Abstract
The Ink4a/Arf locus encodes p16Ink4a and p19Arf and is among the most frequently mutated tumor suppressor loci in human cancer. In mice, many of these effects appear to be mediated by interactions between p19Arf and the p53 tumor-suppressor protein. Because Tp53 mutations are a common feature of the multistep pre-B cell transformation process mediated by Abelson murine leukemia virus (Ab-MLV), we examined the possibility that proteins encoded by the Ink4a/Arf locus also play a role in Abelson virus transformation. Analyses of primary transformants revealed that both p16Ink4a and p19Arf are expressed in many of the cells as they emerge from the apoptotic crisis that characterizes the transformation process. Analyses of primary transformants from Ink4a/Arf null mice revealed that these cells bypassed crisis. Because expression of p19Arf but not p16 Ink4a induced apoptosis in Ab-MLV-transformed pre-B cells, p19Arf appears to be responsible for these events. Consistent with the link between p19Arf and p53, Ink4a/Arf expression correlates with or precedes the emergence of cells expressing mutant p53. These data demonstrate that p19Arf is an important part of the cellular defense mounted against transforming signals from the Abl oncoprotein and provide direct evidence that the p19Arf–p53 regulatory loop plays an important role in lymphoma induction.
Tumor induction is a multistep process involving activation of oncogenes and inactivation of tumor-suppressor genes (1, 2). One experimental system to which this paradigm applies is transformation by Abelson murine leukemia virus (Ab-MLV), an oncogenic retrovirus that encodes the v-Abl protein tyrosine kinase (3). The virus induces mono- or oligoclonal pre-B cell tumors (4, 5), and clonal selection also occurs in vitro. Primary pre-B cell transformants are poorly oncogenic and undergo an apoptotic crisis soon after isolation (6–9). Cells that survive are rapidly growing and highly tumorigenic (6–8). Mutations in the Tp53 tumor-suppressor gene become evident in about half of the surviving clones as they emerge from crisis or soon thereafter (8), and primary Ab-MLV transformants derived from Tp53−/− mice bypass crisis and become established rapidly (I.U., A.R., and N.R., unpublished data). Thus, p53 is one important cellular defense against malignant transformation by Ab-MLV.
p53 is regulated by multiple factors, including p19Arf, a product of the Ink4a/Arf locus (10–15). p19Arf is part of the protective cellular response to oncogenic insults from at least two oncogenes, c-Myc and adenovirus E1A (13, 14). p19Arf stabilizes and activates p53, probably by forming a complex with p53 and the Mdm2 protein (11, 12, 15), a molecule that normally increases p53 ubiquitination and degradation (16, 17). Signals from p19Arf to p53 induce G1 arrest and apoptosis in fibroblast cells and are critical for the normal senescence response (10, 13, 14, 18). Consistent with this mechanism and the importance of p53 in tumorigenesis (reviewed in refs. 19 and 20), mice lacking a functional p19Arf gene develop multiple types of tumors at a high frequency (18).
Although murine models strongly support the idea that p19Arf is a tumor-suppressor protein, evidence for its role in naturally occurring tumors is limited. Indeed, in this arena, attention has focused on the second product of the Ink4a/Arf locus, the p16Ink4a cyclin-dependent kinase inhibitor (21). This protein inhibits the activity of CDK-4 and CDK-6, suppressing their ability to phosphorylate pRb, an event that induces G1 arrest (21–24). p16Ink4a mutations or altered p16Ink4a expression have been documented in many types of tumors, including leukemias and lymphomas (reviewed in ref. 25). Although some of these changes affect both p19Arf and p16Ink4a, others affect only p16 Ink4a (26–30).
We investigated the role of p16Ink4a and p19Arf in Ab-MLV induced pre-B cell transformation to determine whether selection for Tp53 mutation involves the p19Arf–p53 regulatory loop or whether altered expression of p16Ink4a occurs in transformants retaining wt p53. Our studies reveal that primary transformants derived from Ink4a/Arf−/− mice do not undergo crisis. Because overexpression of p19Arf, but not p16 Ink4a, induces apoptosis in a p53-dependent fashion in the transformants, these effects appear to be mediated by p19Arf. These data identify a tumor-suppressor function for p19Arf in Ab-MLV-induced pre-B cell transformation. Furthermore, because elevated expression of p19Arf can precede emergence of cells harboring Tp53 mutations, high levels of p19Arf appear to select for cells expressing mutant forms of p53.
MATERIALS AND METHODS
Cells and Mice.
Ab-MLV-transformed pre-B cell lines and NIH 3T3 cells were grown as described previously (8, 31). The pre-B cell transformants 379-10, 143-2M, and 300-31 express mutant p53, which fails to induce apoptosis after DNA damage and does not activate expression from a promoter containing p53-responsive elements (8); 298-18, 204-3-1, and 38B9 express wild-type (wt) p53 (8). L1-2 is a p53 null cell line (32). 379-10, 298-18, and 38B9 are transformed with Ab-MLV-P160; 143-2M, 300-31, 204-3-1, and L1-2 are transformed with Ab-MLV-P120. Both of these Ab-MLV strains are considered wild type (3). MEL cells are murine erythroleukemic cells that express p16Ink4a and p19Arf (33, 34). For isolating primary transformants, bone marrow cells were infected with Ab-MLV-P160 and plated in soft agar (9). Primary transformants were plated in 24-well plates 10 days later in supplemented RPMI medium (RPMI 1640 medium/2 mM l-glutamine/50 μM 2-mercaptoethanol) containing 20% fetal calf serum. When the cells filled the wells, half of them were transferred to a new well. When the viability exceeded 80%, the cells were transferred to 35-mm dishes and subcultured as before. When the cells consistently maintained a viability of >90% and could be subcultured on regular schedule, they were considered established. In some experiments, pre-B transformants were treated with approximately 1,000-rad γ-irradiation (8); cell viability was scored the following day either by visual inspection, trypan blue staining, or merocyanin 540 (MC540, Molecular Probes) staining. BALB/cByJ and mixed background Ink4a−/− mice (35) were bred at Tufts University.
RNA and Protein Analysis.
Total RNA was prepared by using the RNeasy Mini Kit (Qiagen) and analyzed by Northern blotting (36, 37). The p16Ink4a exon 1α probe was prepared by PCR amplification of bases 6–198 of the p16 cDNA (33); the p19Arf exon 1β probe was prepared by PCR amplification of bases 14–229 of p19Arf cDNA (10). Cell lysates for protein analysis were prepared and analyzed by Western blotting (31) by using anti-p16Ink4a, anti-CDK-4, anti-CDK-6, anti-cyclin D3 (M-156, C-22, C-21, and C-16, respectively, Santa Cruz Biotechnology), anti-p19Arf (ref. 11; R.A.D., unpublished data), and anti-Gag/v-Abl [H548 (38)] antibodies.
Transfections.
To construct p16Ink4a and p19Arf expression plasmids, the EcoRI fragment containing the full-length p16Ink4a cDNA from pKS-mp16 (33) or the ClaI fragment containing the full-length p19Arf cDNA and the codons for the hemagglutinin tag from a pSRα-based retroviral vector (10) were inserted into the pCI vector (Promega). Before transfection, the cells were washed twice in unsupplemented RPMI 1640 and mixed with the p16Ink4a or p19Arf expression plasmids and pCMVEGFP-Spectrin (39) at a ratio of 5:1. pCMVEGFP-Spectrin encodes green fluorescent protein (GFP) fused to the transmembrane domain of spectrin (39). The cells were incubated for 10 min at room temperature and electroporated at 960 μF at 300 V. The cells were incubated on ice for 10 min, plated in supplemented RPMI containing 10% fetal calf serum, and incubated for 24 hr. The cells were harvested, washed twice with Mg2+- and Ca2+-free PBS supplemented with 0.1% BSA, mixed with MC540 at a final concentration of 0.25 μg/ml, and analyzed immediately by flow cytometry. Apoptotic cells are specifically stained with MC540 (40). Gates were set on the GFP-positive cells, and this population was analyzed for live and apoptotic cells.
RESULTS
p16Ink4a and p19Arf Are Expressed in Transformants Expressing Mutant Tp53.
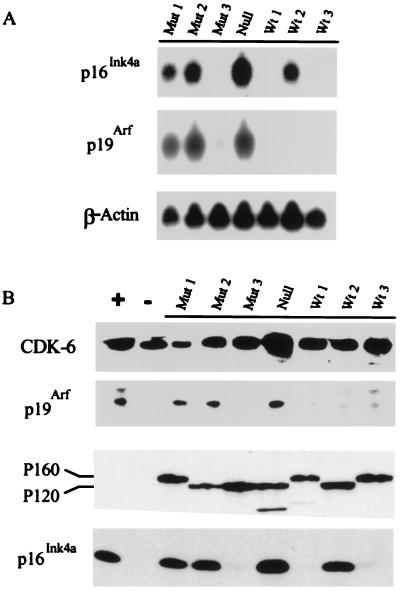
To determine whether a correlation between expression of the Ink4a/Arf locus and Tp53 status exists in Ab-MLV transformed pre-B cells, their expression was examined in a panel of transformants with known Tp53 status (8). Northern analysis revealed that a p53-null cell line (32) and two of the three cell lines expressing mutant p53 expressed readily detectable levels of both RNAs (Fig. 1A). Both RNAs could be detected in samples from Mut3 on prolonged exposure at levels about 10- to 20-fold less than those in Mut1. None of the cell lines expressing wt p53 expressed high levels of both p16Ink4a and p19Arf. Wt2 expressed abundant p16Ink4a RNA, but p19Arf RNA was detectable only on prolonged exposure at about 50-fold-lower levels. Wt1 lacked detectable p16Ink4a RNA, but prolonged exposures revealed about 50-fold-less p19Arf RNA than found in strongly positive samples; Wt3 expressed about 10-fold-less p16Ink4a RNA than the highly positive samples but lacked detectable p19Arf RNA. Western analyses revealed that cells expressing abundant Ink4a/Arf locus RNAs expressed readily detectable levels of the corresponding proteins (Fig. 1B). Consistent with a relationship between Ink4a/Arf locus expression and p53 status, three of four other cell lines expressing mutant p53 expressed both p16Ink4a and p19Arf. Only 1 of 18 cell lines with wt p53 expressed high levels of p19Arf; 4 expressed high levels of p16Ink4a and none expressed high levels of both proteins (data not shown).
Figure 1.
p16Ink4a and p19Arf are expressed in some pre-B cell transformants. (A) Total RNA from established Ab-MLV-transformed pre-B cell lines was analyzed by Northern blotting by using probes specific for exons 1α and 1β and β-actin. Wt1, Wt2, and Wt3 are 298-18, 204-3-1, and 38B9, respectively; Mut1, Mut2, and Mut3 are 379-10, 143-2M, and 300-31, respectively; Null is L1-2. (B) Cell lysates from the same cells were analyzed by Western blotting. Levels of the P120 or P160 v-Abl protein or CDK-6 control for protein loading. Lane +, lysate from MEL cells known to express both p16Ink4a and p19Arf (33, 34); lane −, lysate from NIH 3T3 cells.
p16 and p19Arf Can Be Up-Regulated Before Tp53 Mutations Arise.
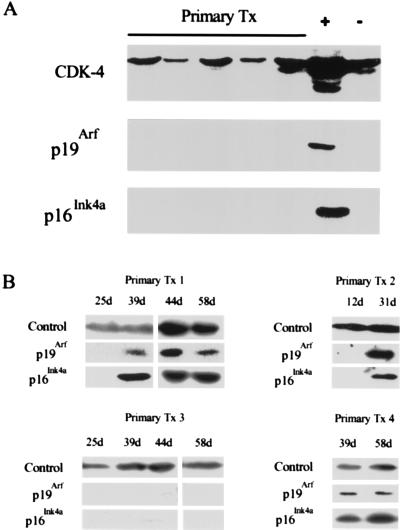
Mutation of Tp53 is usually a late, postcrisis event in Ab-MLV transformation (8). Because the loss of p53 that often accompanies fibroblast cell immortalization is preceded by changes in Ink4a/Arf locus expression (14, 18, 35), we examined primary transformants for expression of Ink4a/Arf locus products and acquisition of Tp53 mutations. None of 10 primary transformants, including the representatives shown (Fig. 2A), expressed detectable levels of p16Ink4a or p19Arf when they were isolated from agar. The primary transformants were cultured, and, as expected (8), crisis began within 2–5 days and usually lasted for 25–35 days. Samples were collected every 10–21 days as the health of the cells allowed. Analyses of serial samples from the 13 primary transformants that became established revealed that 8 had become p16Ink4a/p19Arf-positive as they emerged from crisis; 1 primary transformant expressed only p16Ink4a. In the representative examples shown (Fig. 2B), primary transformants 1, 2, and 4 were considered established at day 35, 40, and 33, respectively; primary transformant 3 never expressed Ink4a locus products but became established at day 38.
Figure 2.
p16Ink4a and p19Arf up-regulation in primary transformants. (A) Primary transformants (Primary Tx) were analyzed for the expression of p16Ink4a and p19Arf by Western blotting 24 hr after explant from agar. The blots also were probed with an anti-CDK-4 antibody to control for protein loading. Lane +, lysate from MEL cells; lane −, lysate from NIH 3T3 cells. (B) Western blots of lysates from four independent primary transformants harvested at different days postexplant were probed with anti-p16Ink4a and p19Arf antibodies. The samples shown are representative of 13 independent primary transformants that became established. Probing with anti-CDK-6 (Primary Tx 1, 4) or anti-cyclin D3 (Primary Tx 2, 3) antibodies controlled for protein loading. d, days after explant from agar.
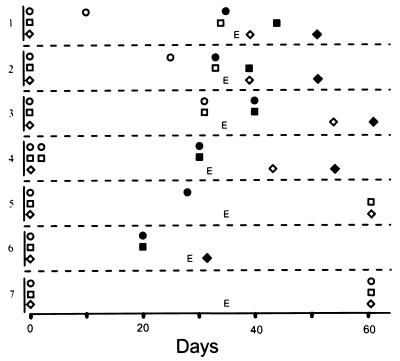
To determine whether any of these cells expressed mutant p53, the susceptibility of the cells to γ-irradiation-induced apoptosis was monitored as soon as the cells emerged from crisis. Our earlier work has shown a strong correlation between resistance to radiation-induced apoptosis and Tp53 mutation in Ab-MLV-transformed pre-B cells (8). By 60 days, all eight of the p16Ink4a/p19Arf-positive cell lines expressed mutant p53 as judged by resistance to irradiation-induced apoptosis; in contrast, all of the other cell lines, including one that expressed only p16Ink4a, underwent rapid apoptosis after irradiation (data not shown). Comparison of the times at which abundant expression of Ink4a/Arf locus products occurred and Tp53 mutations were detected revealed that p16Ink4a/p19Arf expression became evident 10–14 days before Tp53 mutation appeared in four instances (Fig. 3, samples 1–4). In four other cases, none of the available samples distinguished a temporal order for these changes (represented by sample 6, Fig. 3). However, there were no cases in which Tp53 mutation occurred before abundant expression of Ink4a/Arf locus products. These data suggest that Ink4a/Arf locus products play a key role in the crisis period that characterizes the transformation process and that their expression can precede and may select for mutation in Tp53.
Figure 3.
Up-regulation of p16Ink4a and p19Arf occurs as cells emerge from crisis, often before p53 mutation. p16Ink4a and p19Arf expression was analyzed by Western blotting, with day 0 being the day cells were removed from agar. p53 status was monitored by induction of apoptosis after γ-irradiation (8). The data shown are representative of analyses of 13 primary transformants, among which 3 others had patterns similar to sample 6, and 3 others had patterns similar to sample 7. Circles, p16Ink4a; squares, p19Arf; diamonds, p53 status; open symbols, undetectable protein or wt p53; filled symbols, abundant protein or mutant p53; E, the time at which cells became established. Levels of proteins were assigned based on analyses of Western blots similar to those shown in Fig. 1B and Fig. 2B.
Primary Pre-B Cell Transformants from Ink4a/Arf−/− Mice Bypass Crisis.
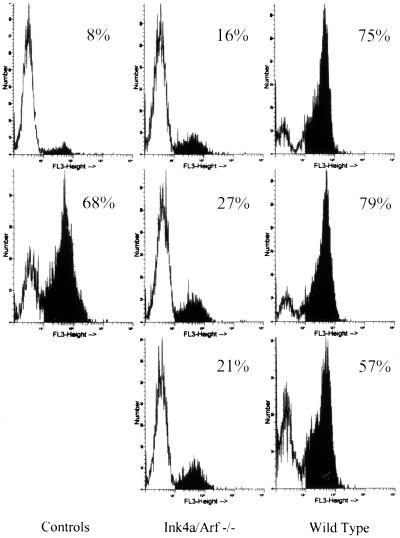
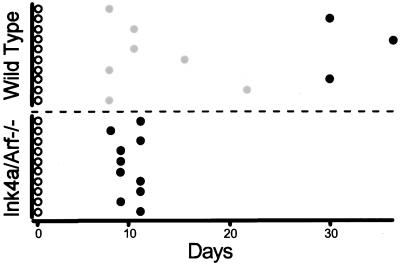
To test the hypothesis that Ink4a/Arf locus products play a role in crisis induction, the outgrowth of primary transformants from BALB/c mice and Ink4a/Arf−/− mice (35) was compared. In both cases, the primary transformants contained >90% viable cells when they were explanted from agar. As expected based on analyses of primary transformants from several strains of inbred mice (I.U., A.R., and N.R., unpublished data and our unpublished data), a high percentage of the cells in all 48 primary transformants from wild-type mice underwent apoptotic crisis beginning 2–5 days after explant and continuing for about a month. MC540 staining of cells from cultures containing viable cells revealed that 50–80% of the cells showed evidence of apoptosis (Fig. 4); 15 of 48 primary transformants survived crisis to become established while all of the cells in the other 33 died by apoptosis during the crisis period (representatives are shown in Fig. 5). In contrast, none of the 48 primary transformants from Ink4a/Arf−/− mice underwent extensive cell death as judged by visual inspection; MC540 staining indicated that fewer than 30% of the cells in these samples showed evidence of apoptosis (Fig. 4). These cells grew exponentially, becoming fully established in less than 10 days (representatives are shown in Fig. 5). Although the genetic background of the control and Ink4a−/− mice used here is not identical, results similar to those obtained with BALB/c cells have been observed in seven other inbred strains and four strains of mixed background similar to that of the Ink4a−/− mice (our unpublished data). Thus, expression of Ink4a/Arf locus products is required for the apoptotic crisis that characterizes Ab-MLV-induced pre-B cell transformation.
Figure 4.
Primary transformants from Ink4a/ARF−/− mice display low levels of apoptosis. Primary pre-B transformants from Ink4a/ARF−/− and wild-type mice were stained with MC540 within a week of culture in liquid medium. Each histogram represents analysis of a single primary transformant. The data shown are representative of analyses of 15 primary transformants from each type of mouse. The controls are healthy, established pre-B cell transformants (Top Left) and pre-B transformants treated with γ-irradiation 8 hr before analysis (Bottom Left). The percentages of MC540-stained cells are indicated.
Figure 5.
Primary pre-B cell transformants from Ink4a/ARF−/− mice are established rapidly. The growth and viability of 10 representative primary pre-B transformants derived from Ink4a/ARF−/− and wild-type mice is illustrated. A total of 48 primary transformants from each strain were analyzed. For wild-type cells, 33 of 48 primary transformants succumbed to crisis between day 7 and day 24; 15 of 48 primary transformants became established between day 28 and day 39. For Ink4a−/− cells, all 48 primary transformants became established between days 9 and 11. Open circles, primary transformants; shaded circles, transformants that succumbed to crisis; solid circles, established cell lines.
Expression of p19Arf Induces p53-Dependent Apoptosis in Transformed Pre-B Cells.
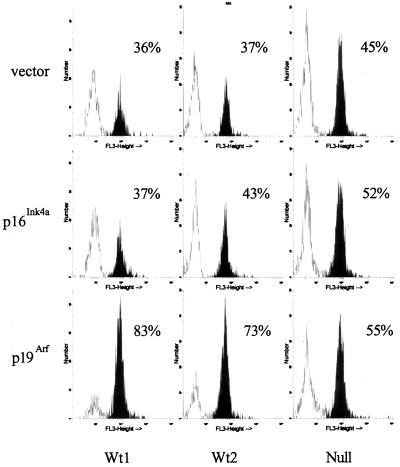
The Ink4a/Arf−/− mice used in our experiments express neither p16Ink4a nor p19Arf (35). To determine the effect of each protein on Ab-MLV-transformed pre-B cells, vectors expressing them were introduced into cells with known p53 status. The plasmids were mixed at a 5-fold excess with a plasmid encoding GFP-spectrin (39) and introduced into the cells by electroporation. GFP-spectrin is retained in cells undergoing apoptosis because the spectrin portion anchors the molecule inside the cell (39). The cells were harvested 24 hr later, stained with MC540, and analyzed by flow cytometry with gates set on the GFP-positive cells to assess the percentage of transfected cells undergoing apoptosis. Expression of either p16Ink4a or p19Arf in p53 null cells (Fig. 6) or cells expressing mutant p53 (not shown) did not affect significantly the percentage of apoptotic cells when these samples were compared with those that had received an equivalent amount of empty vector. However, expression of p19Arf in cells expressing wt p53 increased the percentage of apoptotic cells by about 50% compared with cells receiving empty vector (Fig. 6). The slight increase in apoptotic cells observed in cells receiving the p16Ink4a expression vector and in null cells receiving either the p16Ink4a or p19Arf vector was not reproducible. These results suggest that expression of p19Arf induces p53-dependent apoptosis in Ab-MLV-transformed pre-B cells and that p19Arf is the active Ink4a/Arf locus product responsible for crisis induction in primary Ab-MLV-transformed pre-B cells.
Figure 6.
p19Arf induces apoptosis in pre-B cell transformants expressing wild-type p53. Pre-B cell transformants expressing wild-type p53 (Wt1, 204–3-1) and (Wt2, 38B9) and the p53 null transformant L1–2 (8, 32) were electroporated with pCMVEGFP-spectrin and either vector alone, pCI-p16Ink4a, or pCI-p19Arf and stained with MC540 24 hr later. The samples were analyzed by using a FACScan instrument with gates set on the GFP-positive cells. The data shown are representative of three independent experiments in which these cell lines and two cell lines expressing mutant p53 were analyzed.
DISCUSSION
Our experiments show that the Ink4a/Arf locus plays a major role in Ab-MLV-mediated pre-B cell transformation and identify these genes as one component of the cellular defense mounted against v-Abl-mediated transformation. Unlike pre-B transformants derived from normal mice (6–9), those from Ink4a/Arf−/− mice bypass the crisis that characterizes the transition from primary transformant to established, fully malignant cell line. In addition, all of the primary transformants become established in a very short time. In these respects, the primary Ab-MLV transformants are similar to mouse embryo cells derived from the null animals; these fibroblasts do not senesce in vitro and give rise to a high frequency of established cell lines rapidly (10, 13, 14, 18, 35). In addition, these cells can be transformed by a single, mitogenic oncogene such as Ras without the collaboration of immortalizing oncogenes like Myc and E1A (10, 11, 13, 14, 18, 35). Our observations with v-abl-stimulated lymphocytes suggest that these phenomena are not unique to fibroblasts; analogous events probably are responsible for the high frequency and broad spectrum of tumors observed in Ink4a/Arf and Arf null mice (18, 35).
The Ink4a/Arf locus encodes two proteins, p16Ink4a and p19Arf, both of which have tumor-suppressor functions in appropriate settings (10, 18, 21, 30, 35, 41). However, p19Arf appears to be the product involved in crisis induction. The strong correlation between high levels of Ink4a/Arf locus products and the expression of mutant p53 in our transformants mirrors results obtained with fibroblasts lacking Arf (13, 14, 18). Overexpression of p19Arf in cells induces apoptosis in a p53-dependent fashion while overexpression of p16Ink4a does not significantly alter their health or cell cycle status (A.R. and N.R., unpublished data). This latter result may reflect the high levels of CDK-4 and -6 present in the cells; even when all of the p16 present is bound to these proteins, a large proportion of the CDKs remain complexed to cyclin D3 and the activity of cyclin D3 remains high (unpublished data). Finally, the observation that primary transformants from Tp53−/− mice fail to undergo crisis (I.U., A.R., and W.R., unpublished data) is consistent with a critical role for the recently described p19Arf–p53 regulatory loop (13–15).
A key event in the p19Arf-mediated response is up-regulation of p19Arf expression (14, 18). In fibroblasts, overexpression of c-Myc, E1A, or probably other immortalizing oncogenes can increase p19Arf expression, at least in part through effects on E2F transcription factors (13, 14). The link between v-Abl expression and c-Myc activation is well established (42–45), suggesting a mechanism by which p19Arf expression can be activated. However, about half of all primary transformants do not seem to up-regulate p19Arf, even though activation of c-Myc appears to be an obligatory step in v-Abl-mediated transformation (42, 43, 45). Analyses of 10 transformants that do not express abundant Ink4a/Arf locus products reveal that all of them retain at least one copy of the Ink4a/Arf locus. In addition, these copies usually lack evidence of methylation (A. Halgren, A.R., and N.R., unpublished data). These transformants may have defects in other cellular components required to activate expression of the locus. Interestingly, these transformants still undergo crisis, suggesting that at least one other cellular defense pathway can mediate this response.
At least some primary transformants display high levels of p19Arf expression before cells expressing mutant p53 emerge. Because Tp53 mutations initially were detected by using resistance to radiation-induced apoptosis, survival of as few as 1–5% of the treated cells can be monitored readily. This feature and the strong correlation between resistance to radiation-induced apoptosis and mutant p53 in Ab-MLV-transformed pre-B cells (8) make it unlikely that large numbers of cells harboring mutant forms of p53 were present before their detection. These data suggest that one consequence of high p19Arf expression is selection of cells carrying Tp53 mutations. Consistent with this idea, only 2 of 40 transformants tested express mutant p53 in the absence of high p19Arf levels and only 1 expressed high p19Arf and wt p53. Fibroblasts from mice heterozygous for Arf or Tp53 usually lose expression or function of one but not both of these proteins as they emerge from crisis (18). In addition, tumor-associated mutational inactivation of p53 has not been observed in many melanomas and fibrosarcomas derived from Ink4a/Arf null mice (ref. 41; L. Chin and R.A.D., unpublished observations) and occurs rarely in Arf null mice (18). Finally, a revisited analysis of several types of human cancers showed that dual inactivation of ARF and TP53 is exceptionally rare (11).
The effects of p19Arf expression coupled with analyses of Tp53 null mice (I.U., A.R., and N.R., unpublished data) and patterns of Tp53 mutation in Ab-MLV-transformed pre-B cells suggest a model in which v-Abl expression, through activation of c-Myc, leads to p19Arf up-regulation. p19Arf activates p53 (11, 12, 15), inducing the apoptotic crisis observed in primary transformants. In parallel with these events, v-Abl expression also activates Ras, the product of a classical mitogenic oncogene (46, 47), along with other growth-stimulatory pathways (reviewed in refs. 3 and 48); such signals, known to temper p53-mediated apoptosis in other settings (18, 35, 49–52), counter-balance the protective cellular response and allow some cells to survive crisis. However, as noted earlier, the p19Arf–p53 regulatory loop does not appear to be the only mechanism by which crisis is induced in primary transformants. Identifying this alternative pathway and determining its role, along with that of the p19Arf–p53 regulatory loop in tumor induction, represents a next step toward understanding the multistep process by which tumors arise in this and other systems.
Acknowledgments
We thank Charles Sherr and Andrew Beavis for gifts of reagents, Henry Wortis and Allen Parmelee for assistance with flow cytometry, and Anne Halgren for technical support. This work was supported by grants EY 11267 to R.A.D. and CA 33771 to N.R. from the National Institutes of Health.
ABBREVIATIONS
- Ab-MLV
Abelson murine leukemia virus
- MC540
merocyanin 540
- GFP
green fluorescent protein
- wt
wild type
Footnotes
To whom reprint requests should be addressed at: SC315, Tufts University School of Medicine, 136 Harrison Avenue, Boston, MA 02111. e-mail: nrosenbe@opal.tufts.edu.
References
- 1. Weinberg R A. Cell. 1997;88:573–575. doi: 10.1016/s0092-8674(00)81897-8. [DOI] [PubMed] [Google Scholar]
- 2.Sherr C J. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg N, Witte O N. In: Adv. Virus Res. Shatkin A, editor. Vol. 35. New York: Academic; 1988. pp. 39–81. [DOI] [PubMed] [Google Scholar]
- 4.Green P L, Kaehler D A, Risser R. J Virol. 1987;61:2192–2197. doi: 10.1128/jvi.61.7.2192-2197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green P L, Kaehler D A, Bennett L M, Risser R. J Virol. 1989;63:1989–1994. doi: 10.1128/jvi.63.5.1989-1994.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitlock C A, Ziegler S F, Witte O N. Mol Cell Biol. 1983;3:596–604. doi: 10.1128/mcb.3.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitlock C A, Witte O N. J Virol. 1981;40:577–584. doi: 10.1128/jvi.40.2.577-584.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thome K, Radfar A, Rosenberg N. J Virol. 1997;77:8149–8156. doi: 10.1128/jvi.71.11.8149-8156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenberg N, Baltimore D. J Exp Med. 1976;143:1453–1463. doi: 10.1084/jem.143.6.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quelle D E, Zindy F, Ashmun R A, Sherr C J. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 11.Pomerantz J, Schreiber-Agus N, Liegeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H W, Cordon-Cardo C, DePinho R A. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Xiong Y, Yarbrough W G. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 13.de Stanchina E, McCurrach M E, Zindy F, Shieh S-Y, Ferbeyre G, Samuelson A V, Prives C, Roussel M F, Sherr C J, Lowe S W. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zindy F, Eischen C M, Randle D H, Kamijo T, Cleveland J L, Sherr C J, Roussel M F. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamijo T, Weber J D, Zambretti G, Zindy F, Roussel M F, Sherr C J. Proc Natl Acad Sci USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubbutat M H, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 17.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 18.Kamijo T, Zindy F, Roussel M F, Quelle D E, Downing J R, Ashmun R A, Grosveld G, Sherr C J. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 19.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 20.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 21.Serrano M, Hannon G J, Beach D. Nature (London) 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 22.Lukas J, Parry D, Aagaard L, Mann D J, Bartkova J, Strauss M, Peters G, Bartek J. Nature (London) 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 23.Medema R H, Herrera R E, Lam F, Weinberg R A. Proc Natl Acad Sci USA. 1995;92:6289–6293. doi: 10.1073/pnas.92.14.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koh J, Enders G H, Dynlacht B D, Harlow E. Nature (London) 1995;375:506–510. doi: 10.1038/375506a0. [DOI] [PubMed] [Google Scholar]
- 25.Hall M, Peters G. Adv Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- 26.Walker G J, Hussussian C J, Flores J F, Glendening J M, Haluska F G, Dracopoli N C, Hayward N K, Fountain J W. Hum Mol Genet. 1995;4:1845–1852. doi: 10.1093/hmg/4.10.1845. [DOI] [PubMed] [Google Scholar]
- 27.Gruis N A, van der Velden P A, Sandkuijl L A, Prins D E, Weaver-Feldhaus J, Kamb A, Bergman W, Frants R R. Nat Genet. 1995;10:351–353. doi: 10.1038/ng0795-351. [DOI] [PubMed] [Google Scholar]
- 28.Holland E A, Beaton S C, Becker T M, Grulet O M, Peters B A, Rizos H, Kefford R F, Mann G J. Oncogene. 1995;11:2289–2294. [PubMed] [Google Scholar]
- 29.Quelle D E, Cheng M, Ashmun R A, Sherr C J. Proc Natl Acad Sci USA. 1997;94:669–673. doi: 10.1073/pnas.94.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haber D A. Cell. 1997;91:555–558. doi: 10.1016/s0092-8674(00)80441-9. [DOI] [PubMed] [Google Scholar]
- 31.Parmar K, Rosenberg N. J Virol. 1996;70:1009–1015. doi: 10.1128/jvi.70.2.1009-1015.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolf D, Rotter V. Mol Cell Biol. 1984;4:1402–1410. doi: 10.1128/mcb.4.7.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quelle D E, Ashmun R A, Hannon G J, Rehberger P A, Trono D, Richter K H, Walker C, Beach D, Sherr C J, Serrano M. Oncogene. 1995;11:635–645. [PubMed] [Google Scholar]
- 34.Zindy F, Quelle D E, Roussel M F, Sherr C J. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
- 35.Serrano M, Lee H-W, Chin L, Cordon-Cardo C, Beach D, DePinho R A. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 36.Wang L C, Rosenberg N. Mol Cell Biol. 1993;13:3890–3899. doi: 10.1128/mcb.13.7.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y-Y, Wang L C, Huang M S, Rosenberg N. Genes Dev. 1994;8:688–697. doi: 10.1101/gad.8.6.688. [DOI] [PubMed] [Google Scholar]
- 38.Chesebro B, Wehrly K, Cloyd M, Britt W, Portis J, Collins J, Nishio J. Virology. 1981;112:131–144. doi: 10.1016/0042-6822(81)90619-x. [DOI] [PubMed] [Google Scholar]
- 39.Kalejta R, Shenk T, Beavis A. Cytometry. 1997;29:286–291. doi: 10.1002/(sici)1097-0320(19971201)29:4<286::aid-cyto4>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 40.Reid S, Cross R, Snow E. J Immunol Methods. 1996;192:43–54. doi: 10.1016/0022-1759(96)00004-x. [DOI] [PubMed] [Google Scholar]
- 41.Chin L, Pomerantz J, Polsky D, Jacobson M, Cohen C, Cordon-Cardo C, Horner J W, II, DePinho R A. Genes Dev. 1997;11:2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong K K, Zou X, Merrell K T, Patel A J, Marcu K B, Chellappan S, Calame K. Mol Cell Biol. 1995;15:6535–6544. doi: 10.1128/mcb.15.12.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zou X, Rudchenko S, Wong K-K, Calame K. Genes Dev. 1997;11:654–662. doi: 10.1101/gad.11.5.654. [DOI] [PubMed] [Google Scholar]
- 44.Cleveland J L, Dean M, Rosenberg N, Wang J Y J, Rapp U R. Mol Cell Biol. 1989;9:5685–5695. doi: 10.1128/mcb.9.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sawyers C L, Callahan W, Witte O N. Cell. 1992;70:901–910. doi: 10.1016/0092-8674(92)90241-4. [DOI] [PubMed] [Google Scholar]
- 46.Sawyers C L, McLaughlin J, Witte O N. J Exp Med. 1995;181:307–313. doi: 10.1084/jem.181.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith M R, DeGudicibus S J, Stacey D W. Nature (London) 1986;320:540–543. doi: 10.1038/320540a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J Y J. Curr Opin Genet Dev. 1993;3:35–43. doi: 10.1016/s0959-437x(05)80338-7. [DOI] [PubMed] [Google Scholar]
- 49.Land H, Parada L F, Weinberg R A. Nature (London) 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 50.Hicks G C, Egan S A, Greenberg A H, Mowat M A. Mol Cell Biol. 1991;11:1344–1352. doi: 10.1128/mcb.11.3.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newbold R F, Overell R O. Nature (London) 1983;304:648–651. doi: 10.1038/304648a0. [DOI] [PubMed] [Google Scholar]
- 52.Lin H-J, Eviner V, Prendergast G C, White E. Mol Cell Biol. 1995;15:4536–4544. doi: 10.1128/mcb.15.8.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]