Abstract
Human physical capability is influenced by many environmental and genetic factors, and it is generally accepted that physical capability phenotypes are highly polygenic. However, the ways in which relevant polymorphisms combine to influence the physical capability of individuals and populations are unknown. Initially, the literature was searched to identify associations between 23 genetic polymorphisms and human endurance phenotypes. Next, typical genotype frequencies of those polymorphisms in the general population were obtained from suitable literature. Using probability calculations, we found only a 0.0005% chance of a single individual in the world having the ‘preferable’ form of all 23 polymorphisms. As the number of DNA variants shown to be associated with human endurance phenotypes continues to increase, the probability of any single individual possessing the ‘preferable’ form of each polymorphism will become even lower. However, with population turnover, the chance of such genetically gifted individuals existing increases. To examine the polygenic endurance potential of a human population, a ‘total genotype score’ (for the 23 polymorphisms) was calculated for each individual within a hypothetical population of 1000 000. There was considerable homogeneity in terms of genetic predisposition to high endurance potential, with 99% of people differing by no more than seven genotypes from the typical profile. Consequently, with population turnover world and Olympic records should improve even without further enhancement of environmental factors, as more ‘advantageous’ polygenic profiles occasionally, though rarely, emerge. More broadly, human potential appears limited by the similarity of polygenic profiles at both the ‘elite sport’ and ‘chronic disorder’ ends of the performance continuum.
Achievement at the limit of human physical performance is exemplified in the Olympic Games by elite athletic (track and field specialties) performance, and ultimately by Olympic and world records (http://www.iaaf.org/statistics/records/index.html). Human physical performance is multifactorial and determined by a range of environmental (physical training, nutrition and technological aids) and genetic factors. For example, heritability estimates for the maximal rate of oxygen uptake (a key endurance phenotype) are typically around 50% both in sedentary individuals and in terms of their response to training (Klissouras, 1971; Bouchard et al. 1986, 1998, 1999; Fagard et al. 1991). This heritability is regarded as primarily due to genetic as opposed to non-genetic factors. Indeed, it has been proposed that endurance running ability in the genus Homo evolved about two million years ago (Bramble & Lieberman, 2004), possibly including the enlargement of the gluteus maximus muscle (Lieberman et al. 2006), implicating a positive natural selection for endurance running ability (and thus endurance phenotypes such as the maximal rate of oxygen uptake) in the ancestors of modern humans.
In addition to the maximal rate of oxygen uptake, at least two other endurance phenotypes (economy of movement and lactate/ventilatory threshold) also contribute to the endurance performance phenotype (time taken to travel a given distance) seen in elite competition. Current opinion also includes a fourth, related (but arguably discrete) phenotype, namely oxygen uptake kinetics (Jones & Carter, 2000). While the latter two endurance phenotypes listed (lactate/ventilatory threshold and oxygen uptake kinetics) are yet to be associated with specific genetic polymorphisms in a healthy adult population (and are therefore suitable targets for new association and case-control studies), each of the four endurance phenotypes will have genetic components, which collectively form the genetics of endurance performance.
To date, over 150 DNA polymorphisms have been associated with some form of human physical performance or a health-related fitness phenotype (Rankinen et al. 2006a). For many of the polymorphisms associated with human performance, there has only been a single positive association with a relevant phenotype. Notable exceptions to this statement include the polymorphisms of the ACE (angiotensin I-converting enzyme (peptidyl-dipeptidase A) 1) and ACTN3 (actinin, α3) genes that have been studied by several research groups, using a variety of experimental designs and population types. Replication of association with endurance performance or an endurance phenotype has occurred for just six polymorphisms (ACE, ACTN3, ADRB2 (adrenergic, β-2-, receptor, surface), AMPD1 (adenosine monophosphate deaminase 1 (isoform M)), APOE (apolipoprotein E) and BDKRB2 (bradykinin receptor B2)) and replication of the associations reported for all polymorphisms associated with human performance would obviously increase confidence in the previously reported findings. These observations are indicative of a field of research in its infancy. Nevertheless, the weight of evidence suggests, and indeed it is widely accepted, that physical performance phenotypes are highly polygenic (Rankinen et al. 2006a; Spurway, 2006).
One issue that has not yet been examined, however, is the way in which the relevant polymorphisms combine to influence physical performance potential. The prediction of risk for common complex disease via a combination of relevant polymorphisms has recently been addressed in principle (Yang et al. 2005) and using coronary heart disease risk as a specific focus (Drenos et al. 2007). However, such predictions have yet to be tested experimentally for clinical use. In contrast to common diseases, the potential applicability of such a combinatory polymorphic approach in elite sport would, by definition, focus on probabilities related to the elite end of the human physical performance continuum. However, the probability of individuals carrying the ‘preferable’ genotype for each polymorphism linked to endurance (or indeed any) performance has not been calculated. As many of the ‘preferable’ genotypes of polymorphisms linked to endurance performance are quite rare (< 25% of the population) it seems probable that very few individuals will possess the preferable genotype for each of even a small number of polymorphisms. If these ‘perfect’ genetic individuals (from an endurance perspective) are very rare, the genetic predisposition for endurance of the vast majority of the human population depends on the number of advantageous and disadvantageous genotypes they may possess. Consequently, the first purpose of this study was to calculate, using current knowledge regarding polymorphic associations with endurance performance, whether individuals are likely to exist who possess all ‘preferable’ genotypes for endurance. The second purpose was to assess the distribution of genetic endurance potential for an entire population, via the generation of a ‘total genotype score’ dependent upon the nature and frequencies of all known relevant polymorphisms.
Methods
Overview
Initially, common genetic polymorphisms associated with human endurance performance were identified from the literature, and their typical genotype frequencies established. Next, the probability of a given individual having the ‘preferable’ genotype for each of the polymorphisms linked with endurance phenotypes was calculated, and the results also applied to population statistics. Finally, in order to examine the human genetic heterogeneity for endurance potential, a ‘total genotype score’ (for those polymorphisms linked to endurance phenotypes) was calculated for each individual within a hypothetical population, and the distribution of total genotype scores examined.
Inclusion and exclusion criteria
Initially, we searched the literature for common genetic polymorphisms (minor allele frequency in the general population ≥ 0.01) associated with human endurance exercise performance or a related endurance phenotype, either in gene association studies or case-control studies between elite endurance athletes and other individuals. The search was primarily based on journals indexed in Medline (a National Library of Medicine publication database) using various combinations of key words (e.g. genotype, physical fitness, performance). We also used the most recently published human gene map for performance phenotypes (Rankinen et al. 2006a) and searching of personal libraries. For inclusion, a polymorphism needed to show at least one statistically significant association with endurance performance or an endurance phenotype in healthy Caucasians (data on Caucasians predominate in this field). We felt that this key objective criterion of published statistical significance was consistent with other approaches (Rankinen et al. 2006a,b) and avoided the difficult task of repeatedly passing subjective judgement upon the value of one published study versus another at this relatively early stage of research in this field. Age and sex were not exclusion criteria, but disease and smoking were. Rare genetic polymorphisms related to severely impaired muscle function were not included in our analysis, e.g. muscular dystrophies (Schessl et al. 2006) or disrupted mitochondrial function (Tarnopolsky & Raha, 2005). This process of literature searching and considering published studies against our criteria produced a list of 23 polymorphisms eligible for further analysis.
Following these objective criteria may have led to the inclusion of some polymorphisms that are controversial. For example, some studies have found an association between the R577X polymorphism of the ACTN3 gene and elite endurance athlete status (Yang et al. 2003; Niemi & Majamaa, 2005), whilst three more recent studies reported no evidence of an association with endurance (Lucia et al. 2006; Moran et al. 2007; Paparini et al. 2007). Nevertheless the ACTN3 R577X polymorphism meets our criterion of at least one published statistically significant association with endurance performance or an endurance phenotype, and consequently it was included in our analysis.
Another key decision was not to include some polymorphisms because the reported associations with endurance phenotypes were based on studies that did not use healthy, non-smoking, Caucasian adults as participants. For example, the G-174C polymorphism of the IL6 gene has been strongly associated (P= 0.002) with an endurance phenotype in smokers (Ortlepp et al. 2003), but in the same study there was no association between that polymorphism and the endurance phenotype within a comparable group of non-smokers (P= 0.937). Thus, the IL6 G-174C polymorphism may interact with smoking practices to influence physical endurance, but does not appear relevant to our analysis of endurance phenotype in healthy, non-smoking, Caucasian adults and therefore was not included in our analysis.
Typical genotype frequencies
Data regarding typical genotype frequencies within the 23 polymorphisms in the general population were obtained from suitable literature, generally using the studies with the largest samples. Data from Caucasian populations were again used, primarily UK Caucasians for consistency, occasionally other European (and European descent) populations. In some cases, the typical frequency data were obtained from the same study that provided the statistically significant association with endurance because that study provided the largest, most appropriate, or even the only source of typical frequency data.
Occurrence of individuals with an ‘optimal’ polygenic profile for endurance
What is the probability of an individual having the ‘optimal’ polygenic profile for endurance? To answer this question, we calculated the probability of any given individual possessing every ‘optimal’ genotype for 1 up to all 23 polymorphisms. We selected the polymorphisms in the alphabetical sequence of their official symbols as recommended by the Human Gene Nomenclature Committee (HGNC), which, though arbitrary, ultimately did not notably bias the results. Then, based on the typical frequencies of the ‘optimal’ genotypes, we produced a scale of decreasing probability that any given individual will possess an increasing number of ‘optimal’ genotypes.
Applying data to population statistics
The probability data for the occurrence of individuals with ‘optimal’ genetic potential were then related to population statistics. Hence, we predicted the number of individuals with a given number of optimal genotypes, or indeed the probability that one individual exists, in a medium-sized nation (e.g. 60 million in the UK (http://www.statistics.gov.uk/cci/nugget.asp?id=6)) and the 6.5 billion world population (http://www.census.gov/ipc/www/idb/worldpopinfo.html). Using the polymorphisms selected in sequence of their official HGNC symbols, we applied our data to every possible number of polymorphisms from 1 to 23. As the probability of even a single individual possessing a large number of selected optimal genotypes was so low, we also calculated the probability that an individual possesses any 22 of the 23 optimal genotypes and then extended that to any 21 of the 23.
Polygenic potential for endurance of a whole population
To examine the genetic potential for endurance of all individuals in a wider population (rather than just the upper extreme of the population, as in the previous sections), it was first necessary to allocate scores to each genotype within a particular polymorphism. Typically, the polymorphisms identified were bi-allelic, providing three possible genotypes. The homozygote genotype associated with the better endurance phenotype was given a ‘score’ of 2, with a linear trend applied such that the heterozygotes were scored 1 and the other homozygotes, 0. In studies including only homozygotes (e.g. Williams et al. 2000), heterozygotes were given an intermediate score of 1. For the APOE polymorphism there are three alleles, and thus six genotypes. Based on two studies (Hagberg et al. 1999; Thompson et al. 2004), the ‘optimal’APOE allele for endurance phenotypes was determined as E4, so E4/E4 and E3/E4 scored 2, E3/E3 and E2/E4 scored 1, E2/E2 and E2/E3 scored 0. For the four mitochondrial DNA polymorphisms (Dionne et al. 1991), there were only two possible morphs in each case, scored 2 or 0. The maximum and minimum available values were chosen for the mitochondrial DNA polymorphisms to ensure equivalent weighting to the chromosomal polymorphisms. The genotype scores allocated to each polymorphism can be viewed in the Supplemental material.
To quantify the combined influence of all 23 polymorphisms currently associated with endurance phenotypes, an algorithm was created to incorporate all 23 genotype scores for any given individual in a simple additive model. The total score was then transformed mathematically to lie within the range 0–100 (to be intuitively meaningful) and labelled the ‘total genotype score’ (TGS). Considering the 23 genotype scores GS1, GS2… GS23, the TGS can be calculated as:
A TGS of 100 represents a ‘perfect’ polygenic profile for endurance and a TGS of 0 represents the ‘worst’ possible profile for endurance. The word ‘perfect’ is, of course, only to be interpreted within the context of this paper – when other polymorphisms are shown to be associated with endurance in the future, as we strongly expect, the ‘perfect’ profile will of course change (and actually become less likely to occur spontaneously). To examine the nature of the distribution of total genotype scores in the general population, we created a data set of 1000 000 hypothetical individuals, each with a randomly generated genetic profile (for all 23 polymorphisms) based on the typical frequency of each genotype. Finally, we evaluated the distribution of TGSs within this population, and calculated the degrees of kurtosis and skewness using the ‘Descriptives’ facility within the statistical software package SPSS (v.11).
Results
The complete list of the 23 polymorphisms associated with endurance phenotypes, including full names, typical genotype frequencies and the sources of all information, can be accessed in the Supplemental material. The list in alphabetical order of the official HGNC symbols is also shown in Table 1.
Table 1.
Probability of perfect genetic profile by number of polymorphisms
| Probability of possessing a ‘perfect’ profile | ||||
|---|---|---|---|---|
| Number of polymorphisms influencing endurance performance | New gene included at each stage | Typical frequency of optimal genotype (%) | % chance | Approximate odds ratio |
| 1 | ACE | 21 | 21.0 | 1 : 5 |
| 2 | ACTN3 | 18 | 3.78 | 1 : 25 |
| 3 | ADRA2 | 62 | 2.34 | 1 : 40 |
| 4 | ADRB2 | 35 | 0.82 | 1 : 120 |
| 5 | AMPD1 | 80 | 0.66 | 1 : 150 |
| 6 | APOE | 24 | 0.16 | 1 : 600 |
| 7 | ATP1A2* | 81 | 0.13 | 1 : 800 |
| 8 | ATP1A2 | 5 | 6.4 × 10−3 | 1 : 16 000 |
| 9 | BDKRB2 | 15 | 9.6 × 10−4 | 1 : 100 000 |
| 10 | CKM | 49 | 4.7 × 10−4 | 1 : 200 000 |
| 11 | EPAS1* | 33 | 1.5 × 10−4 | 1 : 600 000 |
| 12 | EPAS1 | 19 | 2.9 × 10−5 | 1 : 3 million |
| 13 | HFE | 4 | 1.2 × 10−6 | 1 : 85 million |
| 14 | HIF1A | 77 | 9.1 × 10−7 | 1 : 110 million |
| 15 | HLA-A | 2 | 1.8 × 10−8 | 1 : 5.5 billion |
| 16 | MT-ND5* | 93 | 1.7 × 10−8 | 1 : 6 billion |
| 17 | MT-ND5 | 7 | 1.2 × 10−9 | 1 : 85 billion |
| 18 | MT-ND5 | 7 | 8.3 × 10−11 | 1 : 1.2 trillion |
| 19 | MT-TT | 7 | 5.8 × 10−12 | 1 : 17 trillion |
| 20 | PPARA | 70 | 4.0 × 10−12 | 1 : 25 trillion |
| 21 | PPARGC1A | 40 | 1.6 × 10−12 | 1 : 62 trillion |
| 22 | UCP2 | 17 | 2.7 × 10−13 | 1 : 364 trillion |
| 23 | VEGFA | 30 | 8.2 × 10−14 | 1 : 1212 trillion |
There is an exponential decrease in the probability of an individual possessing a ‘perfect’ genetic profile as the number of polymorphisms included increases. The probability of any given individual possessing a high number of genotypes optimal for endurance performance is extremely slim.
Repeated entries of a gene reflects the inclusion of more than one polymorphism of that gene.
Occurrence of individuals with an ‘optimal’ polygenic profile for endurance
For the 23 polymorphisms identified, typical genotype frequencies in the general population ranged from 1% (for the TT genotype of the AMPD1 gene (Rico-Sanz et al. 2003) found on chromosome 1) to 93% (for some mitochondrial DNA morphs; Dionne et al. 1991). The typical frequency of the ‘optimal’ genotype of each polymorphism is shown in column 3 of Table 1. Given those data, how likely is it that a given individual possesses a ‘perfect’ genetic profile for a given number of polymorphisms? Considering one polymorphism only (ACE I/D; Williams et al. 2000), the optimal genotype for endurance performance (II) has a typical frequency of approximately 21% in the wider Caucasian population (Mastana et al. 2003). If a second gene (ACTN3; Yang et al. 2003) is also considered, an individual's probability of possessing the optimal ACE and ACTN3 (XX genotype; ∼18% typical frequency) genotypes is already considerably reduced to approximately 4%. Adding a third gene (ADRA2; Wolfarth et al. 2000) with a typical frequency of the optimal genotype (6.7/6.7) of about 62% further reduces an individual's chance of possessing all three optimal genotypes to just 2%. The probability of a given individual possessing all optimal genotypes continues to decrease exponentially until, when all 23 polymorphisms are considered, the probability is just 8.2 × 10−14%, an enormous odds ratio of 1 in 1212 trillion (Table 1).
Applying data to population statistics
Applying the probabilities calculated via the typical frequencies to population statistics, we calculated that there is just a 0.0005% (1 in 200 000) probability that even a single individual exists in the world possessing the optimal DNA variant for endurance performance of all 23 polymorphisms. The probability only increases to approximately 0.1% when seeking an individual with any 22 of the 23 optimal genotypes, and the odds ratio (about 1 in 1000) still strongly suggests that no such individual exists. When any 21 of the 23 optimal genotypes are included, the probability is approximately 10% that an individual exists somewhere in the world with such a genetic profile.
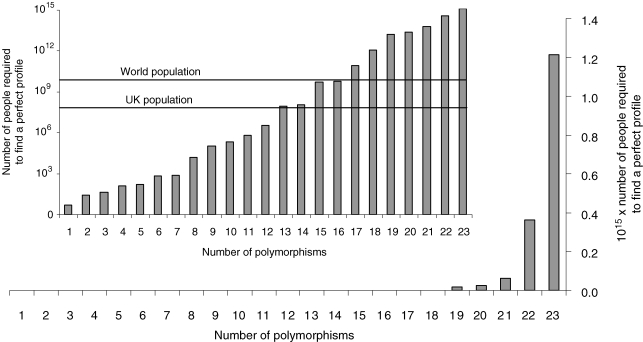
When considering a medium-sized nation (e.g. the approximately 60 million population of the UK), there is only a 0.000 005% (1 in 20 million) chance that an individual exists with all 23 optimal DNA variants. Even for any 21 of the 23 optimal genotypes, the probability of an individual existing remains low at just 0.1%. Figure 1 shows the exponential relationship between the number of genotypes selected as important for endurance and the number of people required to expect one individual to have a ‘perfect’ genetic profile. In the UK, we would expect to find a few individuals with a selected 12 of the 23 optimal genotypes, but probably no individuals with 13 or more. In the world, we would expect to find a few individuals with a selected 16 of the 23 optimal genotypes, but probably no individuals with 17 or more.
Figure 1. Population size required to find a ‘perfect’ genetic profile, by number of polymorphisms.
Large graph shows the population size required to expect one individual to have a ‘perfect’ genetic profile of up to 23 polymorphisms. For < 19 polymorphisms, the bars exist (number of people < 1.7 × 1013) but are too small to be visible. Therefore, the inset shows the same data on a log scale, with UK and world population levels identified. The population sizes required to expect one individual to have all 23 optimal genotypes clearly exceed UK and even world levels.
Polygenic potential for endurance of a whole population
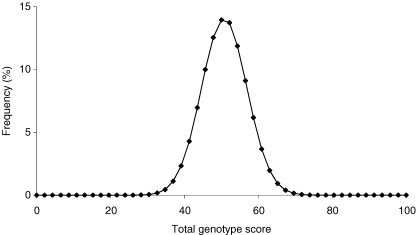
As the probability of an individual possessing a large number of optimal genotypes for endurance performance appeared strikingly low, we examined the distribution of total genotype scores (TGSs) in a population, using the algorithm we developed where each individual was allocated a TGS between 0 and 100. The distribution of the total genotype scores was leptokurtic, i.e. significantly clustered towards the middle (see Fig. 2). In fact, approximately 99% of individuals had a TGS of 37–65, towards the centre of the possible range of 0–100 (the mean TGS was 50.8). The TGS range of 37–65 indicates that 99% of individuals differed by no more than seven genotype scores from the average, demonstrating extremely similar genetic profiles. Furthermore, the total range of TGSs was 22–85, indicating that all individuals differed by at least six genotype scores from the theoretical ‘best’ or ‘worst’ polygenic profiles. It appears that the potential for an individual to possess a ‘perfect’ or ‘near-perfect’ genetic profile for endurance performance is limited by the clustered distribution of total genotype scores. This clustered distribution is a product of the polygenic nature of endurance performance, but is accentuated by the low typical frequencies of some of the optimal genotypes. For example, six of the optimal genotypes had typical frequencies < 10%.
Figure 2. Frequency distribution of total genotype scores.
Frequency distribution of total genotype scores derived from a model sample of 1000 000 randomly generated individuals. Mean score, 50.8; standard deviation, 6.2. The distribution is leptokurtic (i.e. clustered towards the middle; kurtosis statistic 2.5, standard error 0.7; via SPSS v.11), demonstrating that most humans are extremely similar, and thus limited, in terms of their genetic potential for endurance performance. The existence of individuals with very low or very high total genotype scores is clearly restricted, suggesting that any outliers that do exist are extremely unusual individuals.
Readers can see the genetic profiles of almost limitless individuals created on demand by accessing the Supplemental material. Selecting a blank cell in the spreadsheet and then pressing ‘delete’ will generate novel genetic profiles for 1000 randomly generated individuals. If a reader sees a TGS of 100 in column AV on the spreadsheet, then an extreme genetic profile (extremely gifted) in terms of endurance performance has been created. The nature of the distribution of TGSs means that it is extremely unlikely any reader will see a score of 100, or even a score above 90.
Discussion
The main finding of this study is that it is extremely unlikely that even a single individual in the world possesses what could be termed a ‘perfect’ polygenic profile for endurance. Nor, indeed, is it at all likely that a ‘near-perfect’ polygenic profile for endurance exists in a single individual. These observations are based on the current state of published evidence regarding the relevant genes and polymorphisms in the Caucasian population (the population for which there is the most published information). Given the sizeable contribution of genetic factors to human endurance (Bouchard et al. 1998, 1999; Rankinen et al. 2006a; Spurway, 2006), this suggests that there is significant further genetic potential in our species (i.e. that would not require de novo mutations) for endurance performance that is, as yet, unrealized in a single organism. This view concurs with that provided via a prehuman genome modelling approach that predicted a best possible marathon time (about 1 h 58 min) 9 min quicker than the world record of the day (Joyner, 1991). In 2007 the world record remains 7 min slower than that predicted best possible time. The second important finding of this study is that using a genetic algorithm, for the first time it has been demonstrated that the polygenic nature of human endurance constrains the genetic endurance potential of humans within rather narrow upper and lower limits.
Our novel genetic algorithm provides a total genotype score (TGS) of 0–100 to indicate genetic potential for endurance. If individuals really exist with TGSs > 90 according to our algorithm, are they (or have they been) elite endurance athletes? Several studies have used a case-control design to associate a total of 10 genes (ACE (Gayagay et al. 1998), ACTN3 (Yang et al. 2003), ADRA2A (Wolfarth et al. 2000), ADRB2 (Moore et al. 2001), AMPD1 (Rubio et al. 2005), BDKRB2 (Williams et al. 2004), EPAS1 (Henderson et al. 2005), HFE (Chicharro et al. 2004), PPARA (Ahmetov et al. 2006), PPARGC1A (Lucia et al. 2005)) directly with some degree of ‘elite’ endurance athlete status, although these discrete associations tend to be quite weak and a maximum of just two genes (and up to just three polymorphisms) has been reported in a single study. Accordingly, it would be interesting to establish whether truly elite endurance athletes (e.g. Olympic medal winners) really do have an advantageous genetic profile across a larger number of polymorphisms, e.g. for a large proportion of the 23 polymorphisms used in our analysis. Our data put the probability of this occurring into perspective. In the entire world, there is approximately a 1 in 10 probability that an individual exists with any 21 of the 23 optimal genotypes. Given the additional requirement for appropriate environmental factors such as nutrition, economics and culture, the data presented here suggest it is extremely unlikely that any individuals – even elite endurance athletes – possess nearly all of the ‘optimal’ genotypes identified in our analysis. Precisely how close to the maximum figure of 23 elite endurance athletes are remains unknown.
As yet unanswered questions that impact upon the interpretation of the data presented here include (i) whether the 23 polymorphisms identified are true genotype–endurance phenotype associations that will be replicated by future work, and (ii) whether these 23 polymorphisms include all or only some of the genetic factors that influence human endurance capacity. Clearly, replications of genotype–phenotype associations are important given the preliminary nature of many of the associations established to date (Chanock et al. 2007). Considering the early stage of this research, future work may strengthen confidence in the associations of some polymorphisms we have included and fail to replicate those of others. However, based on the premise that endurance performance is polygenic in nature, the broad conclusions of this work would not be altered by a small decrease in the number and composition of the polymorphisms related to endurance performance. It may be (though we think it extremely unlikely) that no further polymorphisms will ever be shown to be associated with human endurance, in which case the 23 polymorphisms (if confirmed via replication) account for all the heritability of endurance. If this was the case, the data we have presented can be taken literally, in perpetuity. However, we are extremely confident in predicting that many additional polymorphisms will be shown to be associated with endurance performance or an endurance phenotype and that the 23 polymorphisms identified to date constitute only a fraction of the genetic factors that influence human endurance. In this extremely likely scenario, the probability that even a single individual exists who possesses the optimal DNA variant for endurance performance of all relevant polymorphisms will reduce far below the 0.0005% value calculated in this study. The odds ratio of a given individual possessing all optimal genotypes will become even greater than the 1 in 1212 trillion calculated, and the distribution of total genotype scores will probably become even more leptokurtic. Hence, the principles underlying our calculations and results will still apply, as will the implications for athletic performance – in fact, the principles we have demonstrated will become quantitatively even more important. The perfect genetic profile for endurance will remain extremely unlikely to occur spontaneously.
Similarly, the quantitative contribution to human endurance of each of the 23 polymorphisms identified thus far is unknown. It is likely that some polymorphisms account for a sizeable proportion of the interindividual variation in endurance phenotypes, whereas others contribute substantially less, and this will doubtless be elucidated as our knowledge evolves. For example, gene–gene interactions may occur via which certain polymorphisms may modify the influence of other genes, under certain conditions. Common sense dictates that, where many genes in a given biochemical pathway are each associated with endurance, the contribution of each of them may alter in the presence of the optimal form of several of the others. In addition, relevant polymorphisms may be in linkage disequilibrium and thus the presence of one optimal genotype may occur more or less often than predicted with the presence of other optimal genotypes. However, neither gene–gene interactions nor linkage disequilibria have been demonstrated clearly between the 23 polymorphisms identified in this study. Furthermore, this area of uncertainty again does not affect the principles underlying our calculations and results, nor the broad implications for Olympic-level athletic performance – high endurance will still be associated with the possession of a high proportion of optimal genotypes. In practice, however, future algorithms will probably include appropriate weighting factors that place greater/lesser emphasis on particular polymorphisms, as that knowledge becomes available.
When viewing Olympic and world records in athletic events as indicators of the limits of human physical potential, it should be recognized (as alluded to previously) that a performance record is a function of economic and social opportunity in addition to genetic potential and physical environment. For example, the large populations of India and China may contain individuals with the genetic potential to rewrite the record books, yet this pool of talent has been largely under-represented at elite levels of athletics to date (Sharp, 2004). However, even that increase in size of the talent pool would be modest compared to the overall increase that will occur with population turnover. New births during the passage of time are the most likely means of producing individuals extremely genetically gifted for endurance performance. Consequently, world records should continue to advance, although probably at a steadily reducing rate, purely through an ever-increasing pool of participants. Any advances in relevant technology, physical training, nutrition, etc., will further progress the records, but even if these factors remain at their current levels then, all other things being equal, Olympic and world records should continue to improve.
Individuals from a relatively small geographical area hold many world athletic records. Indeed, there are considerable regional and ethnic variations in typical frequencies of many genotypes. For example, the II genotype of the ACE I/D polymorphism has a higher frequency in some regions of Oceania than in most of Europe (Kyle et al. 2001). Athletes of north and east African descent excel in endurance events while athletes of west African descent excel in sprint events. However, the possibility of a genetic explanation has yet to be supported by research evidence (Pitsiladis & Scott, 2005), and may in fact be secondary to socio-economic factors. Nevertheless, the medal tables at Olympic and World athletic competitions are at times compelling in their composition, and it would be fascinating to establish whether Kenyan, Ethiopian and Moroccan populations have generally higher total genotype scores according to our algorithm which includes the 23 polymorphisms currently associated with human endurance.
The clustered nature of the total genotype scores in our analysis (Fig. 2) also means that the existence of individuals with large numbers of ‘unfavourable’ genotypes is also extremely unlikely. This is probably advantageous to our species, as morbidity and mortality associated with low aerobic fitness (Lee et al. 1999) and chronic disorders would probably prevail in such individuals. Accordingly, continued research into the polygenic nature of health-related phenotypes such as aerobic function may reveal the lower bounds of genetic potential for endurance that constitute a sufficient risk of common complex diseases to warrant individualized lifestyle or pharmaceutical interventions. In that context, it may be interesting to speculate how the pressure of natural selection against alleles associated with low endurance capability over recent millennia may be reduced with continued advances in society, technology and healthcare systems, which could eventually broaden the polygenic distributions we have described, in the future.
Supplemental material
Online supplemental material for this paper can be accessed at:
The supplemental material is a database in Microsoft Excel format comprising a list of the 23 polymorphisms included, the genotype scores allocated to each genotype within each polymorphism, their typical frequencies in the general population and the sources of all data. The database can be used to generate novel polygenic profiles for 1000 hypothetical individuals at a single keystroke and contains an algorithm which calculates the total genotype score (TGS).
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
http://jp.physoc.org/cgi/content/full/jphysiol.2007.141887/DC1 and http://www.blackwell-synergy.com/doi/suppl/10.1113/jphysiol.2007.141887
References
- Ahmetov II, Mozhayskaya I, Flavell D, Astratenkova IV, Komkova AI, Lyubaeva EV, Tarakin P, Shenkman B, Vdovina A, Netreba AI, Popov DV, Vinogradova O, Montgomery H, Rogozkin V. PPARα gene variation and physical performance in Russian athletes. Eur J Appl Physiol. 2006;97:103–108. doi: 10.1007/s00421-006-0154-4. [DOI] [PubMed] [Google Scholar]
- Bouchard C, An P, Rice T, Skinner JS, Wilmore JH, Gagnon J, Perusse L, Leon AS, Rao DC. Familial aggregation of VO2max response to exercise training: results from the HERITAGE Family Study. J Appl Physiol. 1999;87:1003–1008. doi: 10.1152/jappl.1999.87.3.1003. [DOI] [PubMed] [Google Scholar]
- Bouchard C, Daw EW, Rice T, Perusse L, Gagnon J, Province MA, Leon AS, Rao DC, Skinner JS, Wilmore JH. Familial resemblance for VO2max in the sedentary state: the HERITAGE Family Study. Med Sci Sports Exerc. 1998;30:252–258. doi: 10.1097/00005768-199802000-00013. [DOI] [PubMed] [Google Scholar]
- Bouchard C, Lesage R, Lortie G, Simoneau JA, Hamel P, Boulay MR, Perusse L, Theriault G, Leblanc C. Aerobic performance in brothers, dizygotic and monozygotic twins. Med Sci Sports Exerc. 1986;18:639–646. [PubMed] [Google Scholar]
- Bramble DM, Lieberman DE. Endurance running and the evolution of Homo. Nature. 2004;432:345–352. doi: 10.1038/nature03052. [DOI] [PubMed] [Google Scholar]
- Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, Hirschhorn JN, Abecasis G, Altshuler D, Bailey-Wilson JE, Brooks LD, Cardon LR, Daly M, Donnelly P, Fraumeni JF, Jr, Freimer NB, Gerhard DS, Gunter C, Guttmacher AE, Guyer MS, Harris EL, Hoh J, Hoover R, Kong CA, Merikangas KR, Morton CC, Palmer LJ, Phimister EG, Rice JP, Roberts J, Rotimi C, Tucker MA, Vogan KJ, Wacholder S, Wijsman EM, Winn DM, Collins FS. Replicating genotype–phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- Chicharro JL, Hoyos J, Gomez-Gallego F, Villa JG, Bandres F, Celaya P, Jimenez F, Alonso JM, Cordova A, Lucia A. Mutations in the hereditary haemochromatosis gene HFE in professional endurance athletes. Br J Sports Med. 2004;38:418–421. doi: 10.1136/bjsm.2002.003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne FT, Turcotte L, Thibault MC, Boulay MR, Skinner JS, Bouchard C. Mitochondrial DNA sequence polymorphism, VO2max, and response to endurance training. Med Sci Sports Exerc. 1991;23:177–185. [PubMed] [Google Scholar]
- Drenos F, Whittaker JC, Humphries SE. The use of meta-analysis risk estimates for candidate genes in combination to predict coronary heart disease risk. Ann Hum Genet. 2007;71:611–619. doi: 10.1111/j.1469-1809.2007.00359.x. [DOI] [PubMed] [Google Scholar]
- Fagard R, Bielen E, Amery A. Heritability of aerobic power and anaerobic energy generation during exercise. J Appl Physiol. 1991;70:357–362. doi: 10.1152/jappl.1991.70.1.357. [DOI] [PubMed] [Google Scholar]
- Gayagay G, Yu B, Hambly B, Boston T, Hahn A, Celermajer DS, Trent RJ. Elite endurance athletes and the ACE I allele – the role of genes in athletic performance. Hum Genet. 1998;103:48–50. doi: 10.1007/s004390050781. [DOI] [PubMed] [Google Scholar]
- Hagberg JM, Ferrell RE, Katzel LI, Dengel DR, Sorkin JD, Goldberg AP. Apolipoprotein E genotype and exercise training-induced increases in plasma high-density lipoprotein (HDL)- and HDL2-cholesterol levels in overweight men. Metabolism. 1999;48:943–945. doi: 10.1016/s0026-0495(99)90185-3. [DOI] [PubMed] [Google Scholar]
- Henderson J, Withford-Cave JM, Duffy DL, Cole SJ, Sawyer NA, Gulbin JP, Hahn A, Trent RJ, Yu B. The EPAS1 gene influences the aerobic–anaerobic contribution in elite endurance athletes. Hum Genet. 2005;118:416–423. doi: 10.1007/s00439-005-0066-0. [DOI] [PubMed] [Google Scholar]
- Jones AM, Carter H. The effect of endurance training on parameters of aerobic fitness. Sports Med. 2000;29:373–386. doi: 10.2165/00007256-200029060-00001. [DOI] [PubMed] [Google Scholar]
- Joyner MJ. Modeling: optimal marathon performance on the basis of physiological factors. J Appl Physiol. 1991;70:683–687. doi: 10.1152/jappl.1991.70.2.683. [DOI] [PubMed] [Google Scholar]
- Klissouras V. Heritability of adaptive variation. J Appl Physiol. 1971;31:338–344. doi: 10.1152/jappl.1971.31.3.338. [DOI] [PubMed] [Google Scholar]
- Kyle CV, Abbott W, Young RP, Nijmeijer B, Simmons D, Braatvedt GD. Angiotensin-1-converting enzyme and angiotensinogen gene polymorphisms in Maori and Pacific Island people in New Zealand. Intern Med J. 2001;31:116–118. [PubMed] [Google Scholar]
- Lee CD, Blair SN, Jackson AS. Cardiorespiratory fitness, body composition, and all-cause and cardiovascular disease mortality in men. Am J Clin Nutr. 1999;69:373–380. doi: 10.1093/ajcn/69.3.373. [DOI] [PubMed] [Google Scholar]
- Lieberman DE, Raichlen DA, Pontzer H, Bramble DM, Cutright-Smith E. The human gluteus maximus and its role in running. J Exp Biol. 2006;209:2143–2155. doi: 10.1242/jeb.02255. [DOI] [PubMed] [Google Scholar]
- Lucia A, Gomez-Gallego F, Barroso I, Rabadan M, Bandres F, San Juan AF, Chicharro JL, Ekelund U, Brage S, Earnest CP, Wareham NJ, Franks PW. PPARGC1A genotype (Gly482Ser) predicts exceptional endurance capacity in European men. J Appl Physiol. 2005;99:344–348. doi: 10.1152/japplphysiol.00037.2005. [DOI] [PubMed] [Google Scholar]
- Lucia A, Gomez-Gallego F, Santiago C, Bandres F, Earnest C, Rabadan M, Alonso JM, Hoyos J, Cordova A, Villa G, Foster C. ACTN3 genotype in professional endurance cyclists. Int J Sports Med. 2006;27:880–884. doi: 10.1055/s-2006-923862. [DOI] [PubMed] [Google Scholar]
- Mastana S, Lee D, Singh PP, Singh M. Molecular genetic variation in the East Midlands, England: analysis of VNTR, STR and Alu insertion/deletion polymorphisms. Ann Hum Biol. 2003;30:538–550. doi: 10.1080/0301446031000119593. [DOI] [PubMed] [Google Scholar]
- Moore GE, Shuldiner AR, Zmuda JM, Ferrell RE, McCole SD, Hagberg JM. Obesity gene variant and elite endurance performance. Metabolism. 2001;50:1391–1392. doi: 10.1053/meta.2001.28140. [DOI] [PubMed] [Google Scholar]
- Moran CN, Yang N, Bailey ME, Tsiokanos A, Jamurtas A, Macarthur DG, North K, Pitsiladis YP, Wilson RH. Association analysis of the ACTN3 R577X polymorphism and complex quantitative body composition and performance phenotypes in adolescent Greeks. Eur J Hum Genet. 2007;15:88–93. doi: 10.1038/sj.ejhg.5201724. [DOI] [PubMed] [Google Scholar]
- Niemi AK, Majamaa K. Mitochondrial DNA and ACTN3 genotypes in Finnish elite endurance and sprint athletes. Eur J Hum Genet. 2005;13:965–969. doi: 10.1038/sj.ejhg.5201438. [DOI] [PubMed] [Google Scholar]
- Ortlepp JR, Metrikat J, Vesper K, Mevissen V, Schmitz F, Albrecht M, Maya-Pelzer P, Hanrath P, Weber C, Zerres K, Hoffmann R. The interleukin-6 promoter polymorphism is associated with elevated leukocyte, lymphocyte, and monocyte counts and reduced physical fitness in young healthy smokers. J Mol Med. 2003;81:578–584. doi: 10.1007/s00109-003-0471-6. [DOI] [PubMed] [Google Scholar]
- Paparini A, Ripani M, Giordano GD, Santoni D, Pigozzi F, Romano-Spica V. ACTN3 genotyping by real-time PCR in the Italian population and athletes. Med Sci Sports Exerc. 2007;39:810–815. doi: 10.1097/mss.0b013e3180317491. [DOI] [PubMed] [Google Scholar]
- Pitsiladis YP, Scott R. Essay: The makings of the perfect athlete. Lancet. 2005;366(Suppl. 1):S16–S17. doi: 10.1016/S0140-6736(05)67828-2. [DOI] [PubMed] [Google Scholar]
- Rankinen T, Bray MS, Hagberg JM, Perusse L, Roth SM, Wolfarth B, Bouchard C. The human gene map for performance and health-related fitness phenotypes: the 2005 update. Med Sci Sports Exerc. 2006a;38:1863–1888. doi: 10.1249/01.mss.0000233789.01164.4f. [DOI] [PubMed] [Google Scholar]
- Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Perusse L, Bouchard C. The human obesity gene map: the 2005 update. Obesity. 2006b;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- Rico-Sanz J, Rankinen T, Joanisse DR, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C. Associations between cardiorespiratory responses to exercise and the C34T AMPD1 gene polymorphism in the HERITAGE Family Study. Physiol Genomics. 2003;14:161–166. doi: 10.1152/physiolgenomics.00165.2002. [DOI] [PubMed] [Google Scholar]
- Rubio JC, Martin MA, Rabadan M, Gomez-Gallego F, San Juan AF, Alonso JM, Chicharro JL, Perez M, Arenas J, Lucia A. Frequency of the C34T mutation of the AMPD1 gene in world-class endurance athletes: does this mutation impair performance? J Appl Physiol. 2005;98:2108–2112. doi: 10.1152/japplphysiol.01371.2004. [DOI] [PubMed] [Google Scholar]
- Schessl J, Zou Y, Bonnemann CG. Congenital muscular dystrophies and the extracellular matrix. Semin Pediatr Neurol. 2006;13:80–89. doi: 10.1016/j.spen.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Sharp NCC. Mind the gap: women racers are falling behind. Nature. 2004;432:147. doi: 10.1038/432147d. [DOI] [PubMed] [Google Scholar]
- Spurway N. Top-down studies of genetic contribution to differences in physical capacity. In: Spurway N, Wackerhage H, editors. Genetics and Molecular Biology of Muscle Adaptation. London: Churchill Livingstone, Elsevier; 2006. pp. 25–59. [Google Scholar]
- Tarnopolsky MA, Raha S. Mitochondrial myopathies: diagnosis, exercise intolerance, and treatment options. Med Sci Sports Exerc. 2005;37:2086–2093. doi: 10.1249/01.mss.0000177341.89478.06. [DOI] [PubMed] [Google Scholar]
- Thompson PD, Tsongalis GJ, Seip RL, Bilbie C, Miles M, Zoeller R, Visich P, Gordon P, Angelopoulos TJ, Pescatello L, Bausserman L, Moyna N. Apolipoprotein E genotype and changes in serum lipids and maximal oxygen uptake with exercise training. Metabolism. 2004;53:193–202. doi: 10.1016/j.metabol.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Williams AG, Dhamrait SS, Wootton PTE, Day SH, Hawe E, Payne JR, Myerson SG, World M, Budgett R, Humphries SE, Montgomery HE. Bradykinin receptor gene variant and human physical performance. J Appl Physiol. 2004;96:938–942. doi: 10.1152/japplphysiol.00865.2003. [DOI] [PubMed] [Google Scholar]
- Williams AG, Rayson MP, Jubb M, World M, Woods DR, Hayward M, Martin J, Humphries SE, Montgomery HE. The ACE gene and muscle performance. Nature. 2000;403:614. doi: 10.1038/35001141. [DOI] [PubMed] [Google Scholar]
- Wolfarth B, Rivera MA, Oppert JM, Boulay MR, Dionne FT, Chagnon M, Gagnon J, Chagnon Y, Perusse L, Keul J, Bouchard C. A polymorphism in the alpha2A-adrenoceptor gene and endurance athlete status. Med Sci Sports Exerc. 2000;32:1709–1712. doi: 10.1097/00005768-200010000-00008. [DOI] [PubMed] [Google Scholar]
- Yang Q, Khoury MJ, Friedman J, Little J, Flanders WD. How many genes underlie the occurrence of common complex diseases in the population? Int J Epidemiol. 2005;34:1129–1137. doi: 10.1093/ije/dyi130. [DOI] [PubMed] [Google Scholar]
- Yang N, MacArthur DG, Gulbin JP, Hahn AG, Beggs AH, Easteal S, North K. ACTN3 genotype is associated with human elite athletic performance. Am J Hum Genet. 2003;73:627–631. doi: 10.1086/377590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The supplemental material is a database in Microsoft Excel format comprising a list of the 23 polymorphisms included, the genotype scores allocated to each genotype within each polymorphism, their typical frequencies in the general population and the sources of all data. The database can be used to generate novel polygenic profiles for 1000 hypothetical individuals at a single keystroke and contains an algorithm which calculates the total genotype score (TGS).