Abstract
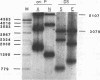
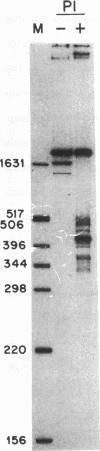
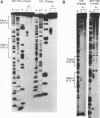
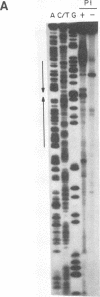
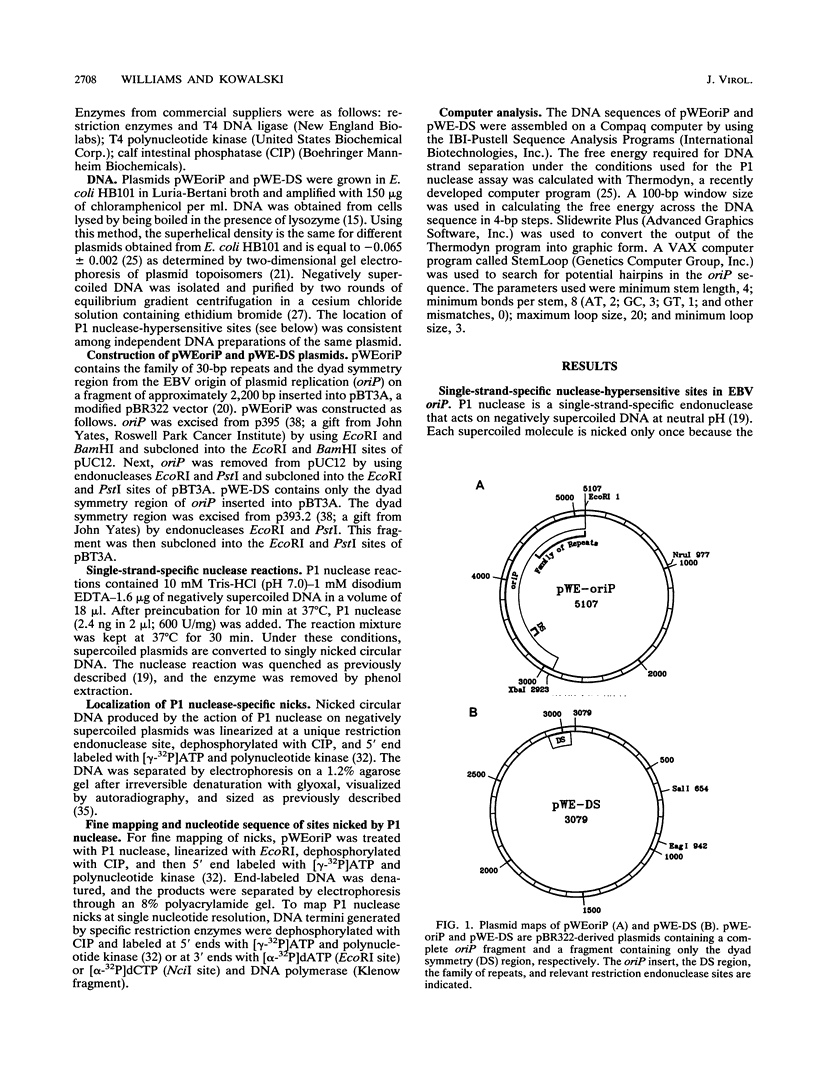
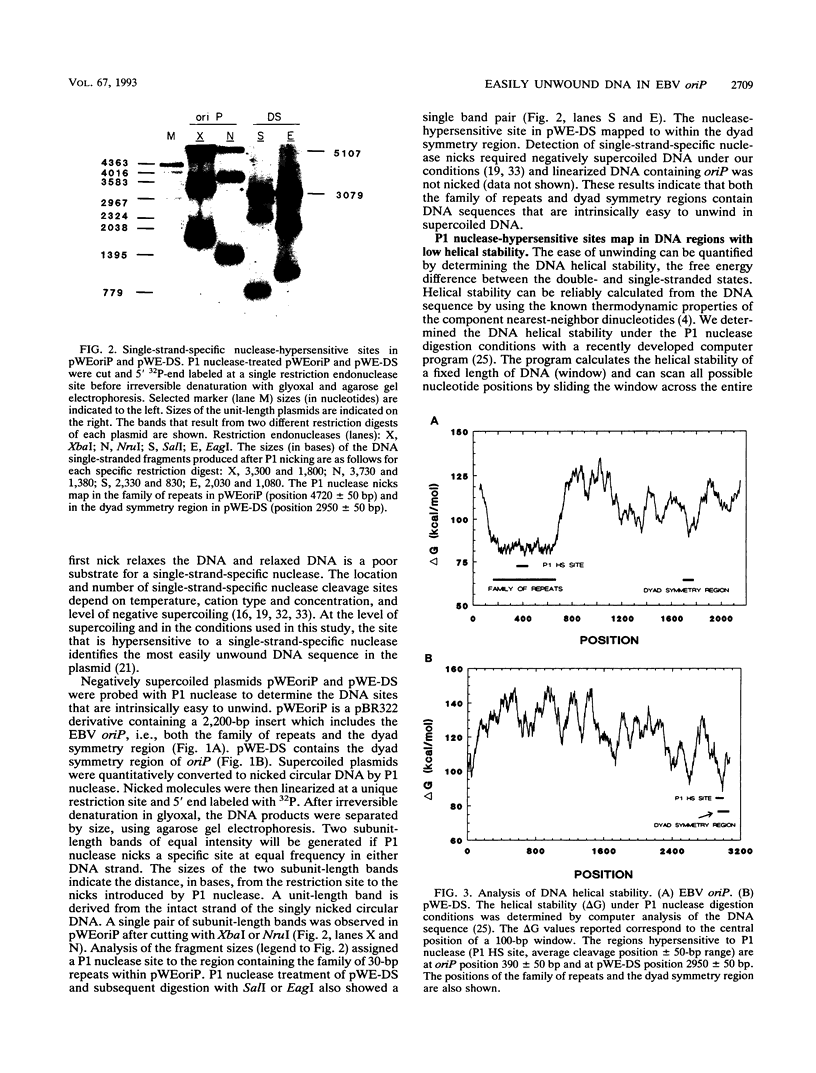
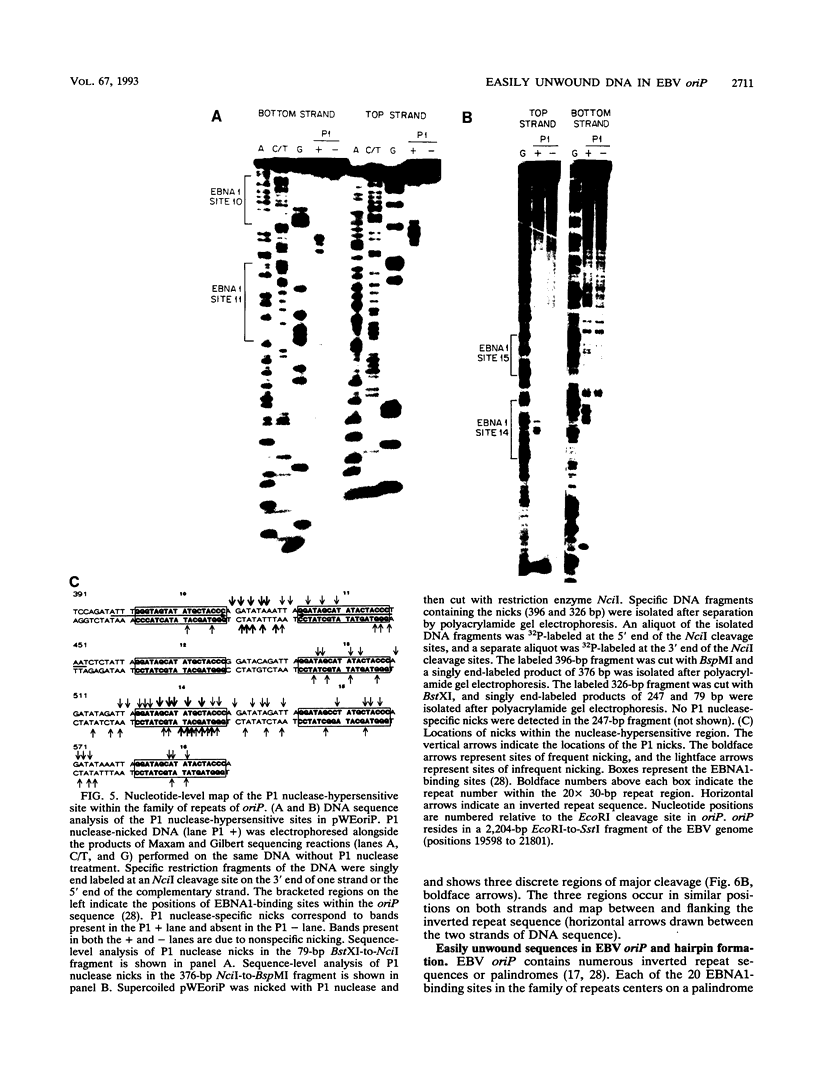
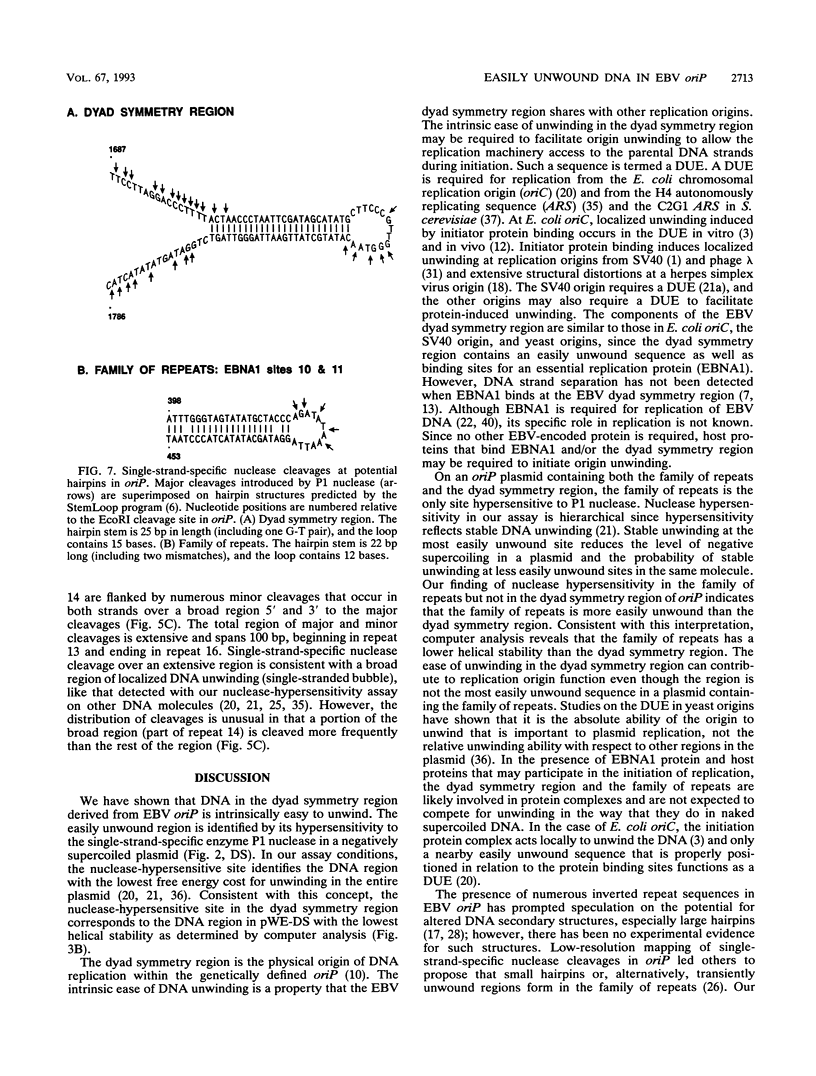
The Epstein-Barr virus (EBV) origin of plasmid replication (oriP) includes two known cis-acting components, the dyad symmetry region and the family of repeats. We used P1 nuclease, a single-strand-specific endonuclease, to probe EBV oriP for DNA sequences that are intrinsically easy to unwind on a negatively supercoiled plasmid. Selective nuclease hypersensitivity was detected in the family of repeats on an oriP-containing plasmid and in the dyad symmetry region on a plasmid that lacks the family of repeats, indicating that the DNA in both cis-acting components is intrinsically easy to unwind. The hierarchy of nuclease hypersensitivity indicates that the family of repeats is more easily unwound than the dyad symmetry region, consistent with the hierarchy of helical stability predicted by computer analysis of the DNA sequence. A specific subset of the family of repeats is nuclease hypersensitive, and the DNA structure deduced from nucleotide-level analysis of the P1 nuclease nicks is a cruciform near a single-stranded bubble. The dyad symmetry region unwinds to form a broad single-stranded bubble containing hairpins in the 65-bp dyad sequence. We propose that the intrinsic ease of unwinding the dyad symmetry region, the actual origin of DNA replication, is an important component in the mechanism of initiation.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borowiec J. A., Hurwitz J. Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J. 1988 Oct;7(10):3149–3158. doi: 10.1002/j.1460-2075.1988.tb03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowater R., Aboul-ela F., Lilley D. M. Large-scale stable opening of supercoiled DNA in response to temperature and supercoiling in (A + T)-rich regions that promote low-salt cruciform extrusion. Biochemistry. 1991 Dec 10;30(49):11495–11506. doi: 10.1021/bi00113a003. [DOI] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988 Mar 11;52(5):743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- Breslauer K. J., Frank R., Blöcker H., Marky L. A. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Wang J. C. Cruciform formation in a negatively supercoiled DNA may be kinetically forbidden under physiological conditions. Cell. 1983 Jul;33(3):817–829. doi: 10.1016/0092-8674(83)90024-7. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappier L., O'Donnell M. EBNA1 distorts oriP, the Epstein-Barr virus latent replication origin. J Virol. 1992 Mar;66(3):1786–1790. doi: 10.1128/jvi.66.3.1786-1790.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappier L., O'Donnell M. Epstein-Barr nuclear antigen 1 mediates a DNA loop within the latent replication origin of Epstein-Barr virus. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10875–10879. doi: 10.1073/pnas.88.23.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahn T. A., Schildkraut C. L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989 Aug 11;58(3):527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- Gellert M., O'Dea M. H., Mizuuchi K. Slow cruciform transitions in palindromic DNA. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5545–5549. doi: 10.1073/pnas.80.18.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille H., Messer W. Localized DNA melting and structural pertubations in the origin of replication, oriC, of Escherichia coli in vitro and in vivo. EMBO J. 1991 Jun;10(6):1579–1584. doi: 10.1002/j.1460-2075.1991.tb07678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearing J., Mülhaupt Y., Harper S. Interaction of Epstein-Barr virus nuclear antigen 1 with the viral latent origin of replication. J Virol. 1992 Feb;66(2):694–705. doi: 10.1128/jvi.66.2.694-705.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henle W., Diehl V., Kohn G., Zur Hausen H., Henle G. Herpes-type virus and chromosome marker in normal leukocytes after growth with irradiated Burkitt cells. Science. 1967 Sep 1;157(3792):1064–1065. doi: 10.1126/science.157.3792.1064. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Iacono-Connors L., Kowalski D. Altered DNA conformations in the gene regulatory region of torsionally-stressed SV40 DNA. Nucleic Acids Res. 1986 Nov 25;14(22):8949–8962. doi: 10.1093/nar/14.22.8949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S. Significant potential secondary structures in the Epstein-Barr virus genome. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6915–6919. doi: 10.1073/pnas.83.18.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koff A., Schwedes J. F., Tegtmeyer P. Herpes simplex virus origin-binding protein (UL9) loops and distorts the viral replication origin. J Virol. 1991 Jun;65(6):3284–3292. doi: 10.1128/jvi.65.6.3284-3292.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D. Changes in site specificity of single-strand-specific endonucleases on supercoiled PM2 DNA with temperature and ionic environment. Nucleic Acids Res. 1984 Sep 25;12(18):7071–7086. doi: 10.1093/nar/12.18.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D., Eddy M. J. The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 1989 Dec 20;8(13):4335–4344. doi: 10.1002/j.1460-2075.1989.tb08620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D., Natale D. A., Eddy M. J. Stable DNA unwinding, not "breathing," accounts for single-strand-specific nuclease hypersensitivity of specific A+T-rich sequences. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9464–9468. doi: 10.1073/pnas.85.24.9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupton S., Levine A. J. Mapping genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol Cell Biol. 1985 Oct;5(10):2533–2542. doi: 10.1128/mcb.5.10.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Middleton T., Sugden B. EBNA1 can link the enhancer element to the initiator element of the Epstein-Barr virus plasmid origin of DNA replication. J Virol. 1992 Jan;66(1):489–495. doi: 10.1128/jvi.66.1.489-495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale D. A., Schubert A. E., Kowalski D. DNA helical stability accounts for mutational defects in a yeast replication origin. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2654–2658. doi: 10.1073/pnas.89.7.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski R., Miller G. Single-stranded structures are present within plasmids containing the Epstein-Barr virus latent origin of replication. J Virol. 1991 Feb;65(2):677–686. doi: 10.1128/jvi.65.2.677-686.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins D. R., Milman G., Hayward S. D., Hayward G. S. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985 Oct;42(3):859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- Reisman D., Sugden B. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol Cell Biol. 1986 Nov;6(11):3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman D., Yates J., Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985 Aug;5(8):1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnos M., Zahn K., Inman R. B., Blattner F. R. Initiation protein induced helix destabilization at the lambda origin: a prepriming step in DNA replication. Cell. 1988 Feb 12;52(3):385–395. doi: 10.1016/s0092-8674(88)80031-x. [DOI] [PubMed] [Google Scholar]
- Sheflin L. G., Kowalski D. Altered DNA conformations detected by mung bean nuclease occur in promoter and terminator regions of supercoiled pBR322 DNA. Nucleic Acids Res. 1985 Sep 11;13(17):6137–6154. doi: 10.1093/nar/13.17.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheflin L. G., Kowalski D. Mung bean nuclease cleavage of a dA + dT-rich sequence or an inverted repeat sequence in supercoiled PM2 DNA depends on ionic environment. Nucleic Acids Res. 1984 Sep 25;12(18):7087–7104. doi: 10.1093/nar/12.18.7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W., Middleton T., Sugden B., Echols H. DNA looping between the origin of replication of Epstein-Barr virus and its enhancer site: stabilization of an origin complex with Epstein-Barr nuclear antigen 1. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10870–10874. doi: 10.1073/pnas.88.23.10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umek R. M., Kowalski D. The DNA unwinding element in a yeast replication origin functions independently of easily unwound sequences present elsewhere on a plasmid. Nucleic Acids Res. 1990 Nov 25;18(22):6601–6605. doi: 10.1093/nar/18.22.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umek R. M., Kowalski D. The ease of DNA unwinding as a determinant of initiation at yeast replication origins. Cell. 1988 Feb 26;52(4):559–567. doi: 10.1016/0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Umek R. M., Kowalski D. Thermal energy suppresses mutational defects in DNA unwinding at a yeast replication origin. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2486–2490. doi: 10.1073/pnas.87.7.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysokenski D. A., Yates J. L. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J Virol. 1989 Jun;63(6):2657–2666. doi: 10.1128/jvi.63.6.2657-2666.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J. L., Warren N., Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. 1985 Feb 28-Mar 6Nature. 313(6005):812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- Yates J., Warren N., Reisman D., Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]