Abstract
Abscisic acid (ABA), an apocarotenoid synthesized from cleavage of carotenoids, regulates seed maturation and stress responses in plants. The viviparous seed mutants of maize identify genes involved in synthesis and perception of ABA. Two alleles of a new mutant, viviparous14 (vp14), were identified by transposon mutagenesis. Mutant embryos had normal sensitivity to ABA, and detached leaves of mutant seedlings showed markedly higher rates of water loss than those of wild type. The ABA content of developing mutant embryos was 70% lower than that of wild type, indicating a defect in ABA biosynthesis. vp14 embryos were not deficient in epoxy-carotenoids, and extracts of vp14 embryos efficiently converted the carotenoid cleavage product, xanthoxin, to ABA, suggesting a lesion in the cleavage reaction. vp14 was cloned by transposon tagging. The VP14 protein sequence is similar to bacterial lignostilbene dioxygenases (LSD). LSD catalyzes a double-bond cleavage reaction that is closely analogous to the carotenoid cleavage reaction of ABA biosynthesis. Southern blots indicated a family of four to six related genes in maize. The Vp14 mRNA is expressed in embryos and roots and is strongly induced in leaves by water stress. A family of Vp14-related genes evidently controls the first committed step of ABA biosynthesis. These genes are likely to play a key role in the developmental and environmental control of ABA synthesis in plants.
In plants, abscisic acid (ABA) is a key hormonal regulator of seed maturation (1) that mediates responses to a variety of stress conditions including water stress (2). Biochemical studies (3) and analyses of mutants that block ABA synthesis (4, 5) indicate that ABA is synthesized from oxidative cleavage of epoxy-carotenoids to produce xanthoxin, which is subsequently converted to ABA via the ABA-aldehyde intermediate (6).
Biochemical studies suggest that cleavage of 9-cis-xanthophylls is the key regulatory step in the ABA biosynthetic pathway (6). The carotenoid precursors are present in very high concentrations relative to ABA, indicating that synthesis of the 9-cis-xanthophylls is unlikely to regulate ABA synthesis in plant tissues (7). The enzyme activities required for conversion of xanthoxin to ABA are constitutively active in most plant tissues (6). However, protein synthesis and transcription inhibitors prevent induction of ABA synthesis in drought-stressed leaves, indicating that stress-induced ABA synthesis requires gene expression (reviewed in ref. 3).
ABA-deficient mutants are known in maize, Arabidopsis, and a few other species (3, 5, 8–12). In maize, ABA-deficient mutants cause precocious seed germination (1). The Arabidopsis aba1 and N. plumbaginifolia aba2 mutants are deficient in the epoxy-carotenoid precursors of ABA (4, 11) and encode a zeaxanthin epoxidase (5). The steps downstream of the cleavage reaction, conversion of xanthoxin to ABA-aldehyde and oxidation of ABA-aldehyde to ABA, are defined in Arabidopsis by the aba2 and aba3 mutants, respectively (9).
Other potential ABA mutants have recently been identified in maize as viviparous seed mutants that have normal carotenoid pigmentation (1). Here we report a biochemical and molecular analysis of one such mutant, vp14. We show that the VP14 protein is related to lignostilbene dioxygenases, bacterial enzymes that catalyze a double-bond cleavage reaction that is chemically similar to the carotenoid cleavage step of ABA biosynthesis (13). Studies of the recombinant protein confirm that VP14 catalyzes carotenoid cleavage reaction (14). In this paper, we show that Vp14 is developmentally regulated and induced in leaves by drought stress.
MATERIALS AND METHODS
The Robertson’s Mutator lines were a gift of Donald S. Robertson, Iowa State University (Ames, IA). The vp14-2274 and vp14-3250 mutant lines were outcrossed to W22 and subsequently maintained by self-pollination of heterozygous plants. The near isogenic wild-type NS-2274 (nonsegregating) and vp14-2274 mutant strains used for biochemical and molecular analyses were extracted from a vp14-2274 heterozygous line.
ABA Determinations and Carotenoid Analysis.
Embryos (5 g per sample) were harvested at 16, 18, and 20 days postpollination (DAP). Extraction and analysis of carotenoids by HPLC were performed as described (4). ABA extractions and quantification by GC with electron capture detector (ECD) were performed as described (8). For determination of stress-induced ABA synthesis, detached leaves of seedlings were allowed to lose 15% of their initial fresh weight by transpiration in room air and then placed in heat-sealed plastic bags and incubated for 5 hr.
Determination of ABA Sensitivity in Culture.
Ears harvested from greenhouse-grown plants at 16 and 18 DAP were surface-sterilized by submersion in a 70% ethanol solution containing 1% dish detergent for 5 min. Embryos were removed aseptically and placed on media containing MS (pH 5.7), 0.2% phytagel, and the indicated concentration of ABA (10). Embryos were cultured at 25°C in a growth chamber, and shoot, root length, and fresh weight were measured after 5 days. The data shown are the means of 15 to 20 embryos for each treatment from 16 DAP. The data for 18 DAP were similar.
Southern and Northern Blot Analysis.
Approximately 10 μg of genomic DNA (15) was digested with the indicated restriction enzymes and resolved by agarose gel electrophoresis. Southern blots on nylon membranes (16) were washed in a 40 mM sodium phosphate/1% SDS solution at 65°C for 2–3 hr for moderate stringency and at 70°C for high stringency.
Total RNA was extracted from maize tissues (1–2 g) in TriZol solution according to the manufacturer’s instructions (GIBCO/BRL) and purified further by precipitation with 2-propanol. Poly(A)+-enriched RNA was prepared by using PolyATtract according to the manufacturer’s instructions (Promega) and quantified spectrophotometrically. One microgram of poly(A)-enriched RNA was resolved in a 1.5% agarose gels containing formaldehyde and probed as described for Southern hybridization (16, 17). DNA probes were radiolabeled with [α-32P]dCTP (3,000 Ci/mmol; 1 Ci = 37 GBq) using the Random Primer DNA Labeling System (GIBCO/BRL).
Construction and Screening of Genomic and cDNA Libraries.
Genomic DNA was digested with restriction enzymes and size-fractionated by centrifugation at 25,000 × g at 4°C for 24 hr through a 10–40% linear sucrose gradient prepared in sterile TE buffer (10 mM Tris⋅HCI/1 mM EDTA, pH 8.0). Selected DNA fractions were ligated into phage cloning vectors (λgt10 or λZAP) and packaged according to the manufacturer’s instructions (Stratagene). A wild-type embryo cDNA library was constructed in λZAP from 5 μg of poly(A) RNA prepared from 18-day postpollination embryos. cDNA was prepared by using the ZAP Express cDNA Synthesis Kit (Stratagene). Libraries were screened by DNA hybridization (16). A 2.5-kb XhoI genomic fragment isolated from vp14-2274 was subcloned in pBluescript (Stratagene) and PCR was used to amplify and clone a 1-kbp sequence flanking Mu1 into pBluescript SK. The 5′ primer derived from terminal repeat of Mu1 (5′-CCATAATGGCAATTATCTC-3′), and the T7 sequencing primer was used as the 3′ (5′-TAATACGACTCACTATAGGG-3′).
Conversion of Xanthoxin to ABA in Cell-Free Extracts.
Embryos (5 g) were homogenized in 0.2 M KPO4, pH 7.5/10 mM DTT. The extract was centrifuged to remove insoluble material, and the supernatant fraction was desalted by passage through a G-25 Sephadex spun column. Enzyme assays contained 1 mM PMSF, 0.25 mM EDTA, and 1 μg cis-xanthoxin and crude enzyme extract. ABA was quantified by GC with ECD (8).
RESULTS
Isolation of vp14.
The vp14-2274 allele was identified in a Robertson’s Mutator strain as a viviparous mutant with weak penetrance under field conditions (1). The mutant embryo shoot axis is elongated relative to the wild type but frequently does not expand sufficiently to rupture the pericarp and initiate germination. Mutant seed that survive desiccation produce fully viable plants. The penetrance of the viviparous phenotype was enhanced under winter greenhouse conditions, suggesting an interaction with environmental factors. Mutant plants were not prone to water stress in the stressful Florida field environment. A second, less penetrant Mutator allele, vp14–3250, was confirmed by complementation testing. The vp14 locus was mapped to chromosome 1L by pollinating vp14-3250 and vp14-2274 plants with the T-B1La, B-A chromosome translocation stock (18). Because B-A chromosomes frequently nondisjoin at the second mitosis in pollen development, a translocation that includes a segment containing Vp14 will produce hybrid embryos that are hemizygous for the mutant allele and are hence viviparous. To test the relationship to other viable viviparous mutants, vp14-2274 plants were crossed to homozygous vp8 (also located on 1L), vp10 (19), and vp*3286 (1) mutant plants. All of these mutants complemented the vp14 alleles.
The vp14 Phenotype Is Rescued by ABA.
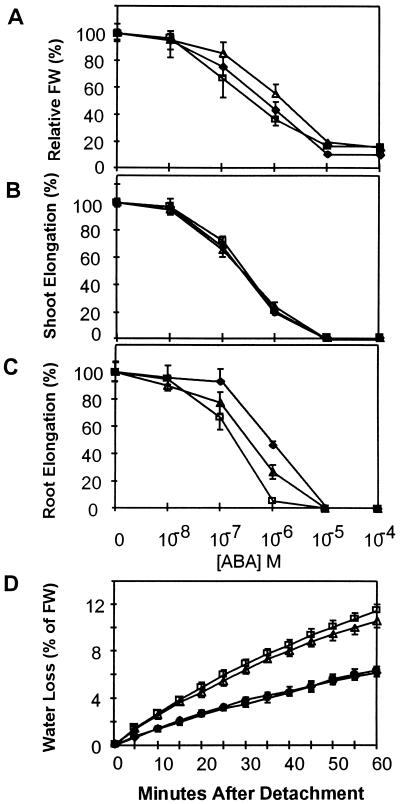
To test whether vp14 had altered sensitivity to ABA, embryos were excised from homozygous mutant and wild-type ears at 16 DAP and incubated for 5 days on media containing various concentrations of ABA. ABA inhibition of shoot elongation and total fresh weight gain was similar for mutants and wild types (Fig. 1A and B). Consistent with previous studies (10) embryos showed significant growth inhibition on 10−7 M ABA and growth was fully inhibited by 10−5 M ABA. Mutant strains showed slightly enhanced sensitivity to ABA inhibition of root growth. Based on these results, the viviparous phenotype could not be attributed to a defect in ABA signal transduction.
Figure 1.
Effect of ABA on embryo fresh weight (A), shoot growth (B), and root growth (C) in culture. Growth of 16-DAP embryos on media containing ABA was measured as percent of the 0 ABA control. (D) Water loss from detached leaves of wild-type and vp14 seedlings. Solid diamond, W22; solid circle, NS-2274; open square, vp14-2272; open triangle, vp14-3250.
ABA Synthesis Is Reduced in vp14 Embryos and Water-Stressed Leaves.
ABA levels were measured in mutant and wild-type embryos at 16, 18, and 20 DAP, a time frame that spans the peak in ABA-regulated gene expression in embryos (20). Table 1 shows that vp14-2274 embryos contained only 28% of the wild-type level of ABA at 16 and 18 DAP and 61% of the wild-type level at 20 DAP. This residual ABA probably accounts for the weak penetrance of the vp14-2274 phenotype. ABA accumulation in detached leaves subjected to a 5-hr water-stress treatment was 45% lower in vp14-2274 leaves than in the NS-2274 wild type (97 ± 5 ng/g fresh weight and 178 ± 12 ng/g fresh weight, respectively). No difference was detected in the ABA content of nonstressed control leaves of mutants and wild types (10 ± 2 ng/g fresh weight and 8 ± 2 ng/g fresh weight, respectively).
Table 1.
ABA levels are reduced in vp14-2274 embryos
| DAP | ABA ng/g fresh weight (%)
|
|
|---|---|---|
| NS-2274 | Vp14-2274 | |
| 16 | 83.3 (100) | 23.3 (27.8) |
| 18 | 127.5 (100) | 36.1 (28.3) |
| 20 | 71.0 (100) | 43.7 (61.5) |
vp14 Impairs Stomatal Regulation in Seedling Leaves.
The ABA-deficient mutants of other species typically cause leaf wilting by affecting stomatal regulation in leaves. Although vp14 plants did not wilt under field conditions, detached leaves of greenhouse-grown mutant seedlings exhibited markedly higher rates of water loss than wild types under ambient conditions (Fig. 1D). This difference was detected within 5 min following detachment, indicating that either stomatal aperture was altered in mutant seedlings prior to detachment or that a very rapid stress response was blocked in the mutant.
Analysis of ABA Pathway Intermediates in vp14 Embryos.
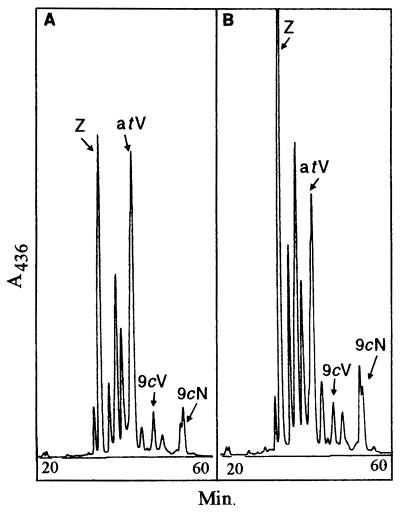
In the proposed ABA biosynthetic pathway, 9-cis-violaxanthin and 9′-cis-neoxanthin are precursors of xanthoxin that is subsequently converted to ABA. Fig. 2 shows HPLC chromatograms of carotenoids extracted from mutant and wild-type embryos at 18 DAP. The relative levels of all-trans-violaxanthin, 9-cis-violaxanthin, and 9′-cis-neoxanthin intermediates were similar in vp14 mutant and wild-type embryos. A relatively low amount of zeaxanthin in the mutant is consistent with the occurrence of precocious leaf development in the viviparous embryos.
Figure 2.
HPLC analysis of carotenoids of vp14 mutant (A) and wild-type (B) 18-day-old embryos. 9cN, 9′-cis-neoxanthin; 9cV, 9-cis-violaxanthin; atV, all-trans-violaxanthin; Z, zeaxanthin. Absorbance was measured at 436 nm.
To assay steps of the pathway downstream of the carotenoid cleavage reaction, the conversion of xanthoxin to ABA was measured in cell-free extracts of mutant and wild-type embryos. As shown in Table 2, the specific activity of xanthoxin conversion to ABA is significantly higher in vp14-2274 than in wild-type embryos. The lack of effects on biochemical steps upstream and downstream of xanthoxin synthesis is consistent with a block at the cleavage step in the vp14 mutant.
Table 2.
Xanthoxin conversion to ABA in vp14-2274 embryos
| Embryos, 20 DAP | Protein, μg | ABA, ng |
|---|---|---|
| NS-2274 | 100 | 18.9 ± 2.1 |
| vp14-2274 | 100 | 35.0 ± 1.4 |
Molecular Analysis of vp14.
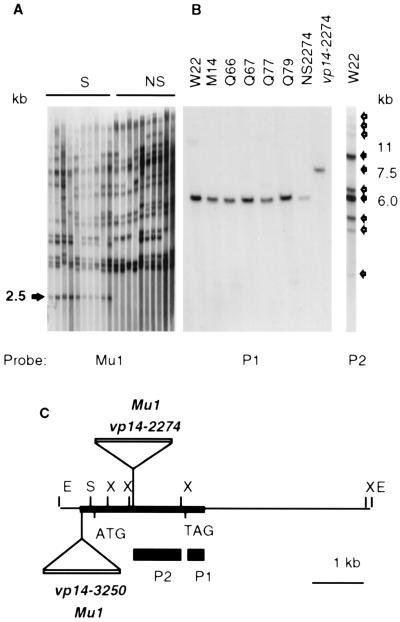
To clone the vp14 gene, a Mu1 transposable element that cosegregated with the vp14-2274 mutant was identified by Southern blot analysis of DNA extracted from plants of a segregating population (Fig. 3A). A cosegregating 2.5-kb XhoI fragment was cloned in λgt10. From this clone, a Mu1 flanking genomic DNA sequence (1 kb) was amplified by PCR and subcloned to obtain a vp14 specific probe that was used subsequently to isolate overlapping wild-type genomic clones and nearly full-length cDNA clones from an embryo cDNA library. The Southern blot shown in Fig. 3B confirmed that the Mu1 insertion in vp14-2274 was not present in any of six inbred strains that founded the Robertson’s Mutator population. To confirm that the cloned sequence was from Vp14, we analyzed the independent Mutator-induced vp14-3250 allele. A 7.5-kb EcoRI genomic DNA fragment containing the vp14-3250 allele was cloned and shown by sequencing to contain a Mu1 insertion 1 kbp upstream of the vp14-2274 Mu1 insertion (Fig. 3C).
Figure 3.
(A) Southern blot analysis of a segregating vp14-2274 family. Lanes contained 20 μg DNA pooled from six seedlings of each line probed with a 1-kbp internal TthIII fragment of Mu1 (27). The arrow indicates a 2.5-kb fragment that cosegregated with vp14. (B) DNA of vp14-2274 mutant and wild-type progenitor lines was hybridized with a vp14 specific probe (P1 indicated in C). On the right, W22 DNA was probed at low stringency with P2, a conserved-sequence region. The solid arrows indicate the 6-kb Vp14 band (on chromosome 1L) and a strongly hybridized 11-kb sequence. Open arrows denote bands detected at low stringency. (C) A physical map of vp14 showing the Mu1 insertions in each mutant. The thick line indicates the single exon. The P1 and P2 probes are indicated as solid rectangles. E, EcoRI; X, XhoI.
RFLP mapping of vp14 probes in recombinant inbred populations (Maize Genome Database, University of Missouri, Columbia, MO) placed vp14 (uFg4a) near the bz2 locus on chromosome 1L. On low-stringency Southern blots, the vp14 cDNA detected at least eight bands in inbred W22 in addition to the 6-kbp Vp14 band (Fig. 3B).
Structure of the Vp14 Gene and Its Relationship with Cleavage Dioxygenases of Bacteria.
Ten nearly full-length cDNA clones were isolated from an 18-DAP W22 embryo cDNA library screened with P2. Comparison of the cDNA and genomic DNA sequences (GenBank accession U95953) detected no introns in Vp14. A putative TATA box was located in the genomic sequence 80 bp upstream of the longest cDNA (data not shown). The cDNA contained a 604-aa ORF that would encode a 65.5-kD protein.
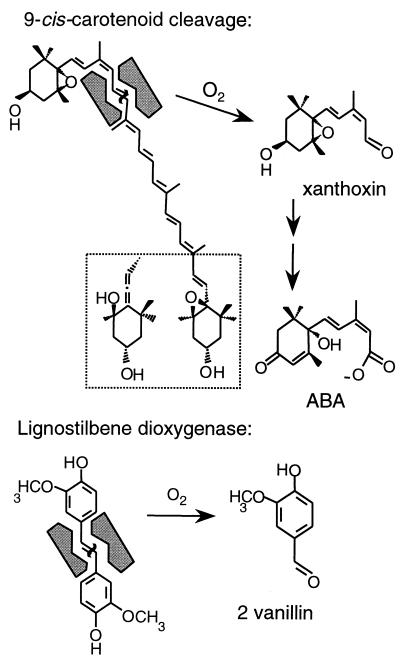
Searches of protein sequence databases detected similarity to lignostilbene dioxygenase (LSD) of Pseudomonas paucimobilis (13), human RPE65 (21), and two sequences in the complete Synechocystis cyanobacterium genome (22). Fig. 4 shows that the protein sequences align closely at the C terminus. Blocks of similarity are clustered around four conserved histidines. In dioxygenases of known structure, conserved histidines are typical ligands of a non-heme iron cofactor and LSD requires non-heme iron (13). LSD catalyzes oxidative cleavage of the central double bond of lignostilbene to yield two molecules of the corresponding aldehyde (vanillin) as illustrated in Fig. 5. This reaction is chemically very similar to the proposed carotenoid cleavage reaction in ABA biosynthesis. The VP14 sequence includes a 100-aa N-terminal extension relative to LSD and RPE65. The N-terminal sequence is consistent with the properties of a chloroplast transit peptide (23) and the evidence that the cleavage reaction occurs in plastids (3).
Figure 4.
Alignment of VP14 with related proteins: LSD, lignostilbene dioxygenase (S65040); RPE65 (HSRPE65S); SYN1 (D90914); and SYN2 (D90914, ORF). Four conserved histidine residues are marked with asterisks. A putative chloroplast-targeting transit peptide located in the N-terminal extension of VP14 is underlined.
Figure 5.
Reactions catalyzed by VP14 and lignostilbene dioxygenase (16). The cleaved bonds are indicated by waved lines.
Vp14 Is Developmentally and Environmentally Regulated.
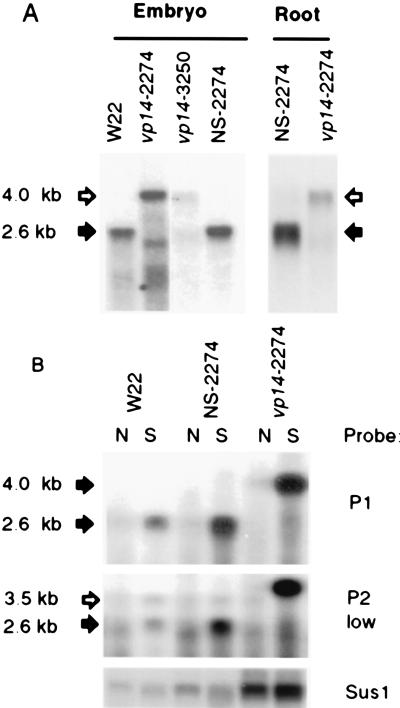
Fig. 6A shows that a 2.6-kb Vp14 transcript is expressed in developing wild-type embryos, whereas size-altered transcripts were detected in vp14-2274 embryos and roots. mRNA levels were greatly reduced in vp14-3250 embryos. The size of the major mutant transcripts is consistent with transcriptional readthrough of the Mu1 insertions. The Northern blot in Fig. 6B probed with the P1 fragment shows that normal seedling leaves contained very low levels of Vp14 transcript and that a water stress treatment (see Materials and Methods) caused accumulation of the Vp14 transcript in detached leaves of seedlings. The induction of the 4.1-kb mutant transcript in the vp14-2274 plants confirmed that Vp14 is regulated by water stress in leaves. At a lower stringency the conserved region of Vp14 (P2) detected related transcripts in nonstressed and stressed leaves of wild-type plants.
Figure 6.
(A) Northern blot analysis of embryo, root, and leaf RNA [1 μg poly(A)-enriched RNA with the P2 probe] was loaded per lane. Arrows indicate the wild-type (solid) and vp14 mutant (open) transcripts. (B) Water-stress induction of vp14 mRNA in detached leaves. Leaves were subjected to water stress as described in the text. Northern blots were probed with the gene-specific P1 probe or P2 at lowered stringency. Each lane was loaded with 1 μg poly(A) RNA. Identical blots were probed with the sus1 (sucrose synthase) gene (28) as a control for poly(A)-RNA loading.
DISCUSSION
Our results show that the vp14 mutant partially blocks ABA synthesis in the developing seed and in water-stressed leaves, suggesting a role for the Vp14 gene in the control of ABA synthesis in maize. The biochemical and molecular analyses of the Vp14 gene support the conclusion that it encodes a dioxygenase responsible for oxidative cleavage of 9-cis-xanthophylls to xanthoxin. Fig. 6B clearly demonstrates induction of the Vp14 mRNA by water stress, so there is environmental control. This result strongly supports the hypothesis that the cleavage reaction is a key regulatory step in the ABA biosynthetic pathway (3).
Evidence that VP14 Is the Cleavage Dioxygenase.
Although the cleavage reaction has not been successfully assayed in plant extracts, deficiencies at other steps in the pathway upstream and downstream of that step appear to be ruled out by our data. The pools of 9-cis-epoxy carotenoid precursors of xanthoxin are not deficient in the mutant, and the enzymes required for conversion of xanthoxin to ABA are fully active in mutant embryo extracts. Although it is possible that only specific cell types in the embryo are affected in these steps, the biochemical phenotype of vp14 clearly differs from the Arabidopsis thaliana and Nicotiana plumbaginifolia mutants that are blocked in the other steps of the pathway (4, 5, 9). Analysis of purified recombinant VP14 has confirmed that it catalyzes oxidative cleavage of 9-cis-epoxy carotenoids to xanthoxin in vitro (14). The all-trans isomers are not substrates of VP14; therefore, the mechanism and possible regulation of the nine-double-bond isomerization reaction remains to be determined. Thus far, no mutants affecting isomerization of carotenoids have been identified in plants.
Our results indicate that the bacterial LSD and VP14 define a new class of dioxygenases that catalyze similar double-bond cleavage reactions. Other related proteins of unknown function are detected in mammals, cyanobacteria, and plants. Organisms within these groups synthesize a variety of physiologically active apo-carotenoids, including retinoids, ionones, and ABA. The similarity of VP14 to human RPE65 is intriguing because the retinal pigment epithelium is very active in retinoid metabolism (21). Elucidation of the mechanism of specific carotenoid cleavage in plants clearly may lead to a better understanding of related reactions in animals.
Overlapping Sources of ABA Synthesis in Plant Development.
There are at least three possible explanations for the residual ABA in developing vp14 seeds: (i) the existing alleles may be leaky, (ii) genes with overlapping function may be expressed at low levels in the seed, and (iii) ABA may be transferred from maternal tissues that express functionally related genes. Leaky expression of a functional protein from the vp14-2274 allele would likely require splicing of the Mu1 insertion from the mRNA. We detect little evidence of correctly spliced mutant transcripts on Northern blots. Suppression of the vp14 phenotype by ABA transferred from maternal tissues might explain the variable penetrance of the viviparous phenotype, because stress-induced ABA synthesis in vegetative tissues is only weakly affected by the mutant. In any case, Vp14 probably accounts for only a subset of the ABA biosynthetic activity in the plant.
Our results imply that other Vp14-related genes are expressed in vegetative tissues of the plant. We detect evidence of related genes on moderate-stringency Southern and Northern blots (Figs. 3 and 6) and have isolated several distinct, but related cDNAs from roots and leaves (B.C.T. and D.R.M., unpublished results). Other related sequences are found in the rice, maize, and Arabidopsis expressed sequence tag collections. We suggest that a family of differentially regulated Vp14-like genes contribute to environmental and developmental control of ABA biosynthesis in plants. Gene families are an important mechanism underlying developmental control of biochemical pathways in plants. Notably, the ACC synthase genes that encode the key regulatory step in ethylene hormone biosynthesis also comprise a differentially regulated gene family (24).
Vp14 Functions in Root and Leaf ABA Synthesis.
The vp14 mutant evidently targets a pool of ABA involved in stomatal regulation in seedling leaves. Previous studies have established that stress-induced changes in bulk ABA synthesis occur too slowly to account for rapid stomatal responses in leaves and that redistribution of preexisting ABA pools can account for stomatal closure (25). It is therefore unlikely that the partial inhibition of stress-induced ABA synthesis in vp14 mutant leaves would account for the impaired stomatal regulation detected within the first 5 min after leaf excision. Moreover, we did not detect any differences in the ABA contents of nonstressed mutant and wild-type leaves that would reflect preexisting variations in ABA pools in the leaf.
For this reason, the properties of vp14 place strong constraints on the possible size and source of the ABA pool responsible for stomatal regulation in response to leaf excision. Given the experimental error of the ABA measurements, the affected pool must comprise less than 20% of the basal ABA content of turgid leaves. Critical sources of ABA may include localized or very rapid synthesis of ABA in cells near the stomatal complex (25) and/or ABA transported from the roots in the transpiration stream (26). Localized or low-level constitutive expression of Vp14 mRNA in leaf tissues is consistent with our Northern blot analysis of nonstressed leaves. On the other hand, that Vp14 is expressed in roots is consistent with the root-source hypothesis. The localization of Vp14 expression at the cellular level will provide important clues to the source and regulation of ABA synthesis in leaves.
Acknowledgments
This work was supported by grants from the National Science Foundation and Department of Energy awarded to D.R.M. and J.A.D.Z. and by the Florida Agricultural Experiment Station (journal series no. R-05960).
ABBREVIATIONS
- ABA
abscisic acid
- DAP
days postpollination
- LSD
lignostilbene dioxygenases
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U95953).
References
- 1.McCarty D R. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:71–93. [Google Scholar]
- 2.Giraudat J. Curr Opin Cell Biol. 1995;7:232–240. doi: 10.1016/0955-0674(95)80033-6. [DOI] [PubMed] [Google Scholar]
- 3.Zeevaart J A D, Creelman R A. Plant Mol Biol. 1988;39:439–473. [Google Scholar]
- 4.Rock C D, Zeevaart J A D. Proc Natl Acad Sci USA. 1991;88:7496–7499. doi: 10.1073/pnas.88.17.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marin E, Nussaume L, Quesada A, Gonneau M, Sotta B, Hugueney P, Frey A, Marion-Poll A. EMBO J. 1996;15:2331–2342. [PMC free article] [PubMed] [Google Scholar]
- 6.Sindhu R K, Walton D C. Plant Physiol. 1987;85:916–921. doi: 10.1104/pp.85.4.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Norman S M, Maier V P, Pon D L. J Agric Food Chem. 1990;38:1326–1334. [Google Scholar]
- 8.Léon-Kloosteriel K M, Gil M A, Ruijs G J, Jacobsen S E, Olszewski N E, Schwartz S H, Zeevaart J A D, Koornneef M. Plant J. 1996;10:655–661. doi: 10.1046/j.1365-313x.1996.10040655.x. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz S H, Leon-Kloosterziel K M, Koornneef M, Zeevaart J A D. Plant Physiol. 1997;114:161–166. doi: 10.1104/pp.114.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robichaud C, Sussex I M. J Plant Physiol. 1986;126:235–242. [Google Scholar]
- 11.Duckham S C, Taylor I B, Linforth R S T, Al-Naieb R J, Marples B A, Bowan W R. Plant Cell Environ. 1991;14:601–606. [Google Scholar]
- 12.Parry A D, Horgan R. Planta. 1992;187:192–197. doi: 10.1007/BF00201937. [DOI] [PubMed] [Google Scholar]
- 13.Kamoda S, Saburi Y. Biosci Biotech Biochem. 1993;57:926–930. doi: 10.1271/bbb.57.926. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz S H, Tan B-C, Gage D A, Zeevaart J A D, McCarty D R. Science. 1997;276:1872–1874. doi: 10.1126/science.276.5320.1872. [DOI] [PubMed] [Google Scholar]
- 15.Dellaporta S. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- 16.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Birchler J A. In: Maize Handbook. Freeling M, Walbot V, editors. New York: Springer; 1994. pp. 330–331. [Google Scholar]
- 19.Smith J D, Neuffer G. Maize Genetics Cooperation News Letter. 1992;66:341. [Google Scholar]
- 20.McCarty D R, Hattori T, Carson C B, Vasil V, Vasil I K. Cell. 1991;66:895–905. doi: 10.1016/0092-8674(91)90436-3. [DOI] [PubMed] [Google Scholar]
- 21.Hamel C P, Tsilou E, Pfeffer B A, Hooks J J, Detrick B, Redmond T M. J Biol Chem. 1993;268:15751–15757. [PubMed] [Google Scholar]
- 22.Kaneko T, Sato H, Kotani A, Tanaka E, Agamizu E, et al. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 23.Archer E K, Keegstra K. J Bioenerg Biomembr. 1990;22:789–810. doi: 10.1007/BF00786931. [DOI] [PubMed] [Google Scholar]
- 24.Liang X, Abel S, Keller J A, Shen N F, Theologis A. Proc Natl Acad Sci USA. 1992;89:11046–11050. doi: 10.1073/pnas.89.22.11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cornish K, Zeevaart J A D. Plant Physiol. 1985;78:623–626. doi: 10.1104/pp.78.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Davies W J. J Exp Bot. 1987;38:2015–23. [Google Scholar]
- 27.Barker R F, Thompson D V, Talbot D R, Swanson J, Bennetzen J L. Nucleic Acids Res. 1984;12:5955–5967. doi: 10.1093/nar/12.15.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarty D R, Shaw J, Hannah L C. Proc Natl Acad Sci USA. 1986;83:9000–9103. doi: 10.1073/pnas.83.23.9099. [DOI] [PMC free article] [PubMed] [Google Scholar]