Abstract
The three-wave migration hypothesis of Greenberg et al. has permeated the genetic literature on the peopling of the Americas. Greenberg et al. proposed that Na-Dene, Aleut-Eskimo and Amerind are language phyla which represent separate migrations from Asia to the Americas. We show that a unique allele at autosomal microsatellite locus D9S1120 is present in all sampled North and South American populations, including the Na-Dene and Aleut-Eskimo, and in related Western Beringian groups, at an average frequency of 31.7%. This allele was not observed in any sampled putative Asian source populations or in other worldwide populations. Neither selection nor admixture explains the distribution of this regionally specific marker. The simplest explanation for the ubiquity of this allele across the Americas is that the same founding population contributed a large fraction of ancestry to all modern Native American populations.
Keywords: migration, Native American, D9S1120, HGDP–CEPH
1. Introduction
There has been extensive debate over the number of migrations into the Americas. Greenberg et al. (1986) hypothesized that Amerind, Na-Dene and Aleut-Eskimo are language phyla which represent three migrations from Asia, occurring in that sequence. This hypothesis stimulated a multitude of genetic investigations into the number and timing of migrations (reviewed in Schurr 2004).
Still, genetic studies have not produced a consensus on the number of migrations into the Americas; we suggest this is because the number of migrations cannot be inferred from genetic data. Migrations may have occurred that have not significantly influenced the current distribution of genetic variation. Distinct migrations from the same source population may produce patterns of variation similar to that produced from a single migration.
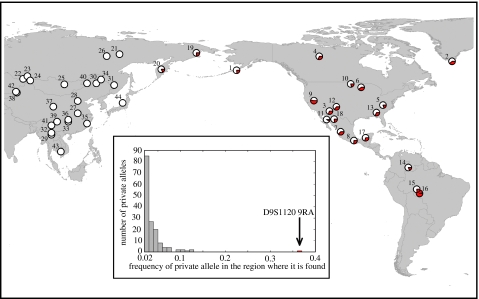
Although the number of migrations might not be inferable from genetic data, whether all Native American populations descend from the same founding population can be addressed if a unique autosomal variant absent from Asian populations is identified throughout the Americas. In their analysis of the HGDP–CEPH human genome diversity panel (henceforth HGDP) genotypes for 377 microsatellites, Zhivotovsky et al. (2003) noted that only in a single instance could a regional group could be distinguished by a private marker. A 275 bp allele at D9S1120 (also known as GATA81C04 or GATA11E11) was observed at high frequencies in all American populations (all of which are Amerind: Pima; Maya; Colombian; Karitiana; and Surui) and was absent from 47 other worldwide populations. This allele had a frequency of 36.5% in the pooled American sample, while no other allele among the 4688 studied was private to a major geographical region (defined as sub-Saharan Africa, Europe and the part of Asia south and west of the Himalayas (including North Africa), East Asia, Oceania and the Americas) with a frequency above 13%. Expansion of the dataset to 783 loci and 9346 alleles (Rosenberg et al. 2005) did not reveal any additional regionally private allele with a frequency above 13% (figure 1, inset).
Figure 1.
Distribution of frequencies of private alleles (with frequency of 2% or above) in the HGDP among 9346 alleles at 783 microsatellites studied by Rosenberg et al. (2005). Inset. Frequency distribution of 9RA (represented by red-shaded area) at D9S1120 by population in Asia and the Americas. Numbers next to pie charts reference table 1.
The 275 bp allele was the smallest one observed at D9S1120 in the HGDP. We have determined through sequencing that this allele contains nine tetranucleotide repeats and is the result of slippage in the repetitive section, as opposed to a deletion elsewhere in the amplicon. Henceforth, we shall refer to alleles at this locus by the corresponding number of repeats and to the 9-repeat allele as ‘9RA’. The next largest allele in the HGDP outside the Americas is 11 repeats and was observed only in three chromosomes. The lack of regionally specific private alleles at a high frequency (figure 1, inset), the striking distribution of 9RA and the rarity of intermediate-sized alleles (table 1) strongly suggests that all or nearly all copies of 9RA descend from a single mutational event.
Table 1.
Frequency of each allele at D9S1120 in all sampled populations. (Three alleles of non-standard fragment size, each observed once in the HGDP (Han, Burusho and Biaka Pygmy), are not represented.)
| population | ref. for figure 1 | N | repeats | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |||
| Aleuta | 1 | 24 | 0.229 | 0.021 | 0.021 | 0.104 | 0.417 | 0.125 | 0.063 | 0.021 | ||||
| Inuit (Greenland)a | 2 | 31 | 0.387 | 0.016 | 0.016 | 0.016 | 0.290 | 0.129 | 0.113 | 0.032 | ||||
| Apachea | 3 | 32 | 0.313 | 0.031 | 0.094 | 0.422 | 0.094 | 0.047 | ||||||
| Dogriba | 4 | 34 | 0.309 | 0.162 | 0.132 | 0.294 | 0.044 | 0.044 | 0.015 | |||||
| Cherokeea | 5 | 25 | 0.200 | 0.020 | 0.020 | 0.200 | 0.420 | 0.120 | 0.020 | |||||
| Chippewa (Mille Lacs)a | 6 | 19 | 0.395 | 0.053 | 0.237 | 0.211 | 0.053 | 0.053 | ||||||
| Huichola | 7 | 16 | 0.375 | 0.188 | 0.438 | |||||||||
| Mixteca | 8 | 67 | 0.261 | 0.008 | 0.067 | 0.485 | 0.134 | 0.045 | ||||||
| Northern Paiutea | 9 | 17 | 0.529 | 0.265 | 0.147 | 0.029 | 0.029 | |||||||
| Sioux (Sisseton/Wahpeton)a | 10 | 26 | 0.212 | 0.039 | 0.019 | 0.154 | 0.289 | 0.077 | 0.096 | 0.115 | ||||
| Seria | 11 | 15 | 0.100 | 0.367 | 0.233 | 0.267 | 0.033 | |||||||
| Jemeza | 12 | 19 | 0.368 | 0.026 | 0.211 | 0.290 | 0.053 | 0.053 | ||||||
| Creeka | 13 | 16 | 0.313 | 0.031 | 0.125 | 0.313 | 0.188 | 0.031 | ||||||
| Colombianb | 14 | 13 | 0.192 | 0.269 | 0.385 | 0.077 | 0.077 | |||||||
| Karitianab | 15 | 24 | 0.250 | 0.250 | 0.125 | 0.125 | 0.250 | |||||||
| Suruib | 16 | 17 | 0.971 | 0.029 | ||||||||||
| Mayab | 17 | 25 | 0.300 | 0.020 | 0.020 | 0.080 | 0.280 | 0.240 | 0.020 | 0.040 | ||||
| Pimab | 18 | 25 | 0.220 | 0.020 | 0.040 | 0.400 | 0.260 | 0.060 | ||||||
| Chukchia | 19 | 21 | 0.238 | 0.024 | 0.024 | 0.191 | 0.238 | 0.238 | 0.048 | |||||
| Koryaksa | 20 | 20 | 0.175 | 0.125 | 0.525 | 0.125 | 0.050 | |||||||
| Evena | 21 | 30 | 0.083 | 0.050 | 0.550 | 0.200 | 0.117 | |||||||
| Southern Altaia | 22 | 39 | 0.013 | 0.038 | 0.013 | 0.308 | 0.346 | 0.244 | 0.026 | 0.013 | ||||
| Northern Altaia | 23 | 11 | 0.091 | 0.227 | 0.409 | 0.227 | 0.045 | |||||||
| Altai Kazakha | 24 | 25 | 0.020 | 0.060 | 0.240 | 0.340 | 0.280 | 0.060 | ||||||
| Mongoliaa | 25 | 24 | 0.042 | 0.229 | 0.458 | 0.167 | 0.063 | 0.042 | ||||||
| Yakutb | 26 | 25 | 0.160 | 0.400 | 0.360 | 0.040 | 0.040 | |||||||
| Hanb | 27 | 34 | 0.015 | 0.147 | 0.500 | 0.309 | 0.015 | |||||||
| Han (North China)b | 28 | 10 | 0.250 | 0.650 | 0.100 | |||||||||
| Daib | 29 | 10 | 0.050 | 0.600 | 0.300 | 0.050 | ||||||||
| Daurb | 30 | 10 | 0.100 | 0.600 | 0.300 | |||||||||
| Hezhenb | 31 | 7 | 0.143 | 0.429 | 0.214 | 0.214 | ||||||||
| Lahub | 32 | 8 | 0.313 | 0.438 | 0.250 | |||||||||
| Miaob | 33 | 10 | 0.150 | 0.450 | 0.400 | |||||||||
| Oroqenb | 34 | 10 | 0.200 | 0.550 | 0.150 | 0.050 | 0.050 | |||||||
| Sheb | 35 | 10 | 0.100 | 0.600 | 0.250 | 0.050 | ||||||||
| Tujiab | 36 | 10 | 0.050 | 0.350 | 0.250 | 0.350 | ||||||||
| Tub | 37 | 10 | 0.250 | 0.550 | 0.100 | 0.100 | ||||||||
| Xibob | 38 | 9 | 0.222 | 0.611 | 0.111 | 0.056 | ||||||||
| Yib | 39 | 10 | 0.200 | 0.500 | 0.300 | |||||||||
| Mongolab | 40 | 8 | 0.063 | 0.125 | 0.500 | 0.313 | ||||||||
| Naxib | 41 | 10 | 0.250 | 0.500 | 0.150 | 0.100 | ||||||||
| Uygurb | 42 | 10 | 0.100 | 0.100 | 0.150 | 0.400 | 0.150 | 0.100 | ||||||
| Cambodianb | 43 | 11 | 0.045 | 0.318 | 0.182 | 0.409 | 0.045 | |||||||
| Japaneseb | 44 | 29 | 0.017 | 0.172 | 0.466 | 0.241 | 0.103 | |||||||
| Orcadianb | n.a. | 16 | 0.063 | 0.219 | 0.500 | 0.156 | 0.063 | |||||||
| Adygeib | n.a. | 17 | 0.029 | 0.235 | 0.647 | 0.059 | 0.029 | |||||||
| Russianb | n.a. | 25 | 0.020 | 0.120 | 0.020 | 0.080 | 0.100 | 0.500 | 0.100 | 0.060 | ||||
| Basqueb | n.a. | 24 | 0.021 | 0.292 | 0.479 | 0.208 | ||||||||
| Frenchb | n.a. | 29 | 0.017 | 0.052 | 0.138 | 0.466 | 0.241 | 0.086 | ||||||
| Italianb | n.a. | 13 | 0.038 | 0.231 | 0.538 | 0.154 | 0.038 | |||||||
| Sardinianb | n.a. | 28 | 0.036 | 0.250 | 0.375 | 0.214 | 0.125 | |||||||
| Tuscanb | n.a. | 8 | 0.063 | 0.125 | 0.438 | 0.313 | 0.063 | |||||||
| Mozabiteb | n.a. | 30 | 0.017 | 0.033 | 0.017 | 0.150 | 0.533 | 0.233 | 0.017 | |||||
| Bedouinb | n.a. | 48 | 0.010 | 0.021 | 0.302 | 0.500 | 0.125 | 0.031 | 0.010 | |||||
| Druzeb | n.a. | 46 | 0.076 | 0.011 | 0.054 | 0.207 | 0.446 | 0.152 | 0.054 | |||||
| Palestinianb | n.a. | 49 | 0.010 | 0.041 | 0.020 | 0.306 | 0.286 | 0.245 | 0.082 | 0.010 | ||||
| Balochib | n.a. | 25 | 0.040 | 0.040 | 0.300 | 0.320 | 0.240 | 0.060 | ||||||
| Brahuib | n.a. | 25 | 0.040 | 0.100 | 0.340 | 0.360 | 0.100 | 0.040 | 0.020 | |||||
| Burushob | n.a. | 25 | 0.020 | 0.040 | 0.220 | 0.380 | 0.260 | 0.060 | ||||||
| Hazarab | n.a. | 24 | 0.063 | 0.167 | 0.438 | 0.250 | 0.083 | |||||||
| Kalashb | n.a. | 25 | 0.020 | 0.320 | 0.460 | 0.100 | 0.060 | 0.040 | ||||||
| Makranib | n.a. | 25 | 0.020 | 0.080 | 0.280 | 0.360 | 0.200 | 0.040 | 0.020 | |||||
| Pathanb | n.a. | 24 | 0.083 | 0.104 | 0.188 | 0.354 | 0.229 | 0.042 | ||||||
| Sindhib | n.a. | 25 | 0.080 | 0.020 | 0.220 | 0.440 | 0.180 | 0.060 | ||||||
| Melanesianb | n.a. | 19 | 0.026 | 0.553 | 0.368 | 0.053 | ||||||||
| Papuanb | n.a. | 17 | 0.059 | 0.029 | 0.118 | 0.471 | 0.265 | 0.059 | ||||||
| Bantu (South Africa)b | n.a. | 8 | 0.063 | 0.313 | 0.250 | 0.250 | 0.125 | |||||||
| Bantu (Kenya)b | n.a. | 12 | 0.042 | 0.042 | 0.208 | 0.500 | 0.167 | 0.042 | ||||||
| Mandenkab | n.a. | 24 | 0.146 | 0.167 | 0.458 | 0.167 | 0.042 | 0.021 | ||||||
| Yorubab | n.a. | 25 | 0.020 | 0.340 | 0.340 | 0.260 | 0.040 | |||||||
| Biaka Pygmyb | n.a. | 32 | 0.031 | 0.125 | 0.531 | 0.219 | 0.078 | |||||||
| Mbuti Pygmyb | n.a. | 15 | 0.067 | 0.067 | 0.333 | 0.200 | 0.267 | 0.067 | ||||||
| Sanb | n.a. | 7 | 0.071 | 0.214 | 0.357 | 0.143 | 0.214 | |||||||
Populations in this study.
Frequencies from populations in the HGDP are from dataset H1048 (Rosenberg 2006).
We hypothesized that if the Aleut-Eskimo, Na-Dene and other indigenous populations throughout the Americas share common ancestry with the American populations in the HGDP, then we would observe 9RA across the Americas. Additionally, if further sampling did not reveal 9RA in putative Asian source populations, then we could conclude that modern Native American populations share more recent common ancestry with each other than with any Asian population.
2. Material and methods
(a) Populations sampled
We sampled two Aleut-Eskimo, two Na-Dene and nine North American Amerind populations (tables 1 and 2) for 9RA. We use the grouping ‘Amerind’ so that our results may be interpreted within the framework of the tripartite migration hypothesis of Greenberg et al. (1986), but note that many historical linguists do not accept Amerind (see Greenberg 1987 and Campbell 1997 for opposing views).
Table 2.
Average frequency of 9RA in linguistic and geographical groups.
| linguistic/geographical group | populations sampled | average population frequency of 9RA |
|---|---|---|
| Aleut-Eskimo | Aleut and Inuit | 0.308 |
| Na-Dene | Dogrib and Apache | 0.311 |
| Amerind | Cherokee, Chippewa, Huichol, Mixtec, Northern Paiute, Sioux, Seri, Jemez, Creek, Colombian, Karitiana, Surui, Maya and Pima | 0.335a |
| Western Beringia | Chukchi and Koryaks | 0.207 |
| North America | Aleut, Inuit, Dogrib, Apache, Cherokee, Chippewa, Huichol, Mixtec, Northern Paiute, Sioux, Seri, Jemez, Creek, Maya and Pima | 0.301a |
| South America | Colombian, Karitiana and Surui | 0.471a |
When the Seri and the Surui are excluded, the frequencies of 9RA in the Amerind, North America and South America are 30.1, 31.5 and 22.1%, respectively.
Populations in the Altaian region of east central Asia are among those thought to be most closely related to modern Native Americans on the basis of Y-chromosome and mtDNA evidence, yet some East Siberian populations also share markers with modern Americans (reviewed in Schurr 2004). Thus, we sampled seven populations in the Altaian region of east central Asia and in East Siberia (table 1).
See the electronic supplementary material for further information on the populations sampled and genotyping of 9RA.
(b) Selection
If the distribution of 9RA across the Americas has significantly been influenced by balancing or positive selection at a linked locus, we would expect D9S1120 to be an outlier when compared with empirical distributions of heterozygosity and FST for neutrally evolving loci. We assume that the majority of the 783 microsatellites for which the American HGDP have been genotyped (Rosenberg et al. 2005) to be selectively neutral. We excluded the Surui from this dataset as an extreme outlier (Zhivotovsky et al. 2003). For each locus, we estimated FST using (Weir & Cockerham 1984) and calculated expected heterozygosity (pooling samples), as given by Weir (1996).
Under mutation–drift equilibrium, a positive correlation is expected between the mean heterozygosity and the number of alleles. For this dataset, mean heterozygosity is not significantly different for loci with eight alleles (which includes D9S1120), compared with nine (see electronic supplementary material). Thus, we created empirical distributions of FST and heterozygosity at neutral loci using 116 microsatellites with eight or nine alleles.
(c) Admixture
Supposing that the Aleut-Eskimo or the Na-Dene descend from different founding populations in which 9RA was not present, we calculated the amount of Amerind admixture required to bring 9RA to the frequencies observed in the Aleut-Eskimo or Na-Dene using Bernstein's (1931) formula , where ph is the observed average frequency of 9RA in the putatively admixed Aleut-Eskimo or Na-Dene; p1 is the observed average frequency of 9RA in the Amerind; and p2 is the frequency of 9RA in the Aleut-Eskimo or the Na-Dene prior to admixture, which is zero.
3. Results
9RA was present in all sampled North and South American populations at an average frequency of 32.9% (figure 1; table 1). It was also observed in the Koryaks and the Chukchi of Western Beringia. 9RA was not observed in putative Asian ancestral populations. The populations most divergent in their frequency of 9RA, the Seri (10.0%) and the Surui (97.1%), have been identified as potential isolates on the basis of linguistic (Campbell 1997) and genetic (Zhivotovsky et al. 2003) data, respectively, and thus the divergent frequencies in these populations probably reflect genetic drift.
The frequencies of the other alleles at D9S1120 in the populations in which 9RA was observed are not outliers. Aside from 9RA, the three most common alleles in the Americas and Western Beringia, the 15-, 16- and 17-repeat alleles, are also those most common in the rest of the world. Relative to populations without 9RA, the average frequencies of these common alleles in the Americas and Western Beringia are reduced by about 30% (between 28.8% and 37.1%).
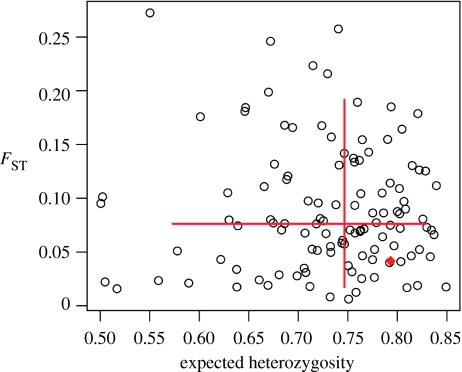
Figure 2 shows FST plotted against expected heterozygosity for 116 microsatellites in four American HGDP populations. D9S1120, located at (0.793, 0.042), is not an outlier with respect to the bivariate distribution of FST and heterozygosity. D9S1120 falls between the 0.2 and 0.8 quantiles of both distributions.
Figure 2.
Plot of FST against heterozygosity for 116 microsatellites in four American HGDP populations. Red bars show the 0.05 and 0.95 quantiles, anchored at the medians. D9S1120 is represented by the red diamond located at (0.793, 0.042).
Under the assumptions of the admixture model, if the Aleut-Eskimo or Na-Dene had acquired 9RA through admixture with the Amerind, the proportions of these populations derived from the Amerind would be 91.9 and 92.8%, respectively. These results are consistent with the similarity of frequencies of 9RA in the Aleut-Eskimo, Na-Dene and Amerind (table 2).
4. Discussion
Irrespective of the evolutionary history of other unlinked loci, the remarkable distribution of 9RA severely constrains the possible evolutionary histories of modern Native American populations. The simplest explanation for the homogeneous frequency of 9RA across the Americas is that the Americas were settled by a single founding population in which 9RA was present and from which all modern Native American populations descend. While homogenization can occur through selection or gene flow, we show it is unlikely either of these processes is solely responsible for the distribution of 9RA.
Were selection, rather than inheritance from a common founding population, responsible for the observed distribution of 9RA, nearly identical selection pressures would be required from the Arctic to the Amazon. Humans in the Americas have coped with remarkable geographical and temporal variations in ecology and hence they have probably been subject to variable selection pressures. In addition, data from four American HGDP populations show that, compared with 115 other microsatellites, D9S1120 is not unusual in FST or heterozygosity. Thus, there is no compelling evidence to suggest that the allele frequency distribution at D9S1120 in the Americas has strongly been influenced by balancing or positive selection.
Despite the simplicity of the admixture model we used, it demonstrates that the frequency of 9RA is too high in the Aleut-Eskimo and Na-Dene to be explained solely by moderate gene flow from the Amerind. If the Aleut-Eskimo or Na-Dene had acquired 9RA through admixture rather than inheritance from a common founding population, then it is probable that their respective gene pools would have been almost completely replaced and that their prehistory cannot be recovered with genetic data. This degree of admixture does not correspond to the traditional concept of multiple founding populations resulting in biologically distinct population expansions.
Hence, the distribution of 9RA is most consistent with the hypothesis that all modern Native Americans descend from a common founding population. The data are not, however, informative about the number of source populations that contributed to this founding population and do not exclude the possibility of small genetic contributions from other populations.
There are a number of possible explanations for the apparent absence of 9RA in putative Asian source populations. 9RA could have (i) arisen in the founding American population, (ii) been sampled from a putative Asian source population, in which it was subsequently lost by genetic drift, and (iii) been segregating in an Asian source population at the time of migration, but that source population has not been identified because it either went extinct or has not been included in modern-day samples. If the allele were segregating in an Asian population, it is improbable that all the copies of 9RA in the Americas descend from more than one ancient sampling event from that population. It is unlikely that an allele at a frequency sufficiently low to destine it for extinction, or an allele the sole source of which is a small, geographically restricted population, would have been included in multiple migratory groups and maintained multiple times.
The presence of 9RA in the Koryaks and Chukchi is consistent with other genetic evidence of shared ancestry between Western Beringians and Native Americans (e.g. Karafet et al. 1997; Lell et al. 1997; Schurr et al. 1999). The observed geographical distribution of 9RA is quite similar to that of two other alleles that descend from unique mutational events, the 16111T and DYS199T transitions which define Native American mtDNA lineage A2 and Y-chromosome lineage Q-M3 (Underhill et al. 1996), respectively. Hence, three independent lines of genetic evidence support the claim (Shields et al. 1993) of an ancient gene pool that included the ancestors of the modern inhabitants of Western Beringia and the Americas.
Acknowledgments
This study was funded by an NSFGRF and a UC Davis Humanities grant to K.B.S., University of Pennsylvania Faculty Research Funds to T.G.S. and NIH grant RR05090 to D.G. Smith. In memory of John McDonough. We thank M.N. Grote, D.A. Bolnick, K. Hunley and R.S. Malhi for helpful comments.
Supplementary Material
Populations sampled, genotyping of 9RA, and mean heterozygosity for loci grouped by number of alleles
References
- Bernstein F. Die geographische Verteilung der Blutgruppen und ihre anthropologische Bedeutung. In: Ginni C, editor. Comitato Italiano per lo Studio dei Problemi della Populazione. vol. 3. Instituto Poligrafico dello Stato Liberia; Rome, Italy: 1931. pp. 227–245. [Google Scholar]
- Campbell L. Oxford University Press; New York, NY: 1997. American Indian languages: the historical linguistics of Native America. [Google Scholar]
- Greenberg J.H. Stanford University Press; Stanford, CA: 1987. Language in the Americas. [Google Scholar]
- Greenberg J.H, Turner C.G, Zegura S.L. The settlement of the Americas—a comparison of the linguistic, dental, and genetic evidence. Curr. Anthropol. 1986;27:477–497. doi:10.1086/203472 [Google Scholar]
- Karafet T.M, et al. Y chromosome markers and Trans-Bering Strait dispersals. Am. J. Phys. Anthropol. 1997;102:301–314. doi: 10.1002/(SICI)1096-8644(199703)102:3<301::AID-AJPA1>3.0.CO;2-Y. doi:10.1002/(SICI)1096-8644(199703)102:3<301::AID-AJPA1>3.0.CO;2-Y [DOI] [PubMed] [Google Scholar]
- Lell J.T, Brown M.D, Schurr T.G, Sukernik R.I, Starikovskaya Y.B, Torroni A, Moore L.G, Troup G.M, Wallace D.C. Y chromosome polymorphisms in Native American and Siberian populations: identification of Native American Y chromosome haplotypes. Hum. Genet. 1997;100:536–543. doi: 10.1007/s004390050548. doi:10.1007/s004390050548 [DOI] [PubMed] [Google Scholar]
- Rosenberg N.A. Standardized subsets of the HGDP–CEPH human genome diversity cell line panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann. Hum. Genet. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. doi:10.1111/j.1469-1809.2006.00285.x [DOI] [PubMed] [Google Scholar]
- Rosenberg N.A, Mahajan S, Ramachandran S, Zhao C, Pritchard J.K, Feldman M.W. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 2005;1:660–671. doi: 10.1371/journal.pgen.0010070. doi:10.1371/journal.pgen.0010070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurr T.G. The peopling of the New World: perspectives from molecular anthropology. Annu. Rev. Anthropol. 2004;33:551–583. doi:10.1146/annurev.anthro.33.070203.143932 [Google Scholar]
- Schurr T.G, Sukernik R.I, Starikovskaya Y.B, Wallace D.C. Mitochondrial DNA variation in Koryaks and Itel'men: population replacement in the Okhotsk Sea–Bering Sea region during the Neolithic. Am. J. Phys. Anthropol. 1999;108:1–39. doi: 10.1002/(SICI)1096-8644(199901)108:1<1::AID-AJPA1>3.0.CO;2-1. doi:10.1002/(SICI)1096-8644(199901)108:1<1::AID-AJPA1>3.0.CO;2-1 [DOI] [PubMed] [Google Scholar]
- Shields G.F, Schmiechen A.M, Frazier B.L, Redd A, Voevoda M.I, Reed J.K, Ward R.H. MtDNA sequences suggest a recent evolutionary divergence for Beringian and northern North American populations. Am. J. Hum. Genet. 1993;53:549–562. [PMC free article] [PubMed] [Google Scholar]
- Underhill P.A, Jin L, Zemans R, Oefner P.J, Cavalli-Sforza L.L. A pre-Columbian Y chromosome-specific transition and its implications for human evolutionary history. Proc. Natl Acad. Sci. USA. 1996;93:196–200. doi: 10.1073/pnas.93.1.196. doi:10.1073/pnas.93.1.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B.S. Sinauer Associates; Sunderland, MA: 1996. Genetic data analysis II: methods for discrete population genetic data. [Google Scholar]
- Weir B.S, Cockerham C.C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. doi:10.2307/2408641 [DOI] [PubMed] [Google Scholar]
- Zhivotovsky L.A, Rosenberg N.A, Feldman M.W. Features of evolution and expansion of modern humans, inferred from genomewide microsatellite markers. Am. J. Hum. Genet. 2003;72:1171–1186. doi: 10.1086/375120. doi:10.1086/375120 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Populations sampled, genotyping of 9RA, and mean heterozygosity for loci grouped by number of alleles