Abstract
Mitochondrial DNA (mtDNA) is the traditional workhorse for reconstructing evolutionary events. The frequent use of mtDNA in such analyses derives from the apparent simplicity of its inheritance: maternal and lacking bi-parental recombination. However, in hybrid zones, the reproductive barriers are often not completely developed, resulting in the breakdown of male mitochondrial elimination mechanisms, leading to leakage of paternal mitochondria and transient heteroplasmy, resulting in an increased possibility of recombination. Despite the widespread occurrence of heteroplasmy and the presence of the molecular machinery necessary for recombination, we know of no documented example of recombination of mtDNA in any terrestrial wild vertebrate population. By sequencing the entire mitochondrial genome (16 761 bp), we present evidence for mitochondrial recombination in the hybrid zone of two mitochondrial haplotypes in the Australian frillneck lizard (Chlamydosaurus kingii).
Keywords: mitochondrial recombination, hybrid zone, frillneck lizard
1. Introduction
Mitochondrial DNA (mtDNA) is the established molecular tool for reconstructing evolutionary events. The frequent use of mtDNA in such analyses derives from the apparent simplicity of its inheritance: maternal and lacking bi-parental recombination (Slate & Gemmell 2004). However, heteroplasmy, the occurrence of more than one mitochondrial haplotype within an individual, has been suggested to facilitate the creation of mitochondrial recombinants (Städler & Delph 2002). Heteroplasmy might be achieved by paternal leakage; that is, paternal mtDNA is not eliminated during fertilization (Städler & Delph 2002). The production of novel haplotypes through recombination of coexisting mtDNA genomes has been observed in species in which leakage of paternal mtDNA is common (Ladoukakis & Zouros 2001). Thus, interactions between co-occurring genomes following leakage of paternal mtDNA is the most common explanation for the origin of recombinant haplotypes (Ladoukakis & Zouros 2001; Städler & Delph 2002; Jaramillo-Correa & Bousquet 2005).
It has been hypothesized that recurrent hybridization events, such as those observed in contact zones of closely related species, could provide the opportunity for heteroplasmy, and hence the possibility of recombination to occur (reviewed in Rokas et al. 2003). In hybrid zones, the reproductive barriers are not completely developed and, in some cases, the mechanisms inducing the elimination of male mitochondria could have broken down, leading to leakage of paternal mitochondria and transient heteroplasmy (Jaramillo-Correa & Bousquet 2005).
Despite the widespread occurrence of heteroplasmy and the presence of the molecular machinery necessary for recombination (reviewed in Rokas et al. 2003), we know of no documented example of recombination of mtDNA in terrestrial wild vertebrate populations. By sequencing the entire mitochondrial genome (16 761 bp), we present evidence for mitochondrial recombination in the hybrid zone of two mitochondrial haplotypes in the Australian frillneck lizard (Chlamydosaurus kingii).
2. Material and methods
(a) Study area and species
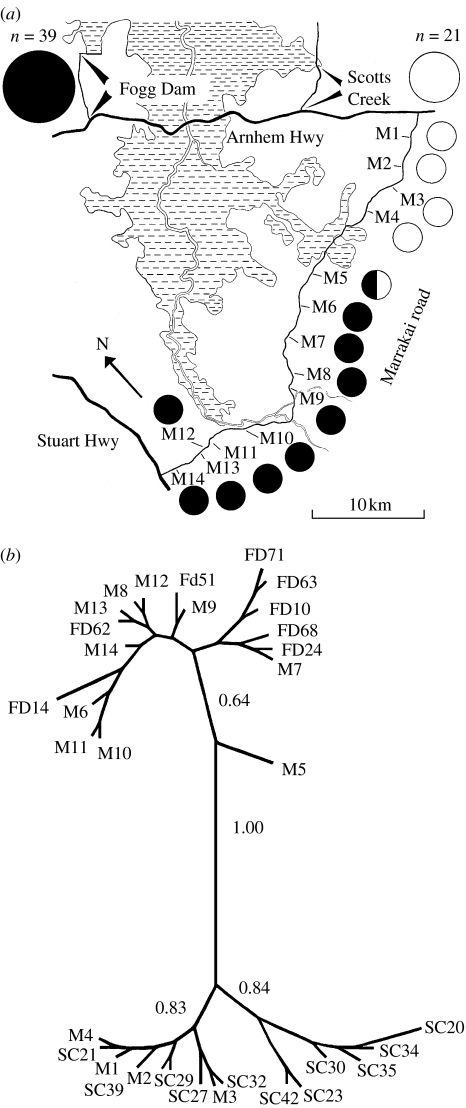
The study area is situated in the wet–dry tropics of Australia, approximately 60 km Southeast of Darwin. Fieldwork was conducted from September to the end of November in 2005. The lizards were captured at three sites: (i) 39 lizards were captured along a 6 km dirt track in Fogg Dam conservation reserve, (ii) 21 lizards along a 4 km dirt track at Scotts Creek, and (iii) 14 lizards along the 50 km long Marrakai road (figure 1a).
Figure 1.
(a) Map of the study area. Stippled areas depict floodplain and unfilled areas savannah woodland. Filled circles denote lizards displaying Fogg Dam ND2–ND4 haplotypes and open circles denote lizards with Scotts Creek ND2–ND4 haplotypes. Lizard M5 is depicted as a mix of the two haplotypes. (b) Unrooted tree based on Bayesian analysis of concatenated mitochondrial ND4 and ND2 genes.
(b) DNA extraction, amplification and sequencing
Genomic DNA was isolated from whole blood by phenol–chloroform extraction. Published sequences of Pogona vitticeps and C. kingii (Macey et al. 2000; Amer & Kumazawa 2005; GenBank accession numbers NC_006922 and AF128469, respectively) were used to design specific primers. The PCRs were performed in a total volume of 20 μl containing 100 ng of total genomic DNA, 1 U of recombinant Taq DNA polymerase (Invitrogen), 0.125 mM of each nucleotide, 2 μl 10×PCR buffer (Invitrogen) and 0.6 μM of each primer. The MgCl2 concentrations were adjusted according to the different primer combinations. The PCR amplifications were conducted as follows: 94°C/3 min plus (94°C/30 s, plus 50–62°C/30 s depending on primer combination, plus 72°C/1 min)×30, plus 72°C/10 min. The PCR products were purified with ExoSap (GE Healthcare) and sequenced from both directions on an ABI 3130xl Genetic Analyzer using BigDye Terminator Kit v. 3.1 (Applied Biosystems). DNA sequences were aligned using BioEdit (Hall 1999) and ClustalW (Chenna et al. 2003). All sequences are deposited in GenBank (accession numbers EF090421–EF090489).
(c) Phylogenetic and recombination analyses
Phylogenetic relationships were assessed by Bayesian inference (Huelsenbeck et al. 2001), using the ND2 and ND4 genes (688 and 672 bp, respectively). Only lizards with polymorphic haplotypes from Fogg Dam (n=8) and Scotts Creek (n=11) populations were used in the analyses, whereas all 14 Marrakai lizards were included. Each gene was aligned separately using ClustalW (Thompson et al. 1994) and the concatenated alignment treated as a single partition. MrModeltest v. 2.0 (Nylander 2004) chose the models: lset nst=2 rates=invgamma (hierarchical likelihood ratio test) and lset nst=2 rates=propinv (Akaike Information Criterion). Trees reported herein were analysed using the latter model, as it has one less parameter (Posada & Crandall 1998). Nevertheless, both the models produced very similar trees; our conclusions are identical no matter which model was used. Four independent runs of 500 000 generations were performed, with trees sampled every 100 generations. Stationarity was assessed by both an examination of the average standard deviation of split frequencies and by examining the potential scale reduction factor. Burnin was evident after 30 000 generations. The majority rule consensus tree was constructed using the pooled trees from four independent runs, after discarding trees prior to convergence (Huelsenbeck & Rannala 2004). We used a Kimura two-parameter distance for the DNA sequence data under a γ distribution (α=0.19) estimated in PUZZLE (Strimmer & Von Haeseler 1996) to calculate mean genetic distance between populations using the program MEGA (Kumar et al. 1993).
The entire mitochondrial genome (16 761 bp) in three lizards (see below) was used to detect putative recombination sites and four recombination tests were conducted: (i) Recombination Detection Program (RDP; Martin & Rybicki 2000), (ii) Geneconv (Sawyer 1989), (iii) Maxchi (Maynard Smith 1992), and (iv) Chimaera (Posada & Crandall 2001). The tests were run using the RDP software, and we set the highest acceptable p-value to 0.05, i.e. the probability that sequences would share high identities by chance. The significance of the Χ2 peaks in Maxchi and Chimaera was determined with 1000 permutations. Sequential Bonferroni corrections were applied to compensate for multiple tests (Rice 1989).
3. Results
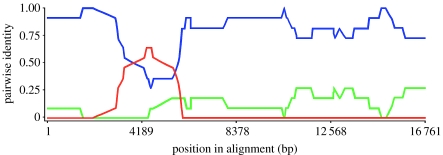
The phylogenetic analyses of the ND2 (688 bp) and ND4 (672 bp) genes revealed two major and very distinct mitochondrial haplotypes (figure 1b, table 1). However, one of the lizards (M5), captured at the contact zone of the two mtDNA groups, displayed a mixed, unique haplotype (figure 1a,b, table 1). Owing to the high similarity within each of the two major haplotype groups, the complete mitochondrial genome was sequenced in one animal from each group and compared with that of lizard M5. All four recombination analyses revealed a highly significant recombination site in lizard M5 (RDP, 8.726×10−8; Geneconv, 5.830×10−7; Maxchi, 8.212×10−3; and Chimaera, 6.501×10−3). Over most of the mitochondrial genome, the RDP analysis showed that lizard M5 displayed very high pairwise identity in common with the Fogg Dam specimen, but very low pairwise identity compared with the Scotts Creek lizard (figure 2). However, the analysis revealed a recombination site between 3827 and 5131 bp (figure 2, table 1), covering the region of the ND2 gene.
Table 1.
All variable nucleotide sites recorded at the ND2 (7 bp) and at the ND4 (13 bp) genes (locations denoted above each bp). At the ND2 gene, lizard M5 exhibited identical nucleotides with the Scotts Creek lizard, whereas at the ND4 gene, lizard M5 exhibited identical nucleotides with the Fogg Dam lizard at 11 out of 13 sites. At the remaining two sites, lizard M5 differed from both the Scotts Creek and the Fogg Dam animals. Apart from site 4407 in the ND2 gene, all substitutions were synonymous.
| ND2 | ND4 | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4274 | 4407 | 4451 | 4553 | 4595 | 4599 | 4686 | 10 038 | 10 209 | 10 413 | 10 516 | 10 557 | 10 584 | 10 653 | 10 827 | 10 848 | 10 869 | 10 881 | 10 968 | 11 043 | |
| Fogg dam | T | G | A | T | C | C | T | G | G | A | C | T | T | G | C | C | A | G | C | A |
| M5 | C | A | G | C | T | T | C | G | G | A | C | T | T | G | C | C | G | G | C | G |
| Scotts creek | C | A | G | C | T | T | C | A | A | G | T | C | C | A | T | T | A | A | T | A |
Figure 2.
Test result for recombination of the complete mitochondrial genome using the general recombination method (RDP). Pairwise identity refers to the average pairwise sequence identity within a 10 nucleotide sliding window moved one nucleotide at a time along an informative site alignment of the three sequences. Blue depicts pairwise identity between lizard M5 and the Fogg Dam lizard, red depicts pairwise identity between lizard M5 and the Scotts Creek lizard and green depicts pairwise identity between the Fogg Dam and the Scotts Creek lizards.
4. Discussion
Our results are unlikely to be an experimental artefact as the recombinant region of lizard M5 was sequenced on three occasions, twice when we first detected its unique haplotype when running the phylogenetic analyses of the ND2 and ND4 genes, each derived from one primer combination, and again when its entire mtDNA genome was sequenced with different primers. All analyses revealed an identical sequence. Furthermore, except for the recombinant region, lizard M5 displayed high pairwise identity in common with the Fogg Dam specimen, but very low pairwise identity compared with the Scotts Creek lizard (figure 2, table 1). Thus, if the high pairwise similarity between lizard M5 and the Scott Creek lizard in the recombinant region was caused by DNA contamination, we would have expected to observe overlapping peaks at all other sites (76 bp) at which the Scotts Creek lizard exhibited differences, but lizard M5 showed similar nucleotides to that of the Fogg Dam sample. No such artefacts were observed, making it highly unlikely that our results were caused by contamination.
Although we did not detect any heteroplasmy among the 74 lizards analysed, other studies have invoked heteroplasmy as the driving force for mtDNA recombination in spite of not detecting heteroplasmic individuals (Städler & Delph 2002; Jaramillo-Correa & Bousquet 2005).
In the contact zone of two conifers, black spruce (Picea mariana) and red spruce (Picea rubens), Jaramillo-Correa & Bousquet (2005) suggested introgressive hybridization to be the major contributing factor to the presence of recombinant haplotypes. Although our results are based on a single species, they mirror those obtained by Jaramillo-Correa & Bousquet (2005). In other words, the only lizard exhibiting a recombinant haplotype was captured in the contact zone between the two major mitochondrial haplotypes (figure 1a). The difference of the mitochondrial genome in lizard M5, compared with the other two mtDNA haplotypes, strongly suggests the recombination was derived via paternal leakage resulting in the fusion of the maternal and paternal mtDNAs.
One explanation for the rare detection of bi-parental inheritance of mitochondrial genomes may be due to the two haplotypes having to be sufficiently dissimilar to be detected during conventional genetic screening (Kvist et al. 2003). The results from the present study imply that recombination in terrestrial vertebrates could be more common than previously suspected.
The importance of recombination in vertebrate mitochondria has broad implications across several fields, ranging from human mitochondrial diseases (Schon 2000) to the compromise of phylogenetic reconstruction (Posada & Crandall 2002), inferences related to demographic history and the application of molecular clocks (Schierup & Hein 2000). With regard to the use of mtDNA for evolutionary studies, our results suggest that we should not draw conclusions about the evolutionary history of the entire mitochondrial genome by only investigating parts of it.
Acknowledgments
The work was supported by grants from the Australian Research Council. We are very grateful to D. Martin for his help in using the RDP program, and for comments provided by three reviewers.
References
- Amer S.A.M, Kumazawa Y. Mitochondrial genome of Pogona vitticepes (Reptilia; Agamidae): control region duplication and the origin of Australasian agamids. Gene. 2005;346:249–256. doi: 10.1016/j.gene.2004.11.014. doi:10.1016/j.gene.2004.11.014 [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson T.J, Higgins D.J, Thompson J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. doi:10.1093/nar/gkg500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Huelsenbeck J.P, Rannala B. Frequentist properties of Bayesian posterior probabilities of phylogenetic trees under simple and complex substitution models. Syst. Biol. 2004;53:904–913. doi: 10.1080/10635150490522629. doi:10.1080/10635150490522629 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P, Ronquist F, Nielsen R, Bollback J.P. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. doi:10.1126/science.1065889 [DOI] [PubMed] [Google Scholar]
- Jaramillo-Correa J.P, Bousquet J. Mitochondrial genome recombination in the zone of contact of two hybridising conifers. Genetics. 2005;171:1951–1962. doi: 10.1534/genetics.105.042770. doi:10.1534/genetics105.042770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. Pennsylvania State University; University Park, PA: 1993. MEGA: molecular evolutionary genetics analysis. 1.01. [Google Scholar]
- Kvist L, Martens J, Nazarenko A.A, Orell M. Paternal Leakage of mitochondrial DNA in the Great Tit (Parus major) Mol. Biol. Evol. 2003;20:243–247. doi: 10.1093/molbev/msg025. doi:10.1093/molbev/msg025 [DOI] [PubMed] [Google Scholar]
- Ladoukakis E.D, Zouros E. Direct evidence for homologous recombination in mussel (Mytilus gallprovincialis) mitochondrial DNA. Mol. Biol. Evol. 2001;18:1168–1175. doi: 10.1093/oxfordjournals.molbev.a003904. [DOI] [PubMed] [Google Scholar]
- Macey J.R, Schulye J.A, Larson A. Evolution and phylogenetic information content of mitochondrial genomic structural features illustrated with acrodont lizards. Syst. Biol. 2000;49:257–277. doi:10.1080/10635159950173843 [PubMed] [Google Scholar]
- Martin D, Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics. 2000;16:562–563. doi: 10.1093/bioinformatics/16.6.562. doi:10.1093/bioinformatics/16.6.562 [DOI] [PubMed] [Google Scholar]
- Maynard Smith J. Analysing the mosaic structure of genes. J. Mol. Evol. 1992;35:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- Nylander J.A.A. Uppsala University; Uppsala, Sweden: 2004. MrModeltest 2.0. Program distributed by the author. [Google Scholar]
- Posada D, Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall K.A. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl Acad. Sci. USA. 2001;98:13 575–13 762. doi: 10.1073/pnas.241370698. doi:10.1073/pnas/241370698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall K.A. The effect of recombination on the accuracy of phylogenetic estimation. J. Mol. Evol. 2002;54:396–402. doi: 10.1007/s00239-001-0034-9. doi:10.1007/s00239-001-0034-9 [DOI] [PubMed] [Google Scholar]
- Rice W.R. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. doi:10.2307/2409177 [DOI] [PubMed] [Google Scholar]
- Rokas A, Ladoukakis E, Zouros E. Animal mitochondrial DNA recombination revisited. Trends Ecol. Evol. 2003;18:411–417. doi:10.10.16/SO169-5347(03)00125-3 [Google Scholar]
- Sawyer S. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 1989;6:526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- Schierup M.H, Hein J. Consequences of recombination on traditional phylogenetic analysis. Genetics. 2000;156:879–891. doi: 10.1093/genetics/156.2.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon E.A. Mitochondrial genetics and disease. Trends Biochem. Sci. 2000;25:555–560. doi: 10.1016/s0968-0004(00)01688-1. doi:10.1016/S0968-0004(00)01688-1 [DOI] [PubMed] [Google Scholar]
- Slate J, Gemmell N.J. Eve ‘n’ Steven: recombination of human mitochondrial DNA. Trends Ecol. Evol. 2004;19:561–563. doi:10.1016/J.tree.2004.09.001 [Google Scholar]
- Strimmer K, Von Haeseler A. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 1996;13:964–969. [Google Scholar]
- Städler T, Delph L.F. Ancient mitochondrial haplotypes and evidence for intragenic recombination in gynodioecious plant. Proc. Natl Acad. Sci. USA. 2002;99:11 730–11 735. doi: 10.1073/pnas.182267799. doi:10.1073/pnas/182267799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D, Higgins D.G, Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. doi:10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]