), O2·− may also serve as a signal for PARP-1 activation.
Generation of O2·− also plays a key role in glutamate excitotoxicity (Patel et al. 1996). Several O2·−-generating pathways have been suggested in both the cytosol and the mitochondria under excitotoxic conditions. The cytosolic pathways include the metabolism of arachidonic acid (AA) (Lafon-Cazal et al. 1993), and the xanthine/xanthine oxidase (X/XO) system (Atlante et al. 1997). Mitochondria have been considered as a major source of ROS in the cell under many pathological conditions (Nicholls & Budd, 2000; Nishikawa et al. 2000; Lesnefsky et al. 2001). The proposed mechanism is the electron leak to oxygen from the respiratory chain to produce O2·− (Brookes et al. 2004). Although mitochondrial ROS generation has been implicated in excitotoxicity (Dugan et al. 1995; Urushitani et al. 2001), no direct evidence has been shown yet. However, abolishment of excitotoxicity by blocking mitochondrial Ca2+ uptake alone indicates that either the amount of ROS produced via cytosolic pathways is not sufficient for cell death, or the ROS produced via mitochondrial pathways exert a ‘privileged’ action on nuclear PARP-1 activation.
In this study we tested the hypothesis that mitochondrial Ca2+ uptake during intense NMDA receptor activation leads to mitochondrial ROS production. This production of mitochondrial ROS is required for the subsequent PARP-1 activation in cultured striatal neurons.
Methods
Chemicals and reagents
Dihydroethidium (DHE), 5-(and 6-)chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate acetyl ester (CM-H2DCFDA), tetramethylrhodamine ethyl ester (TMRE), 2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein acetoxymethyl ester (BCECF AM), LysoTracker Red DND 99, and fura-2 AM were purchased from Molecular Probes (Eugene, OR, USA). Arachidonyltrifluoromethyl ketone (AACOCF3), oligomycin, Ru360, Mn(III)tetrakis(4-benzoic acid) porphyrin chloride (MnTBAP) and anti-PAR rabbit antibody were from Calbiochem (San Diego, CA, USA). Fetal bovine serum was from HyClone (Logan, UT, USA), and culture mediums were from Invitrogen (Carlsbad, CA, USA). Rotenone, carbonyl cyanide p-[trifluoromethoxy]-phenyl-hydrazone (FCCP), glutathione reduced ethyl ester (GSHee) and other reagents were purchased from Sigma (St Louis, MO, USA).
Primary cell culture
Striatal neurons were cultured from E18 embryos from Sprague–Dawley rats based on a published protocol (Peng et al. 1998). All procedures for animal use were in strict accordance with the NIH Guide for the Care and Use of Laboratory Animals, and were approved by the Institutional Animal Care and Use Committee of the University of Rochester. Briefly, pregnant rats were killed by exposure to a rising concentration of CO2 until they stopped breathing for 5 min, and the fetuses were removed. Striata were dissected from embryonic rat brains, and dissociated by gently triturating after incubating with 0.125% trypsin for 10 min at 37°C. Then the suspension was layered onto 5 ml of fetal bovine serum (FBS) and centrifuged at 500 g for 10 min. After centrifugation, the pellet was resuspended in 5 ml of MEM–FBS (Eagle's MEM medium (Gibco) supplemented with 10% (v/v) FBS, 10 mm glucose, 2 mm l-glutamine, 2.5 μg ml−1 fungizone, and 50 μg ml−1 gentamicin). The isolated neurons were then plated onto laminin-coated glass coverslips at a density of 1 × 106 cells per dish and incubated in the MEM–FBS at 37°C in an atmosphere of 5% CO2. After 3 days in vitro (DIV), half of the culture medium was replaced by MEM–FBS containing 5 μm cytosine arabinoside for 48 h to inhibit proliferation of non-neuronal cells. After 6 DIV, cells were fed by half medium changes with serum-free NB medium (Neurobasal Medium (Gibco), N1 supplement (Sigma), 10 mm glucose, and 0.5 mm l-glutamine 27.5 mm NaCl, 2.5 μg ml−1 fungizone, and 50 μg ml−1 gentamicin) for 3 consecutive days. Cultured neurons were maintained in this medium and used for experiments between 11 and 15 DIV.
Measurement of ROS in cultured neurons
The intracellular ROS levels in individual neurons were measured using two redox-sensitive fluorescent indicators: DHE and CM-H2DCFDA. DHE is a blue fluorescent dye and shifts to red emission upon oxidation to ethidium by O2·− (Bindokas et al. 1996). However, DHE cannot provide information about the original location of ROS generation, because ethidium intercalates into DNA and gives fluorescence primarily from the nucleus. To obtain the spatial aspect of ROS generation, we used CM-H2DCFDA, according to a previously reported method (Reynolds & Hastings, 1995). CM-H2DCFDA is membrane permeable so that it can accumulate in intracellular compartments such as mitochondria, where it is cleaved by esterases and then converted into its fluorescent form upon oxidation primarily by hydrogen peroxide (H2O2) and hydroxyl radical (OH·). (Kirkland & Franklin, 2001). Cultured neurons were incubated with either 5 μm DHE or 100 nm CM-H2DCFDA in Hepes buffer (10 mm Hepes, 10 mm glucose, 140 mm NaCl, 5 mm KCl, 2 mm CaCl2, 5 mm NaHCO3, 0.6 mm Na2HPO4, 1.2 mm Na2SO4, pH 7.4) at 37°C for 30 min. After incubation, cells were washed three times with indicator-free Hepes buffer, and then the coverslips were mounted onto the stage of microscope for experiments. For the experiments in which inhibitors or antioxidants were used, cells were pre-incubated with different drugs for 30 min in Hepes buffer, and then drugs were kept in the buffer throughout the experiments (this pretreatment procedure was also used for other measurements).
Single cell images were taken by either fluorescence (TILL Photonics LLC, Pleasanton, CA, USA) or confocal (Leica TCS SP2, Bannockburn, IL, USA) microscopy. The fluorescence imaging system uses a Nikon TE2000s inverted microscope with a ×40 oil objective. The dyes were excited by a 150 W xenon lamp from the illumination unit polychrome V. Excitation and emission wavelength are at 515 nm and 580–630 nm for DHE, and at 488 nm and 495–535 nm for CM-H2DCFDA, respectively. A confocal laser scanning microscope with a FITC filter was also used to acquire images for CM-H2DCFDA. To minimize photo-induced oxidation and photo bleaching, only two scans were acquired for each time point. Images were taken with 2 min intervals and NMDA treatments were added after 10 min recording of baseline. The nuclear region of each single cell was used as the region of interest (ROI) for DHE data analysis due to the DNA-binding property of the oxidized dye. To determine the mitochondrial signal from CM-H2DCFDA fluorescence, cells were simultaneously loaded with 20 nm TMRE. TMRE is a voltage-sensitive indicator that accumulates preferentially into the mitochondria due to their negative membrane potential (ΔΨm), therefore it can be used as a marker for mitochondrial location. The fluorescence patterns of TMRE were chosen as the ROI and then applied onto CM-H2DCFDA images of the same cells to identify the mitochondrial area. Data are shown as the ratio of fluorescence at any given time point (F) to the first time point, 0 min (F0). All average data represent the mean ± s.e.m.
Lysosomes labelling
Lysosomes were labelled by using LysoTracker Red DND 99, which is a fluorescent indicator that specifically accumulates in acidic cellular compartments. Neurons were incubated with 50 nm LysoTracker Red in Hepes buffer for 30 min at 37°C and then were washed 3 times with indicator-free buffer. Single cell images were taken by fluorescence microscopy (TILL), the indicator was excited at 550 nm and fluorescence was collected at 605 ± 25 nm.
Intracellular Ca2+ measurement
The relative intracellular Ca2+ concentration ([Ca2+]i) in individual neurons was measured using the fluorescent probes fura-2 AM and fura-2FF AM according to methods previously reported (Alano et al. 2002). Neurons were incubated with either 4 μm fura-2 AM or 4 μm fura-2FF AM in Hepes buffer for 30 min at 37°C and then were washed 3 times with indicator-free Hepes buffer. Single cell Ca2+ images were taken by fluorescence microscopy (TILL) at room temperature. The dye was excited at 340 and 380 nm, and the emission fluorescence was collected at 510 ± 15 nm. Images were taken every 30 s in a time-lapse fashion and a stable baseline was recorded before any treatment. The fluorescence over whole cell bodies was chosen as ROIs and the results are presented as the fluorescence ratio of 340/380 nm of individual neurons.
pH measurement
Intracellular pH was measured by using the membrane-permeable pH-sensitive dye, BCECF-AM. Cultured cells were loaded with 1 μm BCECF for 30 min in Hepes buffer at 37°C. After loading, the dye was excited at 440 and 488 nm and the emission was collected at 515 ± 20 nm. Single cell images were acquired by fluorescence microscopy (TILL) once per minute during a 60 min time course. The ratio of emission intensity at the two different excitation wavelengths was determined for individual cells. Data are presented as a ratio of fluorescence at any given time point over time 0 (F/F0).
SDS electrophoresis and Western blotting
For Western blots, each assay consisted of approximately 3 × 106 cells. After 15 min NMDA treatments, cultures were lysed in a buffer containing 50 mm Tris·HCl, 10 mm KCl, 1.5 mm MgCl2, 1 mm DTT, 1% (w/v) SDS, 1 mm EDTA, 1 mm EGTA, protease inhibitor cocktail (1 tablet per 50 ml, Roche), pH 7.4. Equal amounts of protein were loaded onto a 4–15% gradient polyacrylamide gel for electrophoresis, and then electrotransferred onto a nitrocellulose membrane (Bio-Rad Laboratories, Hercules, CA, USA). The membranes were blocked with 2% fat-free milk in PBS for 1.5 h followed by incubation with an anti-PAR rabbit polyclonal antibody (1: 2000 dilution) in the blocking buffer for 3 h at room temperature. After washing for 3 times with PBS containing 0.1% (v/v) Tween 20 (0.1% PBST), the membranes were incubated with a 1: 10 000 dilution of peroxidase-labelled goat anti-rabbit IgG antibody in blocking buffer for 1.5 h at room temperature. After the incubation with the secondary antibody, the membranes were washed 3 times again with 0.1% PBST, and then the signal was visualized using a Supersignal ChemiLuminiscence detection kit (Pierce, Rockford, IL, USA).
Comet assay
Comet assays (also called single cell gel electrophoresis assays) were conducted as previously described (Olive et al. 1990) with modifications to evaluate DNA strand breaks. In brief, striatal neurons were cultured in 60 mm dishes with density of 2.5 × 106 cells per dish. Control or treated neurons were washed twice with PBS (Ca2+ and Mg2+ free) and harvested by centrifugation at 700 g for 5 min. Cells were then resuspended in ice-cold PBS at 1 × 105 ml−1. Aliquots of cells (15 μl) were mixed with an equal volume of 1% low-melting-point agarose in PBS, and 20 μl of the cell–gel mixtures were then pipetted onto microscope slides pre-coated with 30 μl of 0.5% normal-melting-point agarose and covered gently with a coverslip. The slides were placed at 4°C in the dark for 30 min for gel solidification. After removing the coverslips, the slides were immersed in ice-cold fresh lysis solution (1% laurosylsarcosinate, 2.5 m NaCl, 100 mm Na2EDTA, 10 mm Tris, 10% DMSO, 1% Triton X-100) for 90 min at 4°C, and then transferred to an alkaline solution (0.3 m NaOH, 1 mm EDTA, pH > 13) for 20 min at 4°C. Electrophoresis was performed in the same alkaline solution at 30 V, 300 mA for 10 min. After electrophoresis, the slides were washed twice with a neutralization buffer (0.4 m Tris-HCl) and stained with 4′,6-diamidino-2-phenylindole (DAPI, 10 μg ml−1). For quantitative measurement, at least 10–20 pictures were taken and 100–300 cells were analysed for each sample using Komet 5.5 software (Kinetic Imaging Ltd, Nottingham, UK). The olive tail moment (defined as the product of the percentage of DNA in the tail multiplied by the tail length) was chosen as the parameter to evaluate DNA damage.
Data analysis
Statistical analyses were performed using one-way ANOVA followed by Tukey's test for multiple comparisons (Origin 7, Origin Lab). Results are considered as either nonsignificant (P > 0.05), significant (P < 0.05), or highly significant (P < 0.01). Each trace from ROS measurements represents the mean value ± s.e.m. of more than 30 individual cells from at least three independent experiments.
Results
Mitochondria are the major sites of NMDA-induced ROS generation
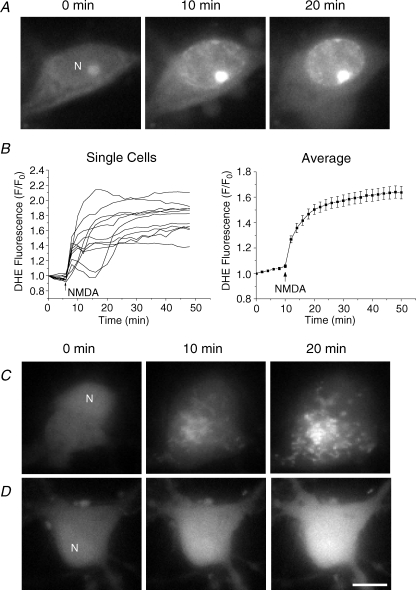
Change of O2·− level from cultured striatal neurons during NMDA receptor activation was first monitored with DHE by fluorescence microscopy. DHE-loaded neurons showed a basal spontaneous ROS generation as evidenced by a slow increase in the baseline fluorescence. After the addition of 100 μm NMDA together with 10 μm glycine, DHE fluorescence increased immediately and reached 161.5 ± 4.7% of baseline values within 30 min, indicating O2·− production induced by NMDA (Fig. 1A and B, n = 37 for the average trace). From the single-cell traces, some cells also showed a delayed secondary rapid phase of ROS increase, which is consistent with a previous study (Vesce et al. 2004). Although these secondary rapid increases in ROS in single-cell traces could be due to DCD, it is difficult for us to draw a precise conclusion since we did not measure Ca2+ dynamics and ROS generation simultaneously as this is not the main focus of the present study. We then used CM-H2DCFDA to assess the spatial profile of the ROS increase. Neurons loaded with CM-H2DCFDA showed homogeneous fluorescence with a slightly higher signal in the nucleus (Fig. 1C, 0 min). Surprisingly, NMDA induced a localized increase in CM-H2DCFDA fluorescence which is completely distinct from that in the DHE signal. There was a gradual appearance of bright fluorescent spots throughout the cells (Fig. 1C, 20 min). This punctate pattern is not due to translocation of the dye upon oxidation, because artificially oxidizing the dye by treating cells with 1 mm H2O2 showed a homogeneous fluorescence increase over the whole cell body (Fig. 1D).
Figure 1. NMDA-induced ROS generation in cultured striatal neurons.
A, fluorescence images illustrate a representative response of a DHE-loaded neuron to NMDA. Images are shown at 0, 10 and 20 min after the treatment of 100 μm NMDA and 10 μm glycine. Note that only the nuclear region (N) shows a significant increase in DHE fluorescence. B, single-cell traces and quantitative analysis of DHE fluorescence during a 50 min time course. Baseline fluorescence was recorded prior to NMDA treatment (arrow). Data are shown as the ratio of fluorescence at each time point (F) to time 0 min (F0). The single-cell traces are from one representative experiment, and the average trace represents the mean ± s.e.m. of 37 cells from at least 3 different experiments. C and D, CM-H2DCFDA-loaded neurons were treated with either 100 μm NMDA and 10 μm glycine (C) or 1 mm H2O2 (D) at time 0 min. Representative images of a single neuron are shown at the indicated time points after treatments. NMDA induced a different localized pattern in CM-H2DCFDA fluorescence. These experiments were performed on at least 5 additional coverslips. N, nucleus. Scale bar: 10 μm.
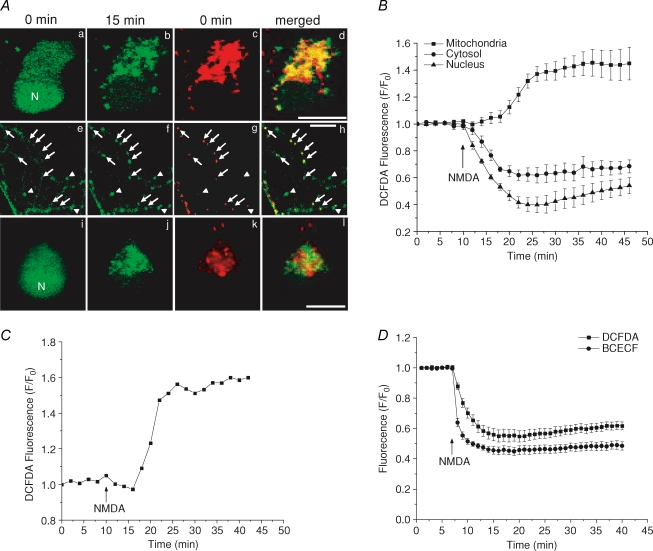
We next investigated the identity of the bright fluorescent spots in CM-H2DCFDA images after NMDA treatment. To test the idea that mitochondria are the source of this signal, neurons were loaded with CM-H2DCFDA and TMRE simultaneously. By merging the CM-H2DCFDA image acquired 15 min after NMDA treatment and the TMRE image acquired prior to NMDA treatment (when ΔΨm is not depolarized by NMDA), we shows that the two images overlapped in both the soma (Fig. 2Aa, b, c and d) and neurites (Fig. 2Ae, f, g and h). Although the overlap is not complete because of mitochondrial movement, fission and fusion during the 15 min time lapse or the heterogeneity of ROS generation, certain spots that appeared later in the CM-H2DCFDA signal in neurites, where individual mitochondria can be easily identified, colocalized exactly to the TMRE signal (Fig. 2Ae, f, g and h, arrows). There are some spots in the CM-H2DCFDA signal that did not colocalize to any TMRE signal (Fig. 2Ae, f, g and h, arrowheads). However, those spots mostly appeared before the NMDA treatment, indicating that those mitochondria might have already depolarized and generated ROS before the treatment. To rule out that the punctate pattern of the CM-H2DCFDA signal was from other organelles such as lysosomes and endosomes, we used LysoTracker Red, which is a fluorescent indicator that specifically labels acidic cellular compartments. LysoTracker Red staining did not produce any colocalized pattern with CM-H2DCFDA signal (Fig. 2Ai, j, k and l). It is possible that lysosomes were able to take up some CM-H2DCFDA. However, due to their lack of ROS generation capability, they were not able to produce CM-H2DCFDA signals as significant as could mitochondria. Indeed, the acidic environment inside lysosomes or endosomes will cause a decrease of CM-H2DCFDA fluorescence due to the pH effect. These data add support to the idea that the NMDA-induced ROS generation is from mitochondria. The TMRE signals were then used to define the mitochondrial ROIs in CM-H2DCFDA images and the fluorescence changes of CM-H2DCFDA from different intracellular compartments were quantitatively analysed. The profiles of CM-H2DCFDA fluorescence were different in nucleus, cytoplasm and mitochondria. Both nucleus and cytoplasm showed a decrease in fluorescence, while mitochondria showed a small transient decrease and then an increase in fluorescence (Fig. 2B, n = 31). A representative time course of mitochondrial CM-H2DCFDA fluorescence from a single neuron is shown in Fig. 2C. The decrease in fluorescence in the cytosol and nucleus was most likely due to the effect of NMDA-induced intracellular acidification (Hartley & Dubinsky, 1993) on CM-H2DCFDA fluorescence (Reynolds & Hastings, 1995). To confirm this in our study, we monitored intracellular pH change induced by NMDA using BCECF. Neurons exposed to NMDA showed a rapid decrease of BCECF fluorescence indicating intracellular acidification (Fig. 2D).
Figure 2. Mitochondria are the source of NMDA-induced ROS production.
Cells loaded with different dyes were exposed to 100 μm NMDA and 10 μm glycine for all experiments. Aa–h, confocal images taken from CM-H2DCFDA (green) and TMRE (red) double stained cells are shown at the indicated time points after NMDA treatment for both the soma (a, b, c and d) and neurites (e, f, g and h). TMRE fluorescence is used to show mitochondrial location. Merged images demonstrate that the pattern of the CM-H2DCFDA signal after NMDA treatment colocalizes with mitochondria. Note that the spots in the CM-H2DCFDA signal that showed up after NMDA treatment in neurites (arrows) colocalize with the TMRE signal, whereas spots already present before the NMDA treatment (arrowheads) do not have TMRE staining. i–l, fluorescence images from neurons double stained with both CM-H2DCFDA (green) and LysoTracker Red (red). Merged image shows that these two signals are not colocalized with each other. N, nucleus. Scale bar: 10 μm. B, time courses of CM-H2DCFDA fluorescence from somatic mitochondria (▪), cytosol (•) and nucleus (▴). C, representative trace of mitochondrial signal from a single cell. D, effect of NMDA on cytosolic CM-H2DCFDA fluorescence (▪) and BCECF fluorescence (•). Traces are normalized in the same way as Fig. 1B. Data represent the mean ± s.e.m. of 30–40 cells in each experiment.
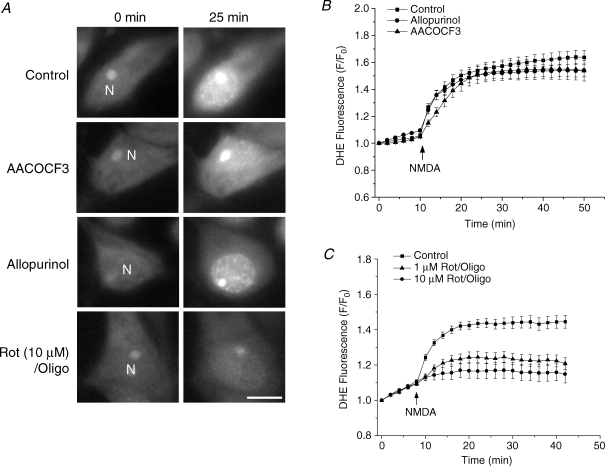
We further examined the contribution of cytosolic pathways to NMDA-induced ROS increase. Two cytosolic ROS-generating pathways have been proposed under excitotoxic conditions. One is the metabolism of arachidonic acid, which is released by phospholipase A2 (PLA2) (Lazarewicz et al. 1990; Lafon-Cazal et al. 1993; Vesce et al. 2004), and the other is the conversion of xanthine to uric acid by XO (Atlante et al. 1997). To evaluate their roles in our study, two inhibitors – arachidonyltrifluoromethyl ketone (AACOCF3, 50 μm), a specific PLA2 inhibitor, and allopurinol (20 μm), a specific XO inhibitor – were used to inhibit these two pathways. Cells were pre-incubated with each inhibitor for 30 min, after which ROS generation was measured by DHE. In control experiments, a dramatic increase in DHE fluorescence was obtained again after NMDA treatment and reached a plateau after about 15 min (n = 37), consistent with the DCFDA data. However, neither AACOCF3 nor allopurinol had any significant inhibitory effect on this ROS generation (10 ± 3.6% inhibition, n = 43, P > 0.3, and 16 ± 4.3% inhibition, n = 36, P > 0.5, respectively, Fig. 3A and B), indicating that these cytosolic pathways did not play a major role in the NMDA-induced ROS generation.
Figure 3. Contribution of cytosolic and mitochondrial pathways to ROS generation induced by NMDA in cultured striatal neurons.
A, DHE fluorescence images from individual neurons are shown at the indicated time points after exposure to 100 μm NMDA and 10 μm glycine either with no pretreatment (first row), or pretreated with: 50 μm AACOCF3 (second row), 20 μm allopurinol (third row) or 10 μm rotenone and 5 μm oligomycin (fourth row). N, nucleus. Scale bar: 10 μm. B, time courses of NMDA-induced DHE fluorescence changes in the presence of inhibitors of the cytosolic ROS generation pathways. Neurons were either with no pretreatment (control, ▪), or pretreated with AACOCF3 (▴) or allopurinol (•). Data represent the mean ± s.e.m. of 37–43 cells. C, time courses of NMDA-induced DHE fluorescence changes in the presence of inhibitors of the mitochondrial respiratory chain. Neurons were either with no pretreatment (control, ▪), or pretreated with 1 μm (▴) or 10 μm (•) rotenone with 5 μm oligomycin. Data represent the mean ± s.e.m. of 30–61 cells.
As a comparison, the contribution of the mitochondrial pathway to this ROS generation was also tested. Studies have shown that inhibiting complex I with rotenone decreases mitochondrial ROS in living cells (Becker et al. 1999; Carriedo et al. 2000; Hongpaisan et al. 2004). In our study, we used rotenone (1 μm and 10 μm) together with complex V inhibitor oligomycin (5 μm), which is used to limit ΔΨm depolarization induced by rotenone and to prevent ATP depletion caused by the reverse mode of ATP synthase (Nicholls & Budd, 2000; Hongpaisan et al. 2004). This was confirmed by monitoring ΔΨm in the neurons treated with rotenone (10 μm) in the presence or absence of oligomycin. A significant drop of ΔΨm was observed in the neurons without oligomycin pretreatment, as indicated by a loss of about 40% of TMRE fluorescence within 15 min. However, in the presence of oligomycin, rotenone only caused a minor depolarization (10% decrease in TMRE fluorescence, data not shown). Then ROS generation in neurons pretreated with rotenone and oligomycin was measured and a rotenone concentration-dependent inhibition of NMDA-induced ROS generation was observed. Rotenone at 1 μm produced a 55.7 ± 8.6% inhibition (n = 61, P < 0.001) and 10 μm rotenone produced an 87.1 ± 11.3% inhibition (n = 30, P < 0.0001) of ROS generation (Fig. 3C). These results suggest that mitochondria are the major sites of ROS generation during the early stage of acute glutamate excitotoxicity.
NMDA-induced mitochondrial ROS generation is Ca2+-dependent
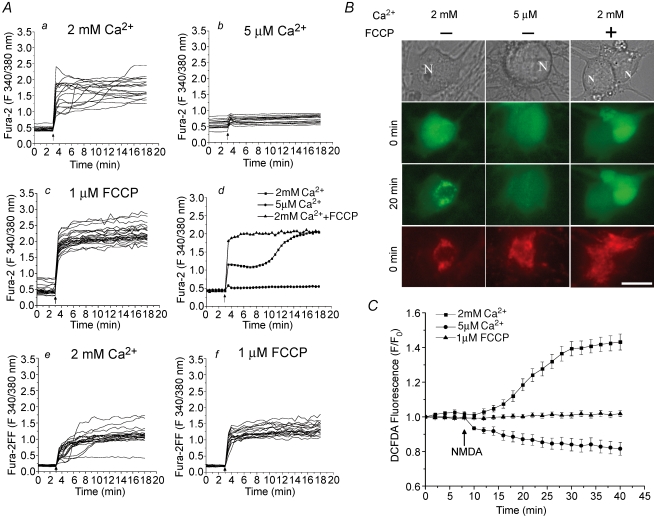
Ca2+ plays a critical role in the excitotoxic cascade, because either removing Ca2+ from extracellular medium (Choi, 1987) or preventing Ca2+ from entering mitochondria by uncouplers (Stout et al. 1998) protects neurons against excitotoxic injury. To test the possible role of Ca2+ in the regulation of mitochondrial ROS generation, we measured ROS generation under these two conditions. An external buffer containing low (5 μm) Ca2+ was used to reduce Ca2+ influx through NMDA receptors. Also, and 1 μm FCCP was co-administrated with NMDA to depolarize mitochondria and therefore prevent mitochondrial Ca2+ uptake in normal Ca2+ buffer (2 mm).
The difference in NMDA-induced Ca2+ influxes was first examined in the conditions of normal Ca2+, low Ca2+ and normal Ca2+ plus FCCP. In normal Ca2+ buffer, cells exhibited an abrupt increase in [Ca2+]i after NMDA treatment, as monitored by the Ca2+-sensitive dye, fura-2 AM. In many cells, a secondary increase in [Ca2+]i was observed within 30 min of NMDA addition, which may correspond to DCD (Fig. 4Aa). In experiments using low Ca2+ buffer, the NMDA-induced [Ca2+]i increase was almost completely abolished (Fig. 4Ab). In the presence of FCCP, the increase in [Ca2+]i was higher, which was expected due to the absence of mitochondrial Ca2+ uptake (Fig. 4Ac). More importantly, none of the cells had the secondary Ca2+ increase in this condition, indicating that DCD may be due to the release of mitochondrial Ca2+ content, which is consistent with our previous study (Alano et al. 2002). To clearly illustrate the difference in the increases of [Ca2+]i under these three conditions, a typical cell response from each condition is shown in Fig. 4Ad. To rule out the possibility that the lack of secondary Ca2+ increases in FCCP-treated cells was due to saturation of fura-2 AM (Kd= 224 nm), we also used another low-affinity Ca2+-sensitive indicator, fura-2FF AM (Kd= 25 μm). However, there was no secondary Ca2+ increase observed in FCCP-treated cells, as with fura-2 AM, except the average signal level for the NMDA-evoked Ca2+ influx was lower in magnitude (Fig. 4Ae and f).
Figure 4. The mitochondrial ROS increase is Ca2+-dependent.
A, NMDA-induced increase in intracellular Ca2+ was measured with either fura-2 AM (a–d) or fura-2FF AM (e and f). Neurons were treated with 100 μm NMDA and 10 μm glycine (indicated by arrows) in the presence of either 2 mm (a) or 5 μm extracellular Ca2+ (b), or with coadministration of 1 μm FCCP in 2 mm Ca2+ buffer (c). Each trace represents the fluorescence change from a single neuron. A typical trace from each condition is shown in the last panel for a clear presentation (d). NMDA-induced Ca2+ influx was also measured using the low affinity indicator fura-2FF AM (e and f) in the same conditions as in a and c, respectively, to rule out the possibility of dye saturation. B and C, ROS generation was measured by CM-H2DCFDA using the same experimental conditions as in A. Fluorescence images (B) from cells double stained with CM-H2DCFDA (green) and TMRE (red, before NMDA and FCCP applications) and time courses of mitochondrial CM-H2DCFDA fluorescence (C) are shown to demonstrate that localized mitochondrial ROS generation occurs only in the 2 mm Ca2+ buffer (▪), but not in the 5 μm Ca2+ buffer (•), or in the presence of FCCP (▴). Data represent mean ± s.e.m. of 30–39 cells. N, nucleus. Scale bar: 10 μm.
When ROS generation was measured in the same conditions, the mitochondrial pattern of the DCFDA signal, which was shown when using the normal Ca2+ buffer, was totally abolished in cells kept in the low Ca2+ buffer (Fig. 4B). A lack of the localized pattern of ROS increase was also found when FCCP was co-administered with NMDA in normal Ca2+ buffer (Fig. 4B and C). In addition, the decrease of CM-H2DCFDA fluorescence was also absent in the presence of the proton ionophore, FCCP, consistent with the idea that the fluorescence decrease was due to pH change.
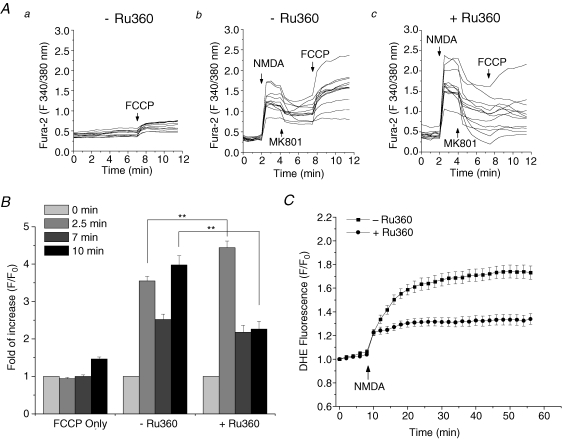
To further test that Ca2+ uptake into mitochondria is required for ROS generation, we inhibited mitochondrial Ca2+ transport during NMDA application. Ruthenium Red (RuR) is a well-known inhibitor of the mitochondrial Ca2+ uniporter. However, studies have shown that it can also inhibit the Ca2+ influx across the plasma membrane (Duchen, 1992). Consistent with this observation, we found that RuR produced a substantial suppression on the NMDA-induced increase in [Ca2+]i (authors' unpublished data). Therefore, we used another more specific inhibitor of mitochondrial Ca2+ uniporter, Ru360, which is a membrane-permeable derivative of RuR. Cells were treated with 10 μm Ru360 for 30 min before and throughout the experiments. The inhibition of mitochondrial Ca2+ uptake by Ru360 was first confirmed by measuring a FCCP-induced increase in [Ca2+]i after NMDA receptor activation, a means of assessing the mitochondrial pool of Ca2+ (Budd & Nicholls, 1996). In control cells, a direct application of FCCP (1 μm) induced a small increase in [Ca2+]i, indicating a release of basal mitochondrial Ca2+ content due to loss of ΔΨm (Fig. 5Aa). When treated with NMDA, cells again showed a robust cytosolic Ca2+ increase, which was attenuated by adding 10 μm MK801 2 min after NMDA treatment. After a short recovery time, addition of FCCP caused a secondary increase in cytosolic Ca2+, which is much higher than the FCCP-induced Ca2+ increase in control cells, indicating a substantial Ca2+ uptake by mitochondria following NMDA receptor activation (Fig. 5Ab). In Ru360-treated cells, the levels of NMDA-induced Ca2+ influxes are higher than the non-treated cells (Fig. 5Ab and c, the first 4 min; and Fig. 5B, 2.5 min), which is similar to the FCCP-treated cells in Fig. 4A (a and c, the first 5 min). However, the FCCP-induced mitochondrial Ca2+ release was significantly diminished when compared to the non-treated cells (Fig. 5Ab and c, after 7 min; and Fig. 5B, 10 min). The statistical analysis of the Ca2+ level at the different time points in these conditions are plotted in Fig. 5B (n = 24–35, **P < 0.001). These data suggest that Ru360 efficiently inhibits mitochondrial Ca2+ uptake, while having no effect on cytosolic Ca2+ influx. When ROS generation was measured by DHE, Ru360 also produced a significant inhibition (59.1 ± 5.6% decrease in fluorescence, n = 35, P < 0.001) compared to control (Fig. 5C, n = 33). These data suggest that not only extracellular influx, but also mitochondrial uptake of Ca2+, is required for ROS production.
Figure 5. Blocking mitochondrial Ca2+ uptake by Ru360 inhibits ROS generation.
A, FCCP-induced increase in cytosolic Ca2+ after NMDA treatment was measured by fura-2 AM. a, control cells were treated only with FCCP (1 μm) to release a basal mitochondrial Ca2+ content. b and c, cells were treated with 100 μm NMDA and 10 μm glycine in the absence (b) or presence (c) of 10 μm Ru630. The NMDA receptor blocker MK801 (10 μm) and FCCP were then added at 2 and 6 min after NMDA application to inhibit extracellular Ca2+ influx and release mitochondrial Ca2+, respectively. The FCCP-induced secondary Ca2+ increase was significantly attenuated in the presence of Ru360. B, the statistical analysis of the Ca2+ level at four different time points, 0, 2.5, 7 and 10 min, that correspond to the four different states of the experiments in A, baseline, after NMDA, after MK801 and after FCCP, respectively. Data are normalized to the fluorescence at time 0. n = 24–35, **P < 0.001. C, effect of Ru360 on the NMDA-induced ROS generation. Time courses of DHE fluorescence are shown in the absence (▪) or presence (•) of Ru630. Data represent mean ± s.e.m. of 33 and 35 cells.
Ca2+-dependency of NMDA-induced PARP-1 activation and DNA damage
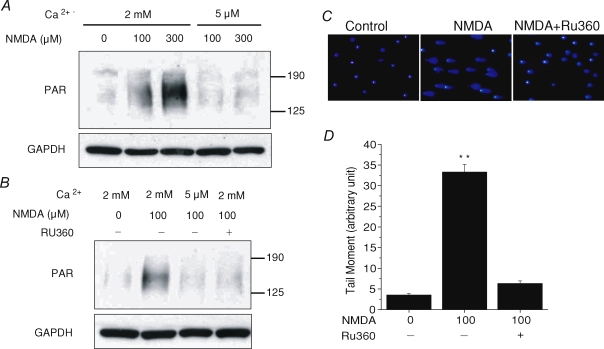
Since PARP-1 activation is another required event for NMDA-induced neuronal death due to its mediation of the translocation of AIF from the mitochondria to the nucleus (Yu et al. 2002), a link between mitochondrial Ca2+ uptake and PARP-1 activation has to exist to connect these two pathways in a unified mechanism. To confirm this point, we next investigated whether PARP-1 activation is also Ca2+-dependent. Cells were treated with NMDA for 15 min in either 2 mm Ca2+ buffer or 5 μm Ca2+ buffer. PARP-1 activation was then examined after NMDA treatment by immunoblot to detect the formation of poly(ADP-ribose) (PAR) polymer, a product of PARP-1 activation. Control cells had a low basal PARP-1 activity (Fig. 6A, first lane). After cells were treated with 100 and 300 μm NMDA in 2 mm Ca2+ buffer, we detected a dramatic concentration-dependent stimulation of PARP-1 activation (Fig. 6A, second and third lane). However, no increase in PARP-1 activation was observed in 5 μm Ca2+ buffer for either concentration of NMDA treatment (Fig. 6A, fourth and fifth lane). To further examine whether the stimulation of PARP-1 activation requires the entry of Ca2+ into mitochondria, neurons were pretreated with Ru360. As shown in Fig. 6B (fourth lane), Ru360 caused a strong inhibition of PARP-1 activation. These data suggest that PARP-1 activation is a downstream event of mitochondrial Ca2+ uptake.
Figure 6. NMDA-induced PARP activation and DNA damage is also Ca2+-dependent.
A and B, PARP-1 activation assayed by immunoblot using an antibody against the PAR polymer. Cells were exposed to the indicated concentrations of NMDA for 15 min in 2 mm or 5 μm Ca2+ buffer and then subjected to immunoblot assay. A significant concentration-dependent PARP-1 activation was detected in the 2 mm Ca2+ buffer, but not in the 5 μm Ca2+ buffer (A) or in the presence of Ru360 (B). Immunoblot for GAPDH was used as a loading control. These experiments were repeated at least three times with similar results. C and D, DNA damage was evaluated by comet assay with or without the pretreatment of Ru360. Representative fluorescence images of comet assays (C) and quantitative analysis of tail moment (D) show that comet tails, caused by NMDA treatment, were strongly attenuated by Ru360. **P < 0.01. Data represent mean ± s.e.m. of 143–289 cells.
However, the signal to activate PARP-1 from mitochondria is still unclear. Since damage to DNA is the cause of PARP-1 activation, the NMDA-induced mitochondrial ROS generation, which could lead to DNA damage and is mitochondrial Ca2+-dependent, stands out as an excellent candidate. To explore this possibility, we first examined the Ca2+-dependency of DNA lesions in NMDA-treated neurons. DNA strand breaks were evaluated by comet assay, which separates damaged DNA from intact DNA. DAPI was used to stain the DNA content of the cells. After electrophoresis, control cells showed bright circular staining indicating intact DNA (Fig. 6C, n = 143). When neurons were treated with 100 μm NMDA for 15 min, we observed obvious comet tails from almost all the cells, representing a significant fraction of damaged DNA (n = 154). Again, Ru360 was used to block mitochondrial Ca2+ uptake during NMDA stimulation, and NMDA-induced comet tails were greatly attenuated in these cells (n = 289). Statistical data are shown in Fig. 6D. Tail moment, which represents both DNA percentage in the tail and the length of the tail, was used as the parameter to evaluate the level of DNA damage. NMDA-induced DNA damage is about 9 times over the non-treated control (**P < 0.01), and reduced to 1.7 times in the Ru360-pretreated cells.
Antioxidants prevent both NMDA-induced DNA damage and PARP-1 activation
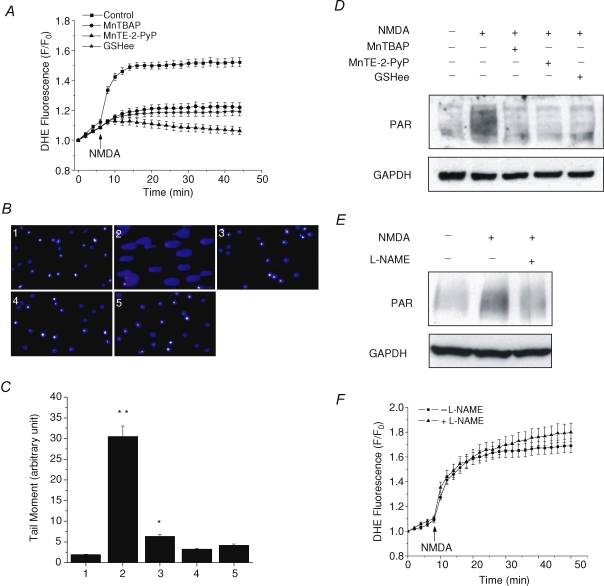
To further test the possibility of mitochondrial ROS increase as the signal to activate PARP-1, the effects of different antioxidants on inhibiting both DNA damage and PARP-1 activation were tested next. Two superoxide dismutase (SOD) mimics, Mn(III)tetrakis(4-benzoic acid)porphyrin (MnTBAP) and Mn(III)tetrakis(N-ethylpyridinium-2yl)porphyrin (MnTE-2-PyP), as well as a cell membrane-permeable form of glutathione, glutathione ethyl ester (GSHee), were used. The ability to scavenge ROS was first confirmed for these drugs. Cells were pretreated with either 200 μm MnTBAP, 20 μm MnTE-2-PyP or 1 mm GSHee for 30 min, and then left in the drug-containing buffers during experiments. ROS generation was measured by DHE. As shown in Fig. 7A, all three drugs produced strong inhibition of NMDA-induced mitochondrial ROS generation (n = 30–47). Since MnTE-2-PyP is a very potent antioxidant, the NMDA-induced cell swelling actually caused a slow decrease of DHE fluorescence due to the dilution of the dye in MnTE-2-PyP-pretreated cells. NMDA-induced Ca2+ influxes were also examined under the same conditions to exclude the possibility that the attenuation of ROS increase by antioxidants was due to the blockage of NMDA receptors. No significant difference was observed between control and antioxidant-treated cells (see online Supplemental Fig. 1). Next, DNA damage was measured by comet assay to determine whether it still occurs in the absence of ROS increase. Again, NMDA induced long bright comet tails in about 90% of cells, and the comet tails were almost completely inhibited by all three antioxidants with slightly different efficiency (Fig. 7B and C, n = 130–175). Lastly, we examined PARP-1 activation after NMDA stimulation in the presence of these antioxidants. Consistent with the ROS measurement and comet assay data, PARP-1 activation was inhibited by all three antioxidants (Fig. 7D). These data strongly suggest that the NMDA-induced mitochondrial ROS generation is the signal to activate PARP-1 via induction of DNA damage. Since evidence has suggested that PARP-1 activation is also NO dependent, as NMDA-induced PAR polymer formation is inhibited in the cell cultures from nNOS knockout mice (Wang et al. 2004), we tested the ability of Nω-nitro-l-arginine methyl ester (l-NAME), a NOS inhibitor, to inhibit PARP-1 activation. Consistent with the previous finding, l-NAME (500 μm) produced a strong inhibition of PARP-1 activation as well (Fig. 7E, n = 3). ROS generation was also measured in the presence or absence of l-NAME, but no significant difference was observed between these two conditions (Fig. 7F, n = 25–53), suggesting that the inhibition of PARP-1 activation was not due to l-NAME causing any reduction of ROS generation. The requirements for both ROS and NO indicate that ONOO− may play a more prominent role in inducing PARP-1 activation.
Figure 7. Antioxidants inhibit NMDA-induced ROS generation, DNA damage and PARP-1 activation.
A, neurons were exposed to 100 μm NMDA and 10 μm glycine, and ROS generation was measured by DHE either with no pretreatment (▪), or pretreated with: 200 μm MnTBAP (•), 20 μm MnTE-2-PyP (▴), or 1 mm GSHee (⋆). All three antioxidants exhibit strong inhibition of the NMDA-induced ROS generation. Data represent mean ± s.e.m. of 30–47 cells. B and C, DNA damage was measured by comet assay under the conditions of: (1) no treatment, (2) NMDA, (3) NMDA with MnTBAP, (4) NMDA with MnTE-2-PyP, and (5) NMDA with GSHee. Representative fluorescence images (B) and quantitative analysis (C) show that all antioxidants effectively eliminated the comet tails induced by NMDA. *P < 0.05, **P < 0.01. Data represent mean ± s.e.m. of 130–175 cells. D and E, PARP-1 activation induced by NMDA is inhibited by all three antioxidants (D), which is consistent with the observations from A and B, or by l-NAME (E). GAPDH was used as a loading control. These experiments were repeated at least two times with similar results. F, effect of l-NAME on the NMDA-induced ROS generation. Time courses of DHE fluorescence are shown in the absence (•) or presence (▴) of l-NAME. Data represent mean ± s.e.m. of 25 and 53 cells.
Discussion
In the current study, we provided novel findings on several points. Firstly, the data from CM-H2DCFDA images and rotenone inhibition showed direct evidence both visually and pharmacologically that ROS generation induced by NMDA receptor activation was mainly from mitochondria during the early stage of acute glutamate excitotoxicity. Secondly, inhibiting the Ca2+ uniporter by Ru360, compared to FCCP-induced depolarization, blocked mitochondrial Ca2+ uptake in a cleaner way, in which both ΔΨm and ATP levels are maintained, and the results showed that mitochondrial Ca2+ uptake is required for NMDA-induced mitochondrial ROS generation. Finally and more importantly, our study demonstrated that this mitochondrial ROS generation served as a signal to activate PARP-1, thus linking mitochondrial Ca2+ uptake and PARP-1 activation as required components in a unified mechanism for excitotoxic neuronal death.
Since every fluorescent indicator currently available for ROS detection has its limitations, in this study we used the two most common dyes, DHE and CM-H2DCFDA. DHE provides better temporal resolution of the initiation of the NMDA-induced ROS generation, whereas CM-H2DCFDA provides superior spatial distribution of ROS generation. We noticed that the rates of ROS generation decreased at the end of both DHE and CM-H2DCFDA time courses. This may reflect a certain level of dye saturation, since we observed a continuously increasing fluorescence when we left DHE in the experimental buffer. However, the kinetic results for the pharmacological manipulations of ROS generation are essentially the same as those we obtained in the dye-washing experiments (Supplemental Fig. 2). Although CM-H2DCFDA has other drawbacks such as pH effect, we were still able to see the localized fluorescence increase, which is consistent with previous reports (Reynolds & Hastings, 1995). When we measured the NMDA-induced pH change using BCECF, which is also able to accumulate into mitochondria (Ruiz-Meana et al. 2003; Costa et al. 2006), there was no punctate increase in the BCECF signal. This indicates that the increase in CM-H2DCFDA fluorescence was not due to localized pH change. However, the NMDA-induced cytosolic acidification caused an overall decrease in CM-H2DCFDA fluorescence in the first 5–10 min after NMDA treatment (Fig. 2D; also see Reynolds & Hastings, 1995). This effect counteracted the ROS-induced CM-H2DCFDA fluorescence increase. For instance, the CM-H2DCFDA fluorescence from a single cell in Fig. 2C shows a biphasic feature with a transient decrease (due to the rapid pH change) followed by a gradual increase. Due to the variations in the magnitude and time course of response in individual cells and in subcellular regions, the averaged time courses of CM-H2DCFDA fluorescence from somatic mitochondria (Fig. 2B, squares) showed a few minutes delay in the increase when compared to DHE data. Nevertheless, the data from either DHE or CM-H2DCFDA measurement cannot directly determine whether the increased fluorescence was due to an increased ROS production or a decreased antioxidant protection. However, this question can be clarified indirectly by the inhibitory effect of rotenone.
The morphological similarity of the localized CM-H2DCFDA pattern to the TMRE staining directly linked the sources of ROS generation to mitochondria. Although the overlay at the two different time points was not 100% in the soma due to mitochondrial dynamics, comparing these two signals from single mitochondria in neurites reveals an exact overlapping pattern, strongly suggesting that the NMDA-induced ROS generation is from mitochondria. Interestingly, mitochondria in the neurites showed a higher degree of heterogeneity in CM-H2DCFDA responses. This may be in part due to some sick mitochondria, which are in transit back to the soma for repair or regeneration; in part due to more heterogeneous mitochondrial Ca2+ uptake as a result of more localized NMDA receptor-mediated Ca2+ microdomains; and in part due to some mitochondria being more likely to undergo the opening of permeability transition pores, even during physiological activities.
The mitochondrial origins of NMDA-induced ROS production are also supported by the results that both inhibition of the mitochondrial electron transport chain (ETC) by rotenone and blockage of mitochondrial Ca2+ uptake by FCCP or Ru360 was able to efficiently inhibit this ROS generation. Furthermore, the inhibition of ROS increase by rotenone indicates that ROS are generated directly from the mitochondrial ETC. Rotenone acts to block electron transfers through complex I to ubiquinone. Hence, it also indicates that the ROS-generating site may be downstream of the ETC. This is relevant because complex III is a source of O2·− (Boveris et al. 1976; Cadenas et al. 1977; Muller et al. 2004). Complex I may not produce much ROS in our conditions, although it has been suggested as a ROS-generating site when inhibited in other studies (Kushnareva et al. 2002; Votyakova & Reynolds, 2005). However, extra caution should be taken when using inhibitors of complexes as they may produce different results with ROS production under different conditions, including different mitochondrial preparations (isolated mitochondria versus mitochondria in intact cells), mitochondria from different tissues, different pharmacological combinations (rotenone with or without oligomycin), or utilization of different respiratory substrates. Moreover, although mitochondria did not depolarize significantly during rotenone pretreatment in our study, we cannot exclude the possibility that the rotenone-inhibited mitochondria more easily lose their membrane potential after NMDA challenge. The loss of mitochondrial membrane potential would lead to a decreased mitochondrial Ca2+ uptake and thus contribute to the rotenone-mediated ROS decrease.
Our data suggest that cytosolic pathways may not play a major role in ROS generation in cultured striatal neurons. Although PLA2-mediated metabolism of AA could produce ROS during glutamate excitotoxicity due to the Ca2+ dependence of PLA2, inhibition of PLA2 with 4-(4-octadecyl)-4-oxobenzene-butanoic acid (OOBB) only produced limited protection against glutamate-induced neuronal death (Ciani et al. 1996). Consistent with this observation, PLA2 inhibitor only caused a partial decrease in the early rate, but not the amplitude of the ROS generation in our study. Moreover, PLA2 is activated by Ca2+ at concentrations below micromolar (Six & Dennis, 2000). However, a detailed study of the relationship of glutamate-induced ROS production and DCD has shown that the increase in ROS production only started after DCD (Vesce et al. 2004). This raises the question of why ROS generation, if mediated by PLA2, is not stimulated by the glutamate-induced initial [Ca2+]i increase, which can be up to micromolar, that is, within the range of concentration that would activate PLA2 (White & Reynolds, 1997; Alano et al. 2002). Inhibition of another cytosolic ROS-generating pathway, the X/XO system, had almost no effect on ROS generation, which is also consistent with Ciani's study that the XO inhibitor, allopurinol, produced no protection against excitotoxic neuronal death. In addition, it is known that NOS, when activated with a low concentration of the substrate l-arginine can produce O2·− (Porasuphatana et al. 2003). However, the contribution of NOS-produced O2·− to NMDA-induced ROS production seems minor, if any, since both the general NOS inhibitor, l-NAME, and a nNOS-specific inhibitor (4S)-N-(4-amino-5[aminoethyl]aminopentyl)-N′-nitro-guanidine (50 μm, authors' unpublished data), failed to significantly change the ROS generation in our study. Recent studies have shown that another O2·−-producing enzyme, NADPH oxidase is also up-regulated in ischaemic brain (Vallet et al. 2005) and involved in ROS production (Abramov et al. 2007). However, this enzyme is unlikely to contribute to ROS production in the experimental settings of the present study, since according to Abramov's study, NADPH oxidase produced ROS after about 40 min, the time when the experiments in the present study were almost completed.
Our results showed that mitochondrial ROS generation is Ca2+-dependent. Moreover, the inhibition of this ROS generation by FCCP and Ru360 indicates that Ca2+ has to be taken up into mitochondria to stimulate ROS production. This is consistent with the central role that mitochondrial Ca2+ plays in excitotoxicity (Krieger & Duchen, 2002; Nicholls, 2004). Although the Ca2+ influx through the uniporter contributes to the majority of mitochondrial Ca2+ uptake, two other Ca2+ uptake mechanisms have been identified: RaM (Sparagna et al. 1995) and the ryanodine receptor (Beutner et al. 2001; Beutner et al. 2005), which account for fast Ca2+ uptakes. This could partially explain why Ru360, which specifically blocks the uniporter, only inhibited about 60% of the ROS increase (Fig. 6B). Interestingly, Ru360 almost completely inhibited NMDA-induced DNA damage and PARP-1 activation in our study. One possible explanation is that there may be a threshold for ROS. Above this threshold, ROS will induce all the damaging events and lead to cell death, but below this threshold, the ROS-induced damage may be minor. In other words, cells are able to handle a certain level of ROS, probably by the antioxidant system. Indeed cells need some ROS as signalling molecules under physiological conditions (Droge, 2002; Poli et al. 2004). In NMDA-treated cells, the ROS levels rise much higher than the threshold and therefore trigger those cell death events. Although Ru360 does not completely inhibit the ROS generation, it could bring the ROS levels below the threshold and therefore, we do not see much increase of the damaging effects.
Although our data showed that the ROS increase requires Ca2+ entry into mitochondria, the exact mechanism by which Ca2+ stimulates ROS production inside mitochondria is still unclear. Several possible mechanisms have been proposed previously (for review, see Brookes et al. 2004). For instance, mitochondrial Ca2+ overload may lead to the inhibition of respiratory activity (Kushnareva et al. 2005), or the releases of cytochrome c from mitochondria via the Ca2+-induced PTP (Newmeyer & Ferguson-Miller, 2003). Studies have shown that a pathological level of Ca2+ can release as much as 45% of the total releasable pool of cytochrome c (Petrosillo et al. 2004). Decreased levels of cytochrome c slow down the electron transfer from complex III to complex IV and therefore enhances ROS generation at the Q cycle. The cytochrome c release-triggered O2·− generation from mitochondria has been shown in both intact cells and isolated mitochondria (Cai & Jones, 1998; Kushnareva et al. 2002; Starkov et al. 2002). Actually, even the dislocation of cytochrome c from mitochondrial inner membrane, which is the first step of the two-step releasing process (Ott et al. 2002), would be enough to disrupt the ETC and induce ROS generation. In other words, ROS generation can occur even earlier than cytochrome c release from mitochondria. This may explain the temporal discrepancy between ROS generation and cytochrome c release, which is usually considered as a delayed event. Moreover, this effect may be enhanced by the activation of several Ca2+-sensitive dehydrogenases in the TCA cycle, which results in an increased production of substrates and thus more electron supply to the ETC (Hajnoczky et al. 1995; Duchen, 2000). This, together with the release of cytochrome c, synergistic increases the probability of reduced states of the chain components, and therefore the probability of electron leak to O2. Another possibility is Ca2+-induced production of NO or its derivatives, such as ONOO− and S-nitrosothiols (SNO), causing inhibitions of complex I and IV activities due to S-nitrosation of certain complex subunits (Stewart & Heales, 2003; Brown & Borutaite, 2004; Zhang et al. 2005). However, since NOS inhibitors had little effect on ROS generation in our study, this possibility is unlikely to be the case.
Our study also showed that mitochondrial Ca2+ uptake is required for PARP-1 activation. This novel finding is important because it suggests that PARP-1 activation is a downstream event of mitochondrial Ca2+ overload. The inhibition of both DNA strand breaks and PARP-1 activation by antioxidants indicates that mitochondrial ROS generation is a signal to activate PARP-1 via DNA damage. As the major reactive oxygen species produced from the ETC, O2·− will be converted to H2O2 by SOD. O2·− can also react with NO, which has been shown to be produced in glutamate excitotoxicity (Stout et al. 1998; Urushitani et al. 1998), to generate ONOO−. Both H2O2 and ONOO− could produce more severe DNA damage than O2·− because of the poor membrane permeability and low intrinsically reactivity of O2·−. Although our results did not directly show which species is more responsible for causing the DNA lesion, it seems that ONOO− is more relevant for the following reasons. Firstly, the SOD mimics, which enhance the dismutation of O2·− to H2O2, produced strong inhibition of DNA damage. This indicates that H2O2 may not induce much DNA damage in our conditions. Secondly, PARP-1 activation during NMDA excitotoxicity was also significantly inhibited by l-NAME, consistent with previous findings from an nNOS knockout mice study (Wang et al. 2004), indicating that NO is also critical for PARP-1 activation. This suggests that ONOO− may be the key signal for PARP-1 activation.
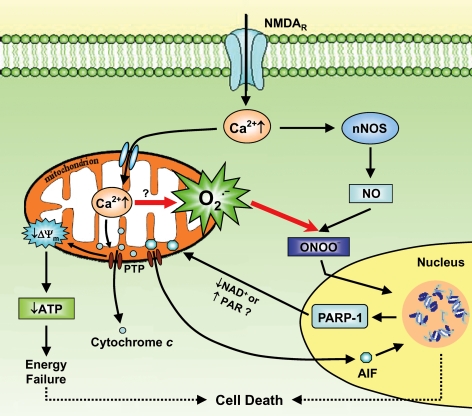
In conclusion, the present observations link mitochondrial Ca2+ overload and nuclear PARP-1 activation as a unified mechanism for excitotoxic neuronal death. This is summarized in Fig. 8. In this scheme, the Ca2+-mediated mitochondrial ROS work along with NO for PARP-1 activation during the progression of excitotoxicity.
Figure 8. Diagram of the excitotoxic cascade showing mitochondrial ROS generation as the link between mitochondrial Ca2+ uptake and PARP-1 activation in excitotoxicity.
Under pathological conditions, the massively activated NMDA receptors cause a strong increase in the cytosolic Ca2+ level, which is then taken up by mitochondria. The mitochondrial Ca2+ overload leads to a loss of ΔΨm and an explosion of O2·− generation, probably through the opening of PTP and the subsequent release of cytochrome c. The elevated cytosolic Ca2+ also activates nNOS and increases NO production. ONOO− is formed from the reaction of O2·− with NO, and then diffuses into the nucleus to cause DNA damage. In response to this DNA damage, PARP-1 is activated, which results in excessive production of PAR polymers and depletion of NAD+. The activation of PARP-1 further induces AIF translocation from the mitochondria to the nucleus, which causes DNA fragmentation. The mechanism for the PARP-1 activation-dependent AIF translocation is still not fully understood, both the depletion of NAD+ and the accumulation of PAR polymer could be the hypothetic signal (Alano et al. 2004; Yu et al. 2006). Finally, the profound DNA damage in combination with the energy failure caused by mitochondrial dysfunction and NAD+ depletion leads to cell death. NAD+, nicotinamide adenine dinucleotide.
Acknowledgments
We thank Dr Gisela Beutner for comments on this manuscript. We also thank Mark Gallagher for the help on striatal neuronal cultures. This work is supported by NIH Grant NS37710, NYS Spinal Cord Injury research programs CO17688 and C020941, and AHA Predoctoral Fellowship 0315306T.
Supplemental material
Online supplemental material for this paper can be accessed at: http://jp.physoc.org/cgi/content/full/jphysiol.2007.145409/DC1 and http://www.blackwell-synergy.com/doi/suppl/10.1113/jphysiol.2007.145409
References
- Abramov AY, Scorziello A, Duchen MR. Three distinct mechanisms generate oxygen free radicals in neurons and contribute to cell death during anoxia and reoxygenation. J Neurosci. 2007;27:1129–1138. doi: 10.1523/JNEUROSCI.4468-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alano CC, Beutner G, Dirksen RT, Gross RA, Sheu S-S. Mitochondrial permeability transition and calcium dynamics in striatal neurons upon intense NMDA receptor activation. J Neurochem. 2002;80:531–538. doi: 10.1046/j.0022-3042.2001.00738.x. [DOI] [PubMed] [Google Scholar]
- Alano CC, Ying W, Swanson RA. Poly (ADP-ribose) polymerase-1-mediated cell death in astrocytes requires NAD+ depletion and mitochondrial permeability transition. J Biol Chem. 2004;279:18895–18902. doi: 10.1074/jbc.M313329200. [DOI] [PubMed] [Google Scholar]
- Atlante A, Gagliardi S, Minervini GM, Ciotti MT, Marra E, Calissano P. Glutamate neurotoxicity in rat cerebellar granule cells: a major role for xanthine oxidase in oxygen radical formation. J Neurochem. 1997;68:2038–2045. doi: 10.1046/j.1471-4159.1997.68052038.x. [DOI] [PubMed] [Google Scholar]
- Becker LB, vanden Hoek TL, Shao ZH, Li CQ, Schumacker PT. Generation of superoxide in cardiomyocytes during ischemia before reperfusion. Am J Physiol Heart Circ Physiol. 1999;277:H2240–H2246. doi: 10.1152/ajpheart.1999.277.6.H2240. [DOI] [PubMed] [Google Scholar]
- Beutner G, Sharma VK, Giovannucci DR, Yule DI, Sheu SS. Identification of a ryanodine receptor in rat heart mitochondria. J Biol Chem. 2001;276:21482–21488. doi: 10.1074/jbc.M101486200. [DOI] [PubMed] [Google Scholar]
- Beutner G, Sharma VK, Lin L, Ryu SY, Dirksen RT, Sheu SS. Type 1 ryanodine receptor in cardiac mitochondria: transducer of excitation-metabolism coupling. Biochim Biophys Acta. 2005;1717:1–10. doi: 10.1016/j.bbamem.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Bindokas VP, Jordan J, Lee CC, Miller RJ. Superoxide production in rat hippocampal neurons: selective imaging with hydroethidine. J Neurosci. 1996;16:1324–1336. doi: 10.1523/JNEUROSCI.16-04-01324.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveris A, Cadenas E, Stoppani AO. Role of ubiquinone in the mitochondrial generation of hydrogen peroxide. Biochem J. 1976;156:435–444. doi: 10.1042/bj1560435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes PS, Yoon Y, Robotham JL, Anders MW, Sheu SS. Calcium, ATP, and ROS: a mitochondrial love-hate triangle. Am J Physiol Cell Physiol. 2004;287:C817–C833. doi: 10.1152/ajpcell.00139.2004. [DOI] [PubMed] [Google Scholar]
- Brown GC, Borutaite V. Inhibition of mitochondrial respiratory complex I by nitric oxide, peroxynitrite and S-nitrosothiols. Biochim Biophys Acta. 2004;1658:44–49. doi: 10.1016/j.bbabio.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Budd SL, Nicholls DG. Mitochondria, calcium regulation, and acute glutamate excitotoxicity in cultured cerebellar granule cells. J Neurochem. 1996;67:2282–2291. doi: 10.1046/j.1471-4159.1996.67062282.x. [DOI] [PubMed] [Google Scholar]
- Cadenas E, Boveris A, Ragan CI, Stoppani AO. Production of superoxide radicals and hydrogen peroxide by NADH-ubiquinone reductase and ubiquinol-cytochrome c reductase from beef-heart mitochondria. Arch Biochem Biophys. 1977;180:248–257. doi: 10.1016/0003-9861(77)90035-2. [DOI] [PubMed] [Google Scholar]
- Cai J, Jones DP. Superoxide in apoptosis. Mitochondrial generation triggered by cytochrome c loss. J Biol Chem. 1998;273:11401–11404. doi: 10.1074/jbc.273.19.11401. [DOI] [PubMed] [Google Scholar]
- Carriedo SG, Sensi SL, Yin HZ, Weiss JH. AMPA exposures induce mitochondrial Ca2+ overload and ROS generation in spinal motor neurons in vitro. J Neurosci. 2000;20:240–250. doi: 10.1523/JNEUROSCI.20-01-00240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DW. Ionic dependence of glutamate neurotoxicity. J Neurosci. 1987;7:369–379. doi: 10.1523/JNEUROSCI.07-02-00369.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DW, Rothman SM. The role of glutamate neurotoxicity in hypoxic-ischemic neuronal death. Annu Rev Neurosci. 1990;13:171–182. doi: 10.1146/annurev.ne.13.030190.001131. [DOI] [PubMed] [Google Scholar]
- Ciani E, Groneng L, Voltattorni M, Rolseth V, Contestabile A, Paulsen RE. Inhibition of free radical production or free radical scavenging protects from the excitotoxic cell death mediated by glutamate in cultures of cerebellar granule neurons. Brain Res. 1996;728:1–6. [PubMed] [Google Scholar]
- Costa AD, Quinlan CL, Andrukhiv A, West IC, Jaburek M, Garlid KD. The direct physiological effects of mitoK (ATP) opening on heart mitochondria. Am J Physiol Heart Circ Physiol. 2006;290:H406–H415. doi: 10.1152/ajpheart.00794.2005. [DOI] [PubMed] [Google Scholar]
- Droge W. Free radicals in the physiological control of cell function. Physiol Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- Duchen MR. Ca2+-dependent changes in the mitochondrial energetics in single dissociated mouse sensory neurons. Biochem J. 1992;283:41–50. doi: 10.1042/bj2830041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchen MR. Mitochondria and calcium: from cell signalling to cell death. J Physiol. 2000;529:57–68. doi: 10.1111/j.1469-7793.2000.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan LL, Sensi SL, Canzoniero LM, Handran SD, Rothman SM, Lin TS, Goldberg MP, Choi DW. Mitochondrial production of reactive oxygen species in cortical neurons following exposure to N-methyl-D-aspartate. J Neurosci. 1995;15:6377–6388. doi: 10.1523/JNEUROSCI.15-10-06377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunter KK, Gunter TE. Transport of calcium by mitochondria. J Bioenerg Biomembr. 1994;26:471–485. doi: 10.1007/BF00762732. [DOI] [PubMed] [Google Scholar]
- Hajnoczky G, Robb-Gaspers LD, Seitz MB, Thomas AP. Decoding of cytosolic calcium oscillations in the mitochondria. Cell. 1995;82:415–424. doi: 10.1016/0092-8674(95)90430-1. [DOI] [PubMed] [Google Scholar]
- Hartley Z, Dubinsky JM. Changes in intracellular pH associated with glutamate excitotoxicity. J Neurosci. 1993;13:4690–4699. doi: 10.1523/JNEUROSCI.13-11-04690.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongpaisan J, Winters CA, Andrews SB. Strong calcium entry activates mitochondrial superoxide generation, upregulating kinase signaling in hippocampal neurons. J Neurosci. 2004;24:10878–10887. doi: 10.1523/JNEUROSCI.3278-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynd MR, Scott HL, Dodd PR. Glutamate-mediated excitotoxicity and neurodegeneration in Alzheimer's disease. Neurochem Int. 2004;45:583–595. doi: 10.1016/j.neuint.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Kennedy LJ, Moore K, Jr, Caulfield JL, Tannenbaum SR, Dedon PC. Quantitation of 8-oxoguanine and strand breaks produced by four oxidizing agents. Chem Res Toxicol. 1997;10:386–392. doi: 10.1021/tx960102w. [DOI] [PubMed] [Google Scholar]
- Kirkland RA, Franklin JL. Evidence for redox regulation of cytochrome c release during programmed neuronal death: antioxidant effects of protein synthesis and caspase inhibition. J Neurosci. 2001;21:1949–1963. doi: 10.1523/JNEUROSCI.21-06-01949.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger C, Duchen MR. Mitochondria, Ca2+ and neurodegenerative disease. Eur J Pharmacol. 2002;447:177–188. doi: 10.1016/s0014-2999(02)01842-3. [DOI] [PubMed] [Google Scholar]
- Kushnareva Y, Murphy AN, Andreyev A. Complex I-mediated reactive oxygen species generation: modulation by cytochrome c and NAD(P)+ oxidation-reduction state. Biochem J. 2002;368:545–553. doi: 10.1042/BJ20021121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnareva YE, Wiley SE, Ward MW, Andreyev AY, Murphy AN. Excitotoxic injury to mitochondria isolated from cultured neurons. J Biol Chem. 2005;280:28894–28902. doi: 10.1074/jbc.M503090200. [DOI] [PubMed] [Google Scholar]
- Lafon-Cazal M, Pietri S, Culcasi M, Bockaert J. NMDA-dependent superoxide production and neurotoxicity. Nature. 1993;364:535–537. doi: 10.1038/364535a0. [DOI] [PubMed] [Google Scholar]
- Lazarewicz JW, Wroblewski JT, Costa E. N-methyl-D-aspartate-sensitive glutamate receptors induce calcium-mediated arachidonic acid release in primary cultures of cerebellar granule cells. J Neurochem. 1990;55:1875–1881. doi: 10.1111/j.1471-4159.1990.tb05771.x. [DOI] [PubMed] [Google Scholar]
- Lesnefsky EJ, Moghaddas S, Tandler B, Kerner J, Hoppel CL. Mitochondrial dysfunction in cardiac disease: ischemia–reperfusion, aging, and heart failure. J Mol Cell Cardiol. 2001;33:1065–1089. doi: 10.1006/jmcc.2001.1378. [DOI] [PubMed] [Google Scholar]
- Mandir AS, Poitras MF, Berliner AR, Herring WJ, Guastella DB, Feldman A, Poirier GG, Wang Z-Q, Dawson TM, Dawson VL. NMDA but not non-NMDA excitotoxicity is mediated by poly (ADP-ribose) polymerase. J Neurosci. 2000;20:8005–8011. doi: 10.1523/JNEUROSCI.20-21-08005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller FL, Liu Y, Van Remmen H. Complex III releases superoxide to both sides of the inner mitochondrial membrane. J Biol Chem. 2004;279:49064–49073. doi: 10.1074/jbc.M407715200. [DOI] [PubMed] [Google Scholar]
- Newmeyer DD, Ferguson-Miller S. Mitochondria: releasing power for life and unleashing the machineries of death. Cell. 2003;112:481–490. doi: 10.1016/s0092-8674(03)00116-8. [DOI] [PubMed] [Google Scholar]
- Nicholls DG. Mitochondrial dysfunction and glutamate excitotoxicity studied in primary neuronal cultures. Curr Mol Med. 2004;4:149–177. doi: 10.2174/1566524043479239. [DOI] [PubMed] [Google Scholar]
- Nicholls DG, Budd SL. Mitochondria and neuronal survival. Physiol Rev. 2000;80:315–360. doi: 10.1152/physrev.2000.80.1.315. [DOI] [PubMed] [Google Scholar]
- Nishikawa T, Du Edelstein DXL, Yamagishi S, Matsumura T, Kaneda Y, Yorek MA, Beebe D, Oates PJ, Hammes HP, Giardino I, Brownlee M. Normalizing mitochondrial superoxide production blocks three pathways of hyperglycaemic damage. Nature. 2000;404:787–790. doi: 10.1038/35008121. [DOI] [PubMed] [Google Scholar]
- Olive PL, Banath JP, Durand RE. Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the ‘comet’ assay. Radiat Res. 1990;122:86–94. [PubMed] [Google Scholar]
- Ott M, Robertson JD, Gogvadze V, Zhivotovsky B, Orrenius S. Cytochrome c release from mitochondria proceeds by a two-step process. Proc Natl Acad Sci U S A. 2002;99:1259–1263. doi: 10.1073/pnas.241655498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M, Day BJ, Crapo JD, Fridovich I, McNamara JO. Requirement for superoxide in excitotoxic cell death. Neuron. 1996;16:345–355. doi: 10.1016/s0896-6273(00)80052-5. [DOI] [PubMed] [Google Scholar]
- Peng TI, Jou MJ, Sheu SS, Greenamyre JT. Visualization of NMDA receptor-induced mitochondrial calcium accumulation in striatal neurons. Exp Neurol. 1998;149:1–12. doi: 10.1006/exnr.1997.6599. [DOI] [PubMed] [Google Scholar]
- Petrosillo G, Ruggiero FM, Pistolese M, Paradies G. Ca2+-induced reactive oxygen species production promotes cytochrome c release from rat liver mitochondria via mitochondrial permeability transition (MPT)-dependent and MPT-independent mechanisms: role of cardiolipin. J Biol Chem. 2004;279:53103–53108. doi: 10.1074/jbc.M407500200. [DOI] [PubMed] [Google Scholar]
- Poli G, Leonarduzzi G, Biasi F, Chiarpotto E. Oxidative stress and cell signalling. Curr Med Chem. 2004;11:1163–1182. doi: 10.2174/0929867043365323. [DOI] [PubMed] [Google Scholar]
- Porasuphatana S, Tsai P, Rosen GM. The generation of free radicals by nitric oxide synthase. Comp Biochem Physiol C Toxicol Pharmacol. 2003;134:281–289. doi: 10.1016/s1532-0456(02)00271-5. [DOI] [PubMed] [Google Scholar]
- Reynolds IJ, Hastings TG. Glutamate induces the production of reactive oxygen species in cultured forebrain neurons following NMDA receptor activation. J Neurosci. 1995;15:3318–3327. doi: 10.1523/JNEUROSCI.15-05-03318.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Meana M, Garcia-Dorado D, Pina P, Inserte J, Agullo L, Soler-Soler J. Cariporide preserves mitochondrial proton gradient and delays ATP depletion in cardiomyocytes during ischemic conditions. Am J Physiol Heart Circ Physiol. 2003;285:H999–H1006. doi: 10.1152/ajpheart.00035.2003. [DOI] [PubMed] [Google Scholar]
- Six DA, Dennis EA. The expanding superfamily of phospholipase A2 enzymes: classification and characterization. Biochim Biophys Acta. 2000;1488:1–19. doi: 10.1016/s1388-1981(00)00105-0. [DOI] [PubMed] [Google Scholar]
- Sparagna GC, Gunter KK, Sheu SS, Gunter TE. Mitochondrial calcium uptake from physiological-type pulses of calcium. A description of the rapid uptake mode. J Biol Chem. 1995;270:27510–27515. doi: 10.1074/jbc.270.46.27510. [DOI] [PubMed] [Google Scholar]
- Starkov AA, Polster BM, Fiskum G. Regulation of hydrogen peroxide production by brain mitochondria by calcium and Bax. J Neurochem. 2002;83:220–228. doi: 10.1046/j.1471-4159.2002.01153.x. [DOI] [PubMed] [Google Scholar]
- Stewart VC, Heales SJ. Nitric oxide-induced mitochondrial dysfunction: implications for neurodegeneration. Free Radic Biol Med. 2003;34:287–303. doi: 10.1016/s0891-5849(02)01327-8. [DOI] [PubMed] [Google Scholar]
- Stout AK, Raphael HM, Kanterewicz BI, Klann E, Reynolds IJ. Glutamate-induced neuron death requires mitochondrial calcium uptake. Nat Neurosci. 1998;1:366–373. doi: 10.1038/1577. [DOI] [PubMed] [Google Scholar]
- Szabo C, Ohshima H. DNA damage induced by peroxynitrite: subsequent biological effects. Nitric Oxide. 1997;1:373–385. doi: 10.1006/niox.1997.0143. [DOI] [PubMed] [Google Scholar]
- Urushitani M, Nakamizo T, Inoue R, Sawada H, Kihara T, Honda K, Akaike A, Shimohama S. N-methyl-D-aspartate receptor-mediated mitochondrial Ca2+ overload in acute excitotoxic motor neuron death: a mechanism distinct from chronic neurotoxicity after Ca2+ influx. J Neurosci Res. 2001;63:377–387. doi: 10.1002/1097-4547(20010301)63:5<377::AID-JNR1032>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Urushitani M, Shimohama S, Kihara T, Sawada H, Akaike A, Ibi M, Inoue R, Kitamura Y, Taniguchi T, Kimura J. Mechanism of selective motor neuronal death after exposure of spinal cord to glutamate: involvement of glutamate-induced nitric oxide in motor neuron toxicity and nonmotor neuron protection. Ann Neurol. 1998;44:796–807. doi: 10.1002/ana.410440514. [DOI] [PubMed] [Google Scholar]
- Vallet P, Charnay Y, Steger K, Ogier-Denis E, Kovari E, Herrmann F, Michel JP, Szanto I. Neuronal expression of the NADPH oxidase NOX4, and its regulation in mouse experimental brain ischemia. Neuroscience. 2005;132:233–238. doi: 10.1016/j.neuroscience.2004.12.038. [DOI] [PubMed] [Google Scholar]
- Vesce S, Kirk L, Nicholls DG. Relationships between superoxide levels and delayed calcium deregulation in cultured cerebellar granule cells exposed continuously to glutamate. J Neurochem. 2004;90:683–693. doi: 10.1111/j.1471-4159.2004.02516.x. [DOI] [PubMed] [Google Scholar]
- Votyakova TV, Reynolds IJ. Ca2+-induced permeabilization promotes free radical release from rat brain mitochondria with partially inhibited complex I. J Neurochem. 2005;93:526–537. doi: 10.1111/j.1471-4159.2005.03042.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Yu S-W, Koh DW, Lew J, Coombs C, Bowers W, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor substitutes for caspase executioners in NMDA-triggered excitotoxic neuronal death. J Neurosci. 2004;24:10963–10973. doi: 10.1523/JNEUROSCI.3461-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor mediates poly (ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci U S A. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S-W, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly (ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- White RJ, Reynolds IJ. Mitochondrial depolarization in glutamate-stimulated neurons: an early signal specific to excitotoxin exposure. J Neurosci. 1996;16:5688–5697. doi: 10.1523/JNEUROSCI.16-18-05688.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ, Reynolds IJ. Mitochondria accumulate Ca2+ following intense glutamate stimulation of cultured rat forebrain neurones. J Physiol. 1997;498:31–47. doi: 10.1113/jphysiol.1997.sp021839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Dawson VL, Dawson TM, Snyder SH. Nitric oxide activation of poly (ADP-ribose) synthetase in neurotoxicity. Science. 1994;263:687–689. doi: 10.1126/science.8080500. [DOI] [PubMed] [Google Scholar]
- Zhang J, Jin B, Li L, Block ER, Patel JM. Nitric oxide-induced persistent inhibition and nitrosylation of active site cysteine residues of mitochondrial cytochrome-c oxidase in lung endothelial cells. Am J Physiol Cell Physiol. 2005;288:C840–C849. doi: 10.1152/ajpcell.00325.2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.