Abstract
Extracellular, intracellular or surface proteins can be used as putative markers to characterize human mesenchymal stem cells (hMSC). However, these markers are also expressed by other cell types and primary cell pools reveal considerable heterogeneity. Therefore, the simultaneous detection of several markers on a single cell appears to be an attractive approach to identify hMSC. Here we demonstrate the specific distinction of human MSC from human osteoblasts via seven-colour fluorescence on the single cell level with simultaneous marker detection of CD44, CD105/endoglin, CD106/VCAM-1, collagen-IV, fibronectin, actin and DAPI nuclear staining. We performed spectral image acquisition using a Sagnac-type interferometer. Subsequent linear unmixing allowed for decomposition of each pixel in its spectral components. Our approach reveals a typical expression profile of the adherent singular cells, allowing the specific distinction between hMSC and osteoblasts on the single cell level.
Keywords: human mesenchymal stem cells, human multipotent mesenchymal stromal cells, multicolour immunofluorescence, osteoblasts, spectral image acquisition
Introduction
Interest in human mesenchymal stem cells or multipotent mesenchymal stromal cells (both abbreviated hMSC; Horwitz et al. 2005) has been increasing since progress in regenerative medicine has revealed possibilities for clinical application in tissue regeneration. hMSC have the ability to self-renew and to differentiate into various tissue types in vitro and in vivo (Bianco et al. 2001; Cancedda et al. 2003; Grove et al. 2004; Caplan & Dennis, 2006). Despite intense research over several years, characterization of hMSC a priori has yet to be achieved and until now the only way to define hMSC is by their differentiation capacity in vitro and in vivo (Kassem, 2006; Prockop et al. 2003). hMSC can be harvested from different tissues, most commonly from bone marrow. These primary cells are very heterogeneous in culture (Colter et al. 2001; Sekiya et al. 2002; Javazon et al. 2004; Vogel et al. 2004) and their morphological appearance ranges from spindle shaped to polygonal and cuboidal in various sizes (Javazon et al. 2004; Smith et al. 2004; Raimondo et al. 2006). However, cultured under standardized culture conditions, the heterogeneity in vitro may reflect different progenitor stages of distinct mesenchymal lineages, for example cells of the osteoblastic lineage (Aubin, 2001), adipocytic lineage or chondrocytic lineage. The largest fraction of hMSC in culture has a fibroblastic morphology and cannot be distinguished from more differentiated cell phenotypes morphologically. Recently, several molecular markers have been presented to distinguish hMSC from fibroblasts (Ishii et al. 2005). Moreover, in cell culture osteoblasts and fibroblasts are morphologically nearly indistinguishable (Ducy et al. 2000).
The antigenic phenotype of hMSC is not unique and no single marker has been found to be specific for them (Conget & Minguell, 1999; Pittenger et al. 1999; Sekiya et al. 2002; Barry & Murphy, 2004; Javazon et al. 2004; Kemp et al. 2005; Kassem, 2006). However, in the past certain antigens, in particular surface proteins, have been used in attempts to characterize hMSC (Haynesworth et al. 1992, 1992; Bruder et al. 1998; Jiang et al. 2002; Gronthos et al. 2003; Majumdar et al. 2003; Otto & Rao, 2004; Kemp et al. 2005; Honczarenko et al. 2006). None of these markers is exclusively expressed by hMSC, but the combination of markers coexpressed on one single cell represents a promising strategy for distinct characterization of hMSC (Kemp et al. 2005). Investigation on the single cell level is also necessary due to the heterogeneity of hMSC as described above (Grove et al. 2004; Kemp et al. 2005). For these reasons we established an immunofluorescence method to detect several characteristic antigens on one single cell using spectral image acquisition (Schieker et al. 2004).
Here we present an improved version of this method with a more specific marker profile and an increased amount of markers simultaneously detected on single cells. By performing seven-colour fluorescence on hMSC and human osteoblasts we can show the specific distinction of these cell types on the single cell level in vitro.
Materials and methods
Cells
hMSC were purchased from Cambrex (USA). The primary cells were isolated from bone marrow by ficoll gradient centrifugation and characterized as described by Pittenger et al. (1999). These hMSC fulfil the minimal criteria defined by the International Society for Cellular Therapy (Dominici et al. 2006) and were cultivated according to the supplier's protocol in hMSC-growth medium (Cambrex). Primary human osteoblasts (hOB; Promocell, Germany) were also cultured as recommended by the supplier in Osteoblast Growth Medium (Promocell). The primary osteoblasts were isolated from human hip bone as described by Kasperk et al. (1995). All cells were plated in T75 flasks (Nunc, USA) and incubated at 37 °C with 5% humidified CO2. To prevent cell culture artefacts, as differentiation due to long-term cell culture, all primary cells were examined before the seventh passage and passaging was carried out before reaching confluence. Fresh complete media were replaced every 3–4 days.
Immunofluorescence
For multicolour immunofluorescence, cells were cultured on uncoated glass slides. Cells were fixed in buffered 3.7% paraformaldehyde and washed in phosphate-buffered saline (PBS). Subsequently, cells were fixed in −20 °C cold acetone and desiccated. The slides were divided into different fields with a hydrophobic pen (Dako, Germany) allowing for up to eight different staining procedures on one slide.
Incubation with Alexa633-conjugated anti-F-actin phalloidin was carried out at a concentration of 10 units per 500 µL for 20 min. Subsequently, cells were blocked with 1% bovine serum albumin (BSA) in order to reduce non-specific antibody binding. For labelling procedures the following primary antibodies raised in different species were used: CD44 (rat), endoglin/CD105 (mouse), VCAM-1/CD106 (rabbit), collagen-IV (goat) and fibronectin (sheep) (see Table 1). A simultaneous detection step for the primary antibodies was made possible, because all secondary antibodies were raised in the same species (donkey), conjugated with different fluorochromes (AMCA, Texas Red, FITC, Alexa546 and Cy2, respectively, Table 1). We omitted the primary antibody to control non-specific binding of secondary antibodies. As additional controls, cells on the same slide were labelled with each primary antibody at the same dilution in a single colour staining step. Nuclear counterstaining with DAPI at a dilution of 1 : 10 000 generated a seventh fluorescent spectrum. All slides were mounted with a polymerizing hydrophilic mounting medium containing an anti-fade reagent (Molecular Probes, USA).
Table 1.
Sources and dilutions of antibodies and stains used for immunofluorescence
| Source | Specification | Dilution | |
|---|---|---|---|
| Primary antibodies | |||
| Rat anti-human CD44 | DSHB | Hermes-1 | 1 : 5 |
| Mouse anti-human endoglin (CD105) | DSHB | P3D1 | 1 : 5 |
| Rabbit anti-human VCAM-1 (CD106) | SantaCruz Biotechnologies | SC-8304 | 1 : 5 |
| Goat anti-human collagen-IV | Accurate Chemical | YMPS063 | 1 : 20 |
| Sheep anti-human fibronectin | Biozol | BZL00279 | 1 : 1200 |
| Secondary antibodies | |||
| AMCA-conjugated donkey anti-rat IgG | Dianova | 712-155-153 | 1 : 25 |
| Fluorescin (FITC)-conjugated donkey anti-rabbit IgG | Dianova | 711-095-152 | 1 : 200 |
| TexasRed-conjugated donkey anti-mouse IgG | Dianova | 715-075-151 | 1 : 25 |
| Alex Fluor546-labelled donkey anti-goat IgG | Molecular Probes | A11056 | 1 : 125 |
| Cy2-conjugated donkey anti-sheep IgG | Dianova | 713-225-147 | 1 : 2000 |
| Stainings | |||
| AlexaFluor633 anti-F-actin phalloidin | Molecular Probes | A22284 | 10 units/500 µL |
| DAPI Nucleic Acid Stain | Molecular Probes | D1306 | 1 : 10 000 |
Spectral data acquisition
The fluorescent spectra were acquired with a Sagnac-type interferometer SpectraCube SD-200 [Applied Spectral Imaging (ASI), Israel] installed on an Axioskop 2 microscope (Zeiss, Germany) which was attached to a CCD camera (Hamamatsu CCD 5880-C, Japan) and a personal computer. A triple-band pass filter set was applied for green, red and infrared spectra (SKY, ASI) as well as a standard filter set which was used for blue spectra (#01, Zeiss, Germany). Details of the principle of data acquisition that allows for demarcation of wavelength ranges of less than 10 nm have been published elsewhere (Malik et al. 1996; Rothmann et al. 1998; Schieker et al. 2004). For image analysis, SpectraView Software (ASI) enabled linear unmixing, a principle based on decomposition of the image in its pure spectral components. The reference spectra, used to analyse the multicolour image, were taken from the single colour labelling of each antibody on the same slide. In addition, images of all antigens in single colour applications were acquired with a digital camera (Cybershot DSC S 75, Sony, Japan), using either a triple band filter for red, green and blue spectra (F61002, AHF, Germany), or the appropriate standard filter sets for each spectrum.
Results
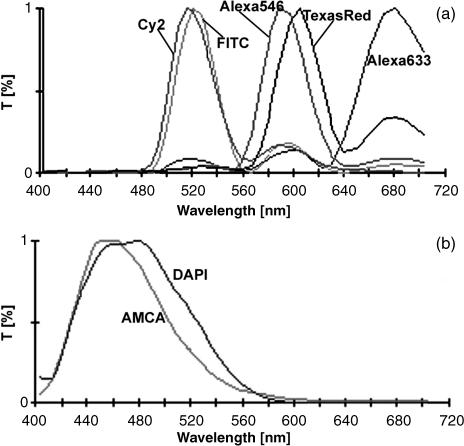
For the decomposition of seven different fluorescent dyes in their pure spectral components two spectral images, obtained with different filter sets, were necessary. Fluorescent spectra of Cy2, FITC, Alexa546, Texas Red and Alexa633 could clearly be separated after image acquisition using the triple band filter set (SKY, ASI) for green, red and infrared in combination with subsequent linear unmixing (Fig. 3a). Detection of AMCA and DAPI blue spectra required the filter set #01 (Zeiss) (Fig. 3b). Although we found a shift of the fluorescent signal for Alexa633 to longer wavelength regions, fluorescent signals of the other dyes were detected within the wavelength regions given by the supplier. Co-localization, in the sense of identical labelling patterns of different antigens, did not occur. Photobleaching during image acquisition was not observed. However, Alexa633 faded after 4 days of storage at 4 °C in darkness.
Fig. 3.
Applied spectra, analysed with SpectraView software. (a) Linear unmixing for Cy2, FITC, Alexa546, Texas Red and Alexa 633 using a triple band filter SKY for green, red and infra red spectra (ASI, Israel). (b) Linear unmixing for AMCA and DAPI using filter #01 for blue spectra (Zeiss, Germany).
hMSC and osteoblasts showed a distinguishable labelling pattern (Table 2) with differences in the labelling of the putative stem cell markers: hMSC showed positive labelling for CD44, CD105/endoglin and VCAM-1, whereas osteoblasts did not label for CD105/endoglin or VCAM-1. The labelling results for all antigens were very homogeneous except for the labelling of collagen-IV, which showed heterogeneous expression patterns with positive and negative staining within the hMSC and hOB cultures.
Table 2.
Labelling profiles of hMSC and osteoblasts
| Endoglin | VCAM-1 | CD44 | col-IV | Fibronectin | F-actin | |
|---|---|---|---|---|---|---|
| hMSC | + | + | + | +/– | + | + |
| Osteoblasts | – | – | + | +/– | + | + |
Summary of labelling profiles. – = no immunofluorescence signal, +/– = heterogeneous labelling profile with positive and negative results, + = labelling of > 85% of cells.
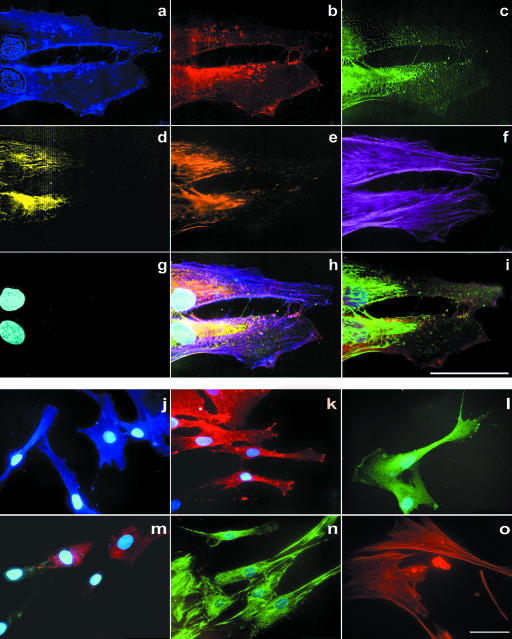
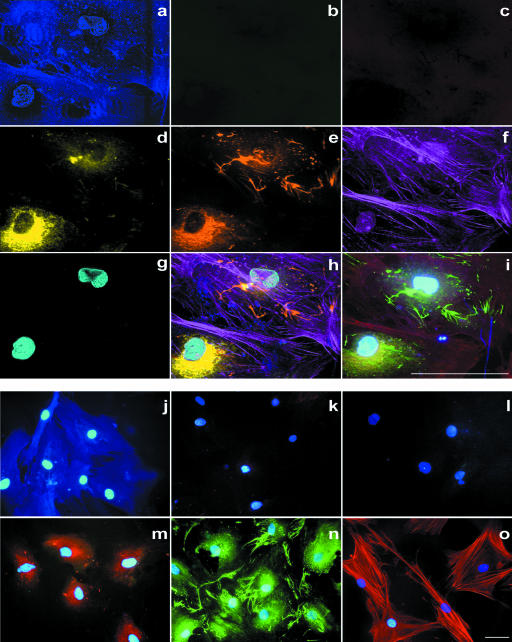
As we performed immunofluorescence on the single cell level we could describe the labelling pattern of the individual antigens. We applied seven-colour labelling of representative cells of each group (Figs 1 and 2). CD105/endoglin was mainly located on the cell surface of hMSC; however, it was also prominent in dense regions of the cell membrane (Fig. 1b,k). VCAM-1 was homogeneously distributed throughout the cytoplasm with a granular labelling pattern and also positive in dense membrane structures (Fig. 1c,l). CD44, detected in hMSC and hOB, revealed homogeneous labelling with accentuation of the cell membrane in hMSC (Figs 1a,j and 2a,j) and appeared to be more intense in regions where fibronectin was also positive. In osteoblasts the accentuation of the cell membrane was less intense (Fig. 2a,j). Cells which labelled positively for collagen-IV showed a granular intracellular pattern and no extracellular labelling (Fig. 1d,m and 2d,m). Fibronectin revealed granular intracellular and a fibrillar extracellular labelling (Figs 1e,n and 2e,n). F-actin showed the typical fibrillar, cytoplasmatic labelling, especially in widespread cells, equally for hMSC and osteoblasts (Fig. 1f,o and 2f,o).
Fig. 1.
Multicolour immunofluorescence of human mesenchymal stem cells. (a–g) Six-colour immunofluorescence plus DAPI nuclear staining with spectral image acquisition. Spectral image after linear unmixing: (a) CD44/AMCA, (b) endoglin (CD105)/TexasRed, (c) VCAM-1 (CD106)/FITC, (d) collagen-IV/Alexa546 (pseudo-coloured yellow), (e) fibronectin/Cy2 (pseudo-coloured brown), (f) f-actin/Alexa633 (pseudo-coloured violet), (g) DAPI nuclear staining. (a,g) Detection with filter #01 for blue spectra (Zeiss, Germany); (b–f) detection with triple band filter SKY for green, red and infrared spectra (ASI, Israel). (h) Digital overlay of a–g. (i) Conventional digital image (without spectral image acquisition) using a triple band filter F61002 for red, green and blue spectra (AHF, Germany). (j–o) Single-colour immunofluorescence plus DAPI nuclear staining of human mesenchymal stem cells with conventional digital image acquisition. (j) Labelling for AMCA/CD44 revealed a homogeneous membrane pattern. (k) Texas Red/endoglin (CD105) exhibited a membrane labelling with accentuation especially of phase-dense membrane regions. (l) FITC/VCAM-1 showed a granular intracellular labelling as well as labelling of phase-dense membrane structures. (m) Labelling for collagen-IV revealed a granular intracellular pattern. (n) Cy2/fibronectin labelled in a granular intracellular and a fibrous extracellular pattern. Cy2 and FITC cannot be distinguished. (o) Alexa633/Phalliodin showed a fibrous actin staining, without DAPI staining. (j) Detection with filter #01 for blue spectra (Zeiss, Germany) and (k–o) triple band filter F61002 for red, green and blue spectra (AHF, Germany). All scale bars = 50 µm.
Fig. 2.
Multicolour immunofluorescence of human osteoblasts. (a–g) Six-colour immunofluorescence plus DAPI nuclear staining with spectral image acquisition and (j–o) single-colour immunofluorescence plus DAPI nuclear staining with conventional digital image acquisition. For each picture (a–o) antibodies and filter sets were used as described in Fig. 1. No labelling could be detected for endoglin (CD105) and VCAM-1 (CD106). All scale bars = 50 µm.
The labelling pattern of the markers in the seven-colour fluorescence was identical with the respective labelling patterns observed in the control single-colour applications (Figs 1j–o and 2j–o).
Discussion
The aim of the study was to characterize hMSC and to distinguish them from mature cell phenotypes on the single cell level. We performed seven-colour immunofluorescence and showed that we can discriminate hMSC from human osteoblasts using this technique.
One of the major difficulties in stem cell research is the identification process of the appropriate cells. Due to the lack of specific markers, hMSC are identified either by analysing their expansion capacity or by performing differentiation assays (Pittenger et al. 1999; Sekiya et al. 2002; Kemp et al. 2005; Kassem, 2006). This is also reflected in the recommendations of the International Society for Cellular Therapy (ISCT) regarding nomenclature (Horwitz et al. 2005) and minimal criteria for defining MSC (Dominici et al. 2006).
Multicolour immunofluorescence allows cell characterization at the single cell level as well as simultaneous detection of several antigens (Tsurui et al. 2000; Schieker et al. 2004). Both features are needed to characterize hMSC because these cells represent a heterogeneous cell population consisting of morphologically and immunocytochemically distinct cell types (Colter et al. 2001; Smith et al. 2004; Vogel et al. 2004; Kemp et al. 2005) without unique antigenic phenotype (Conget & Minguell, 1999; Sekiya et al. 2002; Kassem, 2006). Therefore, this approach may allow us to characterize and identify even very early stages of stem cell differentiation (Gronthos et al. 2001; Schieker et al. 2004).
The surface proteins CD44, CD105/endoglin and CD106/VCAM-1, in particular, have been used to characterize hMSC (Barry et al. 1999; Conget & Minguell, 1999; Pittenger et al. 1999; Ishii et al. 2005; Mareschi et al. 2006). CD44 is one of the first markers to be expressed by hMSC as soon as cells attach to a surface (Zohar et al. 1997) and is reported to be expressed in almost all cells of an hMSC population (Pittenger et al. 1999; Shur et al. 2002; Kotobuki et al. 2004; Mareschi et al. 2006). The distribution pattern may be related to the function of CD44 as a receptor for hyaluronate and other matrix components (Cichy & Pure, 2003). CD105/endoglin is a member of the TGF-beta receptor complex that modulates TGF-beta signalling. Among others, endoglin [SH-2, as described previously (Barry et al. 1999)] was suggested as a putative stem cell marker (Haynesworth et al. 1992; Pittenger et al. 1999; Lodie et al. 2002; Gronthos et al. 2003; Vogel et al. 2004; Dominici et al. 2006). Like CD44, we found CD105/endoglin on almost every hMSC, but the labelling pattern was different to that of CD44. CD105/endoglin was especially located in dense membrane structures, which are involved in the cell migration process. By contrast, CD106/VCAM-1, a membrane protein of the immunoglobulin super family, was predominantly found in the cytoplasm of hMSC, suggesting that further stimuli are needed for its translocation to the membrane. Positive results for labelling of VCAM-1 in hMSC have also been described previously (Gronthos et al. 2003; Pittenger et al. 1999; Honczarenko et al. 2006).
In contrast to the relatively homogeneous results of surface protein labelling, we found heterogeneous (i.e. positive and negative) labelling for collagen-IV in different cells of the same hMSC population. This extracellular matrix component is expressed by hMSC and early progenitors but not by later differentiation stages (Chichester et al. 1993; Deschaseaux & Charbord, 2000). Collagen-IV may therefore be useful in distinguishing between differentiation stages of hMSC.
While fibronectin, an extracellular matrix protein, is expressed by a number of different cell types, hMSC labelled particularly strongly (Vogel et al. 2004). Confirming these studies, our immunocytochemical results showed intra- and extracellular staining for this protein. Phalloidin labelling of the actin cytoskeleton was particularly suitable for assessment of cell morphology.
Although each single marker investigated is not exclusively expressed by hMSC, the simultaneous detection of all these markers by seven-colour fluorescence is a suitable attempt to characterize hMSC on the single cell level. In particular, this enables discrimination of hMSC from other cell types in mixed cell pools as, for example, primary bone marrow aspirates.
Simultaneous detection of several antigens is also possible by flow cytometry on cells in suspension (Tsurui et al. 2000; De Rosa et al. 2003; Mareschi et al. 2006). However, the advantage of immunofluorescence is that it allows the analysis of antigen distribution as well as the investigation of single cells attached to a surface. The importance of the latter fact has been highlighted by other studies: Lodie et al. (2002) demonstrated that two fractions of bone marrow stem cells immunoselected for endoglin showed indistinguishable expression profiles after re-attachment to a surface. Reyes et al. (2001) showed that CD44 expression, in particular, is dependent on the consistency of the surface.
Concluding remarks
In summary, we conclude that a more sophisticated characterization of human mesenchymal stem cells on the single cell level is possible, when seven-colour fluorescence is used. With this technique we were able specifically to distinguish hMSC from osteoblasts. Multicolour immunofluorescence would be especially suitable to the further characterization of subtypes of hMSC (Smith et al. 2004).
Acknowledgments
This work was supported by the Friedrich-Baur Foundation, Munich, and the Bavarian Research Foundation [Bavarian Research Collaboration for Tissue Engineering and Rapid Prototyping (ForTePro)]. The monoclonal antibodies Hermes-1 and P3D1 were obtained from the Developmental Studies Hybridoma Bank maintained by The University of Iowa, Department of Biological Sciences, under contract NO1-HD-7-3263 from the NICDH. This study is part of the doctoral thesis of Florian Haasters and is published with permission of the Ludwig-Maximilians-University, Munich.
References
- Aubin JE. Regulation of osteoblast formation and function. Rev Endocr Metab Disord. 2001;2:81–94. doi: 10.1023/a:1010011209064. [DOI] [PubMed] [Google Scholar]
- Barry FP, Boynton RE, Haynesworth S, Murphy JM, Zaia J. The monoclonal antibody SH-2, raised against human mesenchymal stem cells, recognizes an epitope on endoglin (CD105) Biochem Biophys Res Commun. 1999;265:134–139. doi: 10.1006/bbrc.1999.1620. [DOI] [PubMed] [Google Scholar]
- Barry FP, Murphy JM. Mesenchymal stem cells: clinical applications and biological characterization. Int J Biochem Cell Biol. 2004;36:568–584. doi: 10.1016/j.biocel.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Bianco P, Riminucci M, Gronthos S, Robey PG. Bone marrow stromal stem cells: nature, biology, and potential applications. Stem Cells. 2001;19:180–192. doi: 10.1634/stemcells.19-3-180. [DOI] [PubMed] [Google Scholar]
- Bruder SP, Ricalton NS, Boynton RE, et al. Mesenchymal stem cell surface antigen SB-10 corresponds to activated leukocyte cell adhesion molecule and is involved in osteogenic differentiation. J Bone Miner Res. 1998;13:655–663. doi: 10.1359/jbmr.1998.13.4.655. [DOI] [PubMed] [Google Scholar]
- Cancedda R, Bianchi G, Derubeis A, Quarto R. Cell therapy for bone disease: a review of current status. Stem Cells. 2003;21:610–619. doi: 10.1634/stemcells.21-5-610. [DOI] [PubMed] [Google Scholar]
- Caplan AI, Dennis JE. Mesenchymal stem cells as trophic mediators. J Cell Biochem. 2006;98:1076–1084. [Google Scholar]
- Chichester CO, Fernandez M, Minguell JJ. Extracellular matrix gene expression by human bone marrow stroma and by marrow fibroblasts. Cell Adhes Commun. 1993;1:93–99. doi: 10.3109/15419069309095685. [DOI] [PubMed] [Google Scholar]
- Cichy J, Pure E. The liberation of CD44. J Cell Biol. 2003;161:839–843. doi: 10.1083/jcb.200302098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colter DC, Sekiya I, Prockop DJ. Identification of a subpopulation of rapidly self-renewing and multipotential adult stem cells in colonies of human marrow stromal cells. Proc Natl Acad Sci USA. 2001;98:7841–7845. doi: 10.1073/pnas.141221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conget PA, Minguell JJ. Phenotypical and functional properties of human bone marrow mesenchymal progenitor cells. J Cell Physiol. 1999;181:67–73. doi: 10.1002/(SICI)1097-4652(199910)181:1<67::AID-JCP7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- De Rosa SC, Brenchley JM, Roederer M. Beyond six colors: a new era in flow cytometry. Nat Med. 2003;9:112–117. doi: 10.1038/nm0103-112. [DOI] [PubMed] [Google Scholar]
- Deschaseaux F, Charbord P. Human marrow stromal precursors are alpha 1 integrin subunit-positive. J Cell Physiol. 2000;184:319–325. doi: 10.1002/1097-4652(200009)184:3<319::AID-JCP5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Dominici M, Le Blanc K, Mueller I, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- Ducy P, Schinke T, Karsenty G. The osteoblast: a sophisticated fibroblast under central surveillance. Science. 2000;289:1501–1504. doi: 10.1126/science.289.5484.1501. [DOI] [PubMed] [Google Scholar]
- Gronthos S, Franklin DM, Leddy HA, Robey PG, Storms RW, Gimble JM. Surface protein characterization of human adipose tissue-derived stromal cells. J Cell Physiol. 2001;189:54–63. doi: 10.1002/jcp.1138. [DOI] [PubMed] [Google Scholar]
- Gronthos S, Zannettino AC, Hay SJ, et al. Molecular and cellular characterisation of highly purified stromal stem cells derived from human bone marrow. J Cell Sci. 2003;116:1827–1835. doi: 10.1242/jcs.00369. [DOI] [PubMed] [Google Scholar]
- Grove JE, Bruscia E, Krause DS. Plasticity of bone marrow-derived stem cells. Stem Cells. 2004;22:487–500. doi: 10.1634/stemcells.22-4-487. [DOI] [PubMed] [Google Scholar]
- Haynesworth SE, Baber MA, Caplan AI. Cell surface antigens on human marrow-derived mesenchymal cells are detected by monoclonal antibodies. Bone. 1992;13:69–80. doi: 10.1016/8756-3282(92)90363-2. [DOI] [PubMed] [Google Scholar]
- Honczarenko M, Le Y, Swierkowski M, Ghiran I, Glodek AM, Silberstein LE. Human bone marrow stromal cells express a distinct set of biologically functional chemokine receptors. Stem Cells. 2006;24:1030–1041. doi: 10.1634/stemcells.2005-0319. [DOI] [PubMed] [Google Scholar]
- Horwitz EM, Le Blanc K, Dominici M, et al. Clarification of the nomenclature for MSC: the International Society for Cellular Therapy position statement. Cytotherapy. 2005;7:393–395. doi: 10.1080/14653240500319234. [DOI] [PubMed] [Google Scholar]
- Ishii M, Koike C, Igarashi A, et al. Molecular markers distinguish bone marrow mesenchymal stem cells from fibroblasts. Biochem Biophys Res Commun. 2005;332:297–303. doi: 10.1016/j.bbrc.2005.04.118. [DOI] [PubMed] [Google Scholar]
- Javazon EH, Beggs KJ, Flake AW. Mesenchymal stem cells: paradoxes of passaging. Exp Hematol. 2004;32:414–425. doi: 10.1016/j.exphem.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Jahagirdar BN, Reinhardt RL, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–49. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- Kasperk C, Wergedal J, Strong D, et al. Human bone cell phenotypes differ depending on their skeletal site of origin. J Clin Endocrinol Metab. 1995;80:2511–2517. doi: 10.1210/jcem.80.8.7629252. [DOI] [PubMed] [Google Scholar]
- Kassem M. Stem cells: potential therapy for age-related diseases. Ann NY Acad Sci. 2006;1067:436–442. doi: 10.1196/annals.1354.062. [DOI] [PubMed] [Google Scholar]
- Kemp KC, Hows J, Donaldson C. Bone marrow-derived mesenchymal stem cells. Leuk Lymphoma. 2005;46:1531–1544. doi: 10.1080/10428190500215076. [DOI] [PubMed] [Google Scholar]
- Kotobuki N, Hirose M, Takakura Y, Ohgushi H. Cultured autologous human cells for hard tissue regeneration: preparation and characterization of mesenchymal stem cells from bone marrow. Artif Organs. 2004;28:33–39. doi: 10.1111/j.1525-1594.2004.07320.x. [DOI] [PubMed] [Google Scholar]
- Lodie TA, Blickarz CE, Devarakonda TJ, et al. Systematic analysis of reportedly distinct populations of multipotent bone marrow-derived stem cells reveals a lack of distinction. Tissue Eng. 2002;8:739–751. doi: 10.1089/10763270260424105. [DOI] [PubMed] [Google Scholar]
- Majumdar MK, Keane-Moore M, Buyaner D, et al. Characterization and functionality of cell surface molecules on human mesenchymal stem cells. J Biomed Sci. 2003;10:228–241. doi: 10.1007/BF02256058. [DOI] [PubMed] [Google Scholar]
- Malik Z, Cabib D, Buckwald RA, Talmi A, Garini Y, Lipson SG. Fourier transform multipixel spectroscopy for quantitative cytology. J Microsc. 1996;182:133–140. [Google Scholar]
- Mareschi K, Ferrero I, Rustichelli D, et al. Expansion of mesenchymal stem cells isolated from pediatric and adult donor bone marrow. J Cell Biochem. 2006;97:744–754. doi: 10.1002/jcb.20681. [DOI] [PubMed] [Google Scholar]
- Otto WR, Rao J. Tomorrow's skeleton staff: mesenchymal stem cells and the repair of bone and cartilage. Cell Prolif. 2004;37:97–110. doi: 10.1111/j.1365-2184.2004.00303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger MF, Mackay AM, Beck SC, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- Prockop DJ, Gregory CA, Spees JL. One strategy for cell and gene therapy: harnessing the power of adult stem cells to repair tissues. Proc Natl Acad Sci USA. 2003;100(Suppl. 1):11917–11923. doi: 10.1073/pnas.1834138100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondo S, Penna C, Pagliaro P, Geuna S. Morphological characterization of GFP stably transfected adult mesenchymal bone marrow stem cells. J Anat. 2006;208:3–12. doi: 10.1111/j.1469-7580.2006.00511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes M, Lund T, Lenvik T, Aguiar D, Koodie L, Verfaillie CM. Purification and ex vivo expansion of postnatal human marrow mesodermal progenitor cells. Blood. 2001;98:2615–2625. doi: 10.1182/blood.v98.9.2615. [DOI] [PubMed] [Google Scholar]
- Rothmann C, Bar-Am I, Malik Z. Spectral imaging for quantitative histology and cytogenetics. Histol Histopathol. 1998;13:921–926. doi: 10.14670/HH-13.921. [DOI] [PubMed] [Google Scholar]
- Schieker M, Pautke C, Reitz K, et al. The use of four-colour immunofluorescence techniques to identify mesenchymal stem cells. J Anat. 2004;204:133–139. doi: 10.1111/j.1469-7580.2004.00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya I, Larson BL, Smith JR, Pochampally R, Cui JG, Prockop DJ. Expansion of human adult stem cells from bone marrow stroma: conditions that maximize the yields of early progenitors and evaluate their quality. Stem Cells. 2002;20:530–541. doi: 10.1634/stemcells.20-6-530. [DOI] [PubMed] [Google Scholar]
- Shur I, Marom R, Lokiec F, Socher R, Benayahu D. Identification of cultured progenitor cells from human marrow stroma. J Cell Biochem. 2002;87:51–57. doi: 10.1002/jcb.10267. [DOI] [PubMed] [Google Scholar]
- Smith JR, Pochampally R, Perry A, Hsu SC, Prockop DJ. Isolation of a highly clonogenic and multipotential subfraction of adult stem cells from bone marrow stroma. Stem Cells. 2004;22:823–831. doi: 10.1634/stemcells.22-5-823. [DOI] [PubMed] [Google Scholar]
- Tsurui H, Nishimura H, Hattori S, Hirose S, Okumura K, Shirai T. Seven-color fluorescence imaging of tissue samples based on Fourier spectroscopy and singular value decomposition. J Histochem Cytochem. 2000;48:653–662. doi: 10.1177/002215540004800509. [DOI] [PubMed] [Google Scholar]
- Vogel W, Grunebach F, Messam CA, Kanz L, Brugger W, Buhring HJ. Heterogeneity among human bone marrow-derived mesenchymal stem cells and neural progenitor cells. Haematologica. 2004;88:126–133. [PubMed] [Google Scholar]
- Zohar R, Sodek J, McCulloch CA. Characterization of stromal progenitor cells enriched by flow cytometry. Blood. 1997;90:3471–3481. [PubMed] [Google Scholar]