Abstract
The INK4A/ARF tumor suppressor locus is frequently inactivated in hepatocellular carcinoma (HCC), yet the consequences of this remain unknown. We recently described a HCC mouse model in which loss of the Ink4a/Arf locus accelerates the development of metastasis and enhances tumor cell migration and invasion in cell culture assays. We show here that knockdown of p19Arf in a HCC cell line increases invasion in cell culture assays. Further, reintroduction of p19Arf into HCC cell lines lacking Ink4a/Arf inhibits tumor cell invasion, without affecting cell proliferation, or cell transformation as measured by soft agar colony formation. Inhibition of cell invasion by p19Arf was dependent on its C-terminal binding protein (CtBP) interaction domain, but independent of Mdm2 binding and nucleolar localization. Indeed, RNAi-mediated knockdown of CtBP1 or CtBP2 decreased cell invasion, and ectopic expression of CtBP2 enhanced tumor cell migration and invasion. Thus, our data indicate a novel role for the Arf tumor suppressor protein in regulating phenotypes associated with tumor progression and metastasis in HCC cells.
Keywords: Hepatocellular carcinoma, invasion, Arf, CtBP
Introduction
Worldwide, 626,000 new cases of hepatocellular carcinoma (HCC) are diagnosed each year, with a survival rate of less than 5%, and an average survival of less than one year after diagnosis (1). To date, surgery remains the only effective approach for treatment of small HCCs; unfortunately, there is currently no reliable therapy for most patients with advanced or metastatic HCC that has spread to the lymph nodes, portal vein, or lungs (2).
Epidemiologic and molecular studies have determined that HCC development is associated with chronic hepatitis B virus (HBV) and hepatitis C virus (HCV) infection, alcoholic cirrhosis, metabolic disorders, and environmental factors such as aflatoxin B1 exposure (3). Furthermore, genetic alterations resulting in the activation of the c-myc oncogene; inactivation of tumor suppressor genes such as TP53, RB1, INK4A/ARF; and alterations that stimulate the Wnt signaling pathway are particularly important in hepatocarcinogenesis (4). However, little is known regarding the genes and molecular pathways that influence metastasis. Recent in vivo models and gene expression analyses have begun to shed some light on the tumor and host factors regulating metastasis in HCC (5–10).
To elucidate the mechanisms of hepatocarcinogenesis, several mouse models have been generated through the expression of oncogenes or viral genes, the inactivation of tumor suppressor genes, and chemical carcinogenesis. Given the impact of metastasis on the clinical management of HCC patients, models that allow the dissection of metastasis are of great importance. We have recently described a novel HCC mouse model induced by the somatic and sporadic activation of oncogenes specifically in the liver (8). We have further refined this model to include liver specific deletion of the Trp53 and Ink4a/Arf tumor suppressor loci (Y-W.C. et al, submitted). Our recent data demonstrated that liver-specific Trp53 deletion induces lung metastases, the formation of which can be accelerated by concomitant deletion of Ink4a/Arf. Furthermore, we showed that mouse HCC cell lines lacking both Trp53 and Ink4a/Arf displayed enhanced migration and invasion capabilities, suggesting that the Ink4a/Arf locus may play a role in regulating these processes in liver cancer cells.
The Ink4a/Arf locus encodes two distinct tumor suppressors, the cyclin dependent kinase (Cdk) inhibitor p16Ink4a and a protein translated from an alternative reading frame, Arf, (p14ARF in human and p19Arf in mouse) (11). Through the inhibition of cyclin D/cdk4, p16 maintains pRb in the hypophosphorylated state and regulates the G1 transition of the cell cycle (12). On the other hand, p19Arf inhibits the p53-ubiquitinating function of Mdm2, thereby stabilizing the p53 transcription factor, which regulates genes mediating G1 cell cycle arrest and apoptosis (13). Arf also has p53-independent functions. It mediates a p53-independent cell cycle arrest by targeting both E2F1 and c-Myc to the nucleolus and preventing their transcription activation functions, and also induces p53-independent apoptosis by targeting the transcription factor CtBP2 for degradation (14–16). CtBP transcription factors have been previously shown to induce an epithelial to mesenchymal transition (EMT), and to stimulate cell migration (17–19). Thus, regulation of CtBP activity by Arf may represent an important tumor suppressor function. Significantly, the INK4A/ARF locus is frequently inactivated in HCC, yet its role in the pathogenesis of this disease remains unclear (20–22).
Guided by these findings in HCC and our mouse model, we sought to identify mechanisms by which Arf may regulate HCC metastasis. Using mouse HCC cell lines, we found that RNAi-mediated loss of Arf enhances cell invasion, and conversely, reintroduction of Arf into cell lines deficient for both Trp53 and Ink4a/Arf inhibits tumor cell invasion. The ability of Arf to inhibit invasion is independent of its interaction with Mdm2 and nucleolar localization. However, it is dependent on Arf’s ability to bind to CtBP. Consistent with this, RNAi-mediated depletion of CtBP1 or CtBP2 reduces cell invasion, and ectopic CtBP2 expression enhances tumor cell migration and invasion. Thus, our findings demonstrate a new role for the Arf tumor suppressor in HCC progression, one that may be applicable to other tumor types.
Material and Methods
Cell lines
Immediately after harvest, tumor tissue was minced and dissociated by pipetting in DMEM. Cells were washed once in sterile PBS and plated in DMEM supplemented with 10% fetal bovine serum (FBS) and antibiotics. After 48 hours, non-adherent cells were removed, adherent cells washed with PBS and refed with fresh medium containing FBS.
To generate cells with knockdown of p19Arf, BL185 HCC cells were infected with MLP retroviral vectors encoding an shRNA targeted to Arf-specific exon 1β of the Ink4a/Arf locus (23). Infected cells were then placed in DMEM containing 8μg/ml puromycin (EMD Biosciences). Resistant cells were then analyzed by immunoblot.
To generate cell lines expressing either wild-type or mutant p19Arf proteins, HCC cell lines were infected with pBabe-puro retroviruses and then placed in DMEM containing 8μg/ml puromycin (EMD Biosciences). Resistant cells were then analyzed by immunoblot and immunofluoresence.
Cells with ectopic CtBP2 expression were generated by transfecting MM189 cells with pcDNA3-V5-CtBP2 or pcDNA3-V5-CtBP2, and selection of transfected cells in 4 mg/ml G418 (EMD Biosciences).
RNAi-mediated depletion of CtBP1 and CtBP2 was achieved by infecting HCC cells with pLKO-based lentiviruses encoding shRNAs targeting the appropriate mRNA. Infected cells were selected in 8μg/ml puromycin. To generate knockdown in HCC cells expressing the p19ArfL46D mutant that were already puromycin resistant, infection with shRNA containing lentiviruses was performed twice within 24 hours. Knockdown was confirmed by immunoblot.
Immunoblotting
Cells were collected and lysed in RIPA buffer containing protease inhibitors. Protein concentration was determined with BCA assay kit (Pierce), and equal amounts of protein were loaded per lane of a polyacrylamide gel. After protein transfer, nitrocellulose membranes were blocked with 5% dry milk in tris-buffered saline, 0.1% Tween 20 (TBS-T) for 1 hour. Primary antibodies were incubated overnight at 4°C in 5% bovine serum albumin (BSA, Sigma) in TBS-T, and secondary antibodies for 1 hour at room temperature. Chemiluminescence was performed with Supersignal reagent (Pierce). Primary antibodies: anti-p19Arf (Abcam), anti-CtBP2 (Upstate Biotechnology), anti β-actin (1:5000, Santa Crutz), α-catenin (1:1000, Becton Dickinson (BD)), anti β-catenin (1:1000, BD), anti γ-catenin (1:1000, BD), anti E-cadherin (1:1000, BD), anti α smooth actin clone A14 (1:1000, Sigma), anti N-cadherin (1:1000, BD), anti fibronectin (1:1000, Santa Cruz), anti vimentin (1:1000, Lab Vision), and anti-V5 (1:2000, Covance).
Cell proliferation assay
103 cells were seeded in quadruplicate onto collagen coated 96-well plates and incubated at 37°C under 5% CO2. After 24 hours, viable cell numbers were measured in quadruplicate every period for 4 days using CellTiter 96 Aqueous One Solution Cell proliferation assay (Promega) according to the manufacturer’s instructions. The proliferation curves were constructed by calculating the mean value of optical density measurement at 490 nm using a 96-well plate reader.
Soft agar assay
Soft agar assays were performed as previously described (24). The number of colonies larger than 100 μm in diameter present within 20 microscopic fields was counted under a light microscope.
Migration and invasion assays
2.5 × 104 cells in 0.5 ml of serum-free DMEM were plated into either control or matrigel-coated invasion inserts (BD). Inserts were then placed in wells with 0.75 ml of DMEM containing 10% FBS as a chemoattractant. After culture for 20–24 hours at 37°C, cells were fixed with methanol for 8 minutes at room temperature and stained with Giemsa reagent diluted 5X in H2O. Cells on the upper sides of the inserts were removed with a cotton swab, and the insert membranes removed and mounted on glass slides. Cell numbers for migration and invasion were then determined by counting the number of cells present in 10 microscope fields at 50X magnification per insert. The percent invasion was calculated as the number of invading cells divided by the number of migrating cells. The percent invasion was then normalized to get the invasion index, with the value for the control cell population set to 1. All experiments were performed in duplicate and repeated a minimum of 3 times. Data are shown for representative experiments.
Immunofluorescence
Cells on collagen coated culture slides were fixed with 4% paraformaldehyde for 10 min and permeabilized with 0.1% Triton X-100 in PBS for 15 min at room temperature. After blocking with 10% goat serum in PBS for 1 hr, cells were incubated with primary anti-p19Arf antibody (1:200, Abcam) at 4°C overnight, followed by incubation with appropriate secondary antibodies and Rhodamine-Phalloidin at room temperature. Slides were mounted with antifade and viewed under a fluorescence microscope.
Results
p19Arf inhibits cell invasion
We have previously shown that mouse HCC cell lines lacking both Trp53 and Ink4a/Arf have enhanced migration and invasion activity in Boyden chamber assays compared to a cell line that lacks Trp53 alone (25). To determine whether loss of p19Arf influences tumor cell migration and invasion, we infected the BL185 HCC cell line which lacks Trp53 but retains functional Ink4a/Arf, with retroviral vectors encoding shRNA against p19Arf, or luciferase as a control (23). Knockdown of p19Arf was confirmed by immunoblot (Figure 1A). We then measured the migration and invasion activities of these cells using Boyden chamber assays. We found that depletion of Arf resulted in a 1.5-fold increase in the number of invading cells (Figure 1C), a result confirmed in independent infections (shRNA 1 and 2, Figure 1C), and multiple replications of the invasion assays. These data indicate that acute loss of Arf stimulates the in vitro invasion activity of Trp53 null HCC cells, albeit to a lesser extent than that observed in HCC cells with genetic ablation of Trp53 and Ink4a/Arf (data not shown).
Figure 1.
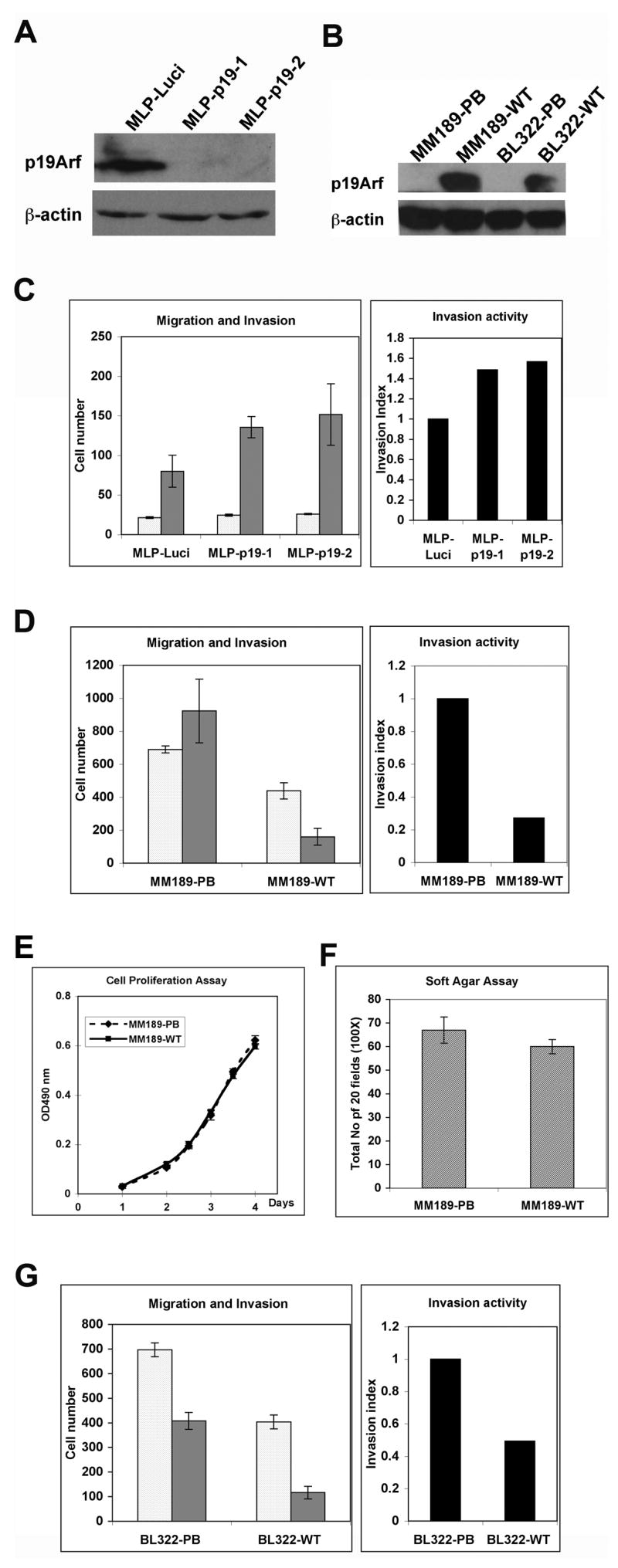
p19Arf inhibits tumor cell invasion. (A) Immunoblot analysis of BL185 HCC cells infected with retroviral vectors targeting luciferase or p19Arf. β-actin serves as a control. (B) Immunoblot detection of p19Arf in HCC cell lines infected with either pBabe puro retrovirus (PB) or pBabe puro retrovirus encoding p19Arf (WT). β-actin serves as a control. (C) Migration (clear bars) and invasion (grey bars) activity of BL185-MLP-Luci and BL185-MLP-p19 cells. Data are from a representative experiment performed in triplicate. Error bars represent standard error of the mean (S.E.M.). The cell number is the number of migrated or invaded cells in five 50X microscopic fields per insert. The invasion index was calculated as described in the methods. (D) Migration (clear bars) and invasion (grey bars) activity of MM189-PB and MM189-WT cells. Data are from a representative experiment performed in duplicate. Error bars represent S.E.M. (E) Cell proliferation assay for MM189-PB and MM189-WT cells. (F) Anchorage-independent growth assay for MM189-PB and MM189-WT cells. The colony number is the total number for twenty 100 X microscopic fields in every plate. Data are from a representative experiment performed in duplicate. Error bars are S.E.M. (G) Migration and invasion assay for BL322-PB and BL322-WT cells. Error bars are S.E.M.
To ascertain whether loss of p19Arf was required for the invasive phenotype of HCC cells lacking Trp53 and Ink4a/Arf, double null HCC cell lines, MM189 and BL322, were infected with a retroviral vector encoding mouse p19Arf, or empty vector as a control, and expression confirmed by immunoblot (Figure 1B). By immunohistochemistry with an anti-p19Arf antibody, positive signals were detected in nearly 70% of retrovirally infected MM189 cells, suggesting that p19Arf was re-expressed in most, but not all, cells (data not shown). Measurement of cell migration and invasion in Boyden chamber assays demonstrated that p19Arf expression reduced the invasion activity of MM189 cells by 80% (Figure 1D). p19Arf expression also led to a more modest decrease in cell migration in most, but not all, experiments (Figure 1D and data not shown). Consistent with this finding, p19Arf expression did not significantly impair cell migration in a wound healing assay (data not shown). Thus, p19Arf specifically inhibits the invasion of HCC cells.
Analysis of vector controls and p19Arf-expressing MM189 cells demonstrated that p19Arf expression did not affect the proliferation of these cells (Figure 1E). Likewise, analysis of cell transformation by soft agar colony formation demonstrated no effect of p19Arf expression (Figure 1F and Supplementary Figure 1). Thus, in MM189 cells, p19Arf appears to regulate cell invasion without affecting any other features related to cell transformation. To confirm that this phenomenon was not restricted to a single HCC cell line, we performed the migration and invasion assays on BL322 HCC cells infected with either p19Arf-expresing retrovirus or vector controls. We found a similar effect on migration and invasion in this cell line suggesting that this phenotype is not cell line specific (Figure 1G).
Tumor cell migration and invasion are often associated with reorganization of the actin cytoskeleton and a phenomenon called the epithelial to mesenchymal transition (EMT) (26, 27). To investigate whether p19Arf-mediated inhibition of cell invasion occurs via the reversal of an EMT, we analyzed p19Arf expressing cells (MM189-WT) and vector controls (MM189-PB) by morphology and expression of epithelial and mesenchymal markers. Immunoblot experiments demonstrated that MM189 cells expressed epithelial proteins, such as α-catenin, β-catenin, γ-catenin and E-cadherin, and the expression of p19Arf did not enhance the levels of these proteins (Figure 2A). Examination of E-cadherin localization by immunofluorescent staining did not reveal any significant differences between MM189-PB and MM189-WT cells (data not shown). Consistent with these findings, MM189 cells did not express mesenchymal markers such as vimentin, a feature unaffected by p19Arf expression (Figure 2A). Analysis of mRNA levels for epithelial and mesenchymal markers demonstrated that these were similarly unaffected by p19Arf expression, although interestingly, the HCC cells expressed vimentin mRNA indicating that regulation occurred at the post-transcriptional level (Supplementary Figure 2). In agreement with the above data, analysis of the morphology of MM189-PB cells and MM189-WT cells showed that the expression of p19Arf did not alter cell morphology (Figure 2B). Finally, analysis of the actin cytoskeleton by phalloidin staining did not demonstrate any differences between p19Arf-expressing cells and vector controls (Figure 2C).
Figure 2.
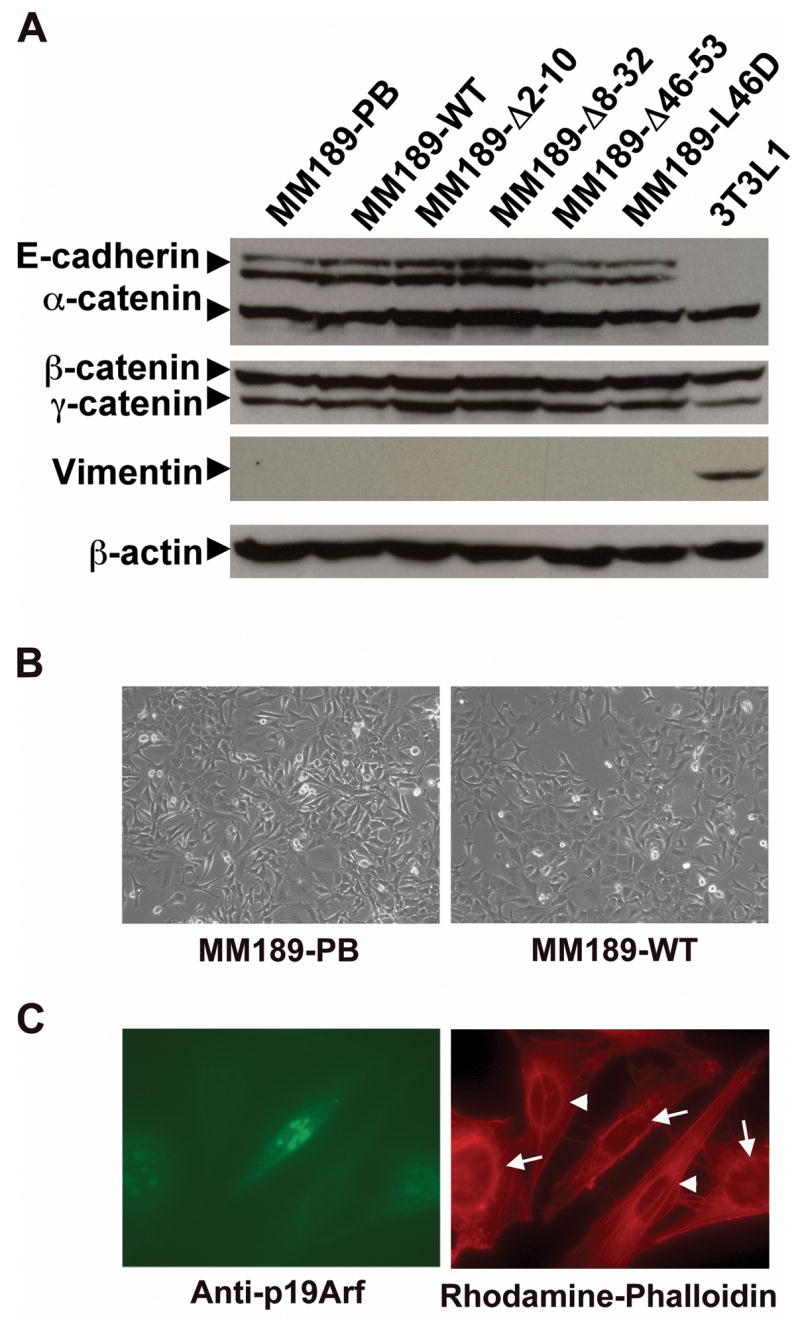
p19Arf does not influence cell morphology or EMT. (A) Immunoblot analysis of epithelial and mesenchymal proteins in MM189 cells expressing either wild-type p19Arf or the indicated p19Arf mutant proteins. 3T3L1 serves as a positive control for expression of the mesenchymal marker vimentin. β-actin serves as a loading control. (B) Phase contrast image of MM189-PB and MM189-WT cells grown for 3 days on a collagen matrix. (C) Immunofluorescent images of MM189 cells expressing p19Arf and vector controls. Left panel: p19Arf positive cells (arrows) are labeled with FITC-conjugated antibodies, while p19Arf negative cells (arrowheads) are not. Right panel: Rhodamine-conjugated phalloidin labels the actin cytoskeleton.
Inhibition of cell invasion by p19Arf requires the CtBP interaction domain
To determine the functional domains of p19Arf that are required for inhibition of cell invasion, we introduced retroviral vectors encoding various p19Arf mutant proteins into MM189 cells. The introduced mutants were Δ2–10 that impairs Mdm2 binding; Δ8–32 that impairs Mdm2 binding and nucleolar localization; Δ46–53 that blocks CtBP binding; and the L46D point mutant that also blocks CtBP binding. The locations of these mutations are shown schematically in Figure 3A. The amino terminal deletions also affect Arf’s interaction with c-Myc, E2F1 and Foxm1b. We confirmed the expression of the p19Arf mutants by immunoblotting with Arf specific antibodies (Figure 3B), and demonstrated that the majority of cells expressed the mutant p19Arf proteins by immunostaining (data not shown).
Figure 3.
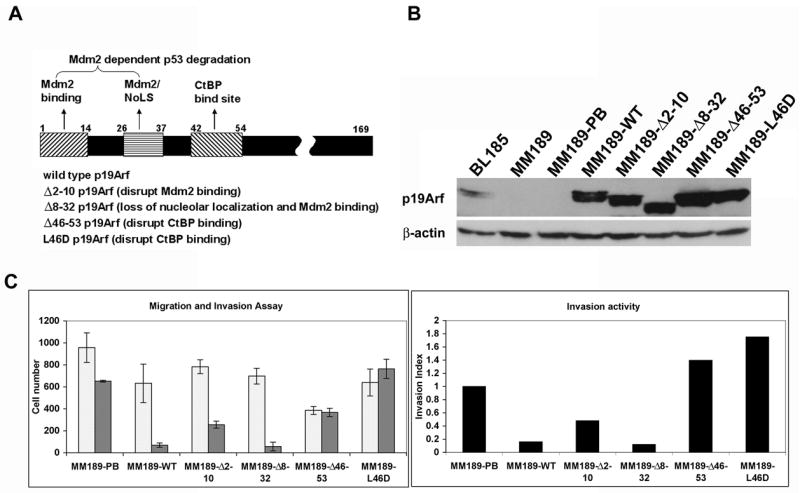
p19Arf inhibition of cell invasion is dependent on the CtBP binding domain. (A) Schematic illustration of the p19Arf mutants used in this study. (B) Immunoblot detection of expression of wild-type or mutant p19Arf proteins in MM189 HCC cells. (C) Migration (clear bars) and invasion (grey bars) activity of MM189 cells infected with retroviruses encoding the indicated p19Arf mutant proteins.
Introduction of the Δ2–10 and Δ8–32 mutants into MM189 cells led to the efficient inhibition of cell invasion (Figure 3C). However, introduction of the Δ46–53 and L46D mutants failed to appreciably impair cell invasion, particularly in comparison to the number of migrating cells (Figure 3C). This resulted in an increased invasion index for cells expressing these two p19Arf mutants (Figure 3C). To rule out the possibility that some effects were due to the random integration of retroviruses, retroviral infections and invasion assays were repeated with independent retroviral stocks, several pools of infected cells with varying levels of p19Arf expression examined, and consistent results obtained. As was observed with expression of wild type p19Arf, expression of the Arf mutants did not affect proliferation of MM189 cells (data not shown). Thus, p19Arf-mediated inhibition of tumor cell invasion is independent of Mdm2 binding and nucleolar localization, but is dependent on the CtBP interaction domain. As was observed after reintroduction of wild-type p19Arf, introduction of mutant Arf proteins did not alter the expression of epithelial and mesenchymal proteins (Figure 2A). Thus, p19Arf-mediated inhibition of cell invasion does not occur via the reversal of an EMT.
CtBP regulates tumor cell invasion
Our finding that the p19Arf Δ46–53 and L46D mutants fail to impair tumor cell invasion suggested that functional inhibition of CtBP proteins by p19Arf might be critical in this process. We therefore determined whether inactivation of CtBP reduces tumor cell invasion. Lentiviral shRNA vectors targeting CtBP1 or CtBP2, or vector control, were introduced into MM189 cells, the cells selected for puromycin resistance, and their invasion activity measured. By immunoblotting, we identified two shRNAs targeting each of the CtBP family members that displayed efficient knockdown (Figure 4A). Measurement of the proliferation of CtBP knockdown cells demonstrated that loss of CtBP proteins did not affect cell proliferation (Supplementary Figure 3). Analysis of the invasion activity demonstrated that cells with CtBP knockdown had lower invasion activity relative to vector controls (Figure 4C). We next determined whether depletion of CtBP1 and CtBP2 in MM189 cells expressing the Arf L46D mutant would similarly result in reduced cell invasion. We introduced shRNAs targeting either CtBP1 or CtBP2 into these cells and confirmed knockdown by immunoblotting with anti-CtBP antibodies (Supplementary Figure 4). We found that depletion of either family member led to a reduced cell invasion index (Figure 4D). Thus, CtBP family members are required for HCC tumor cell invasion.
Figure 4.
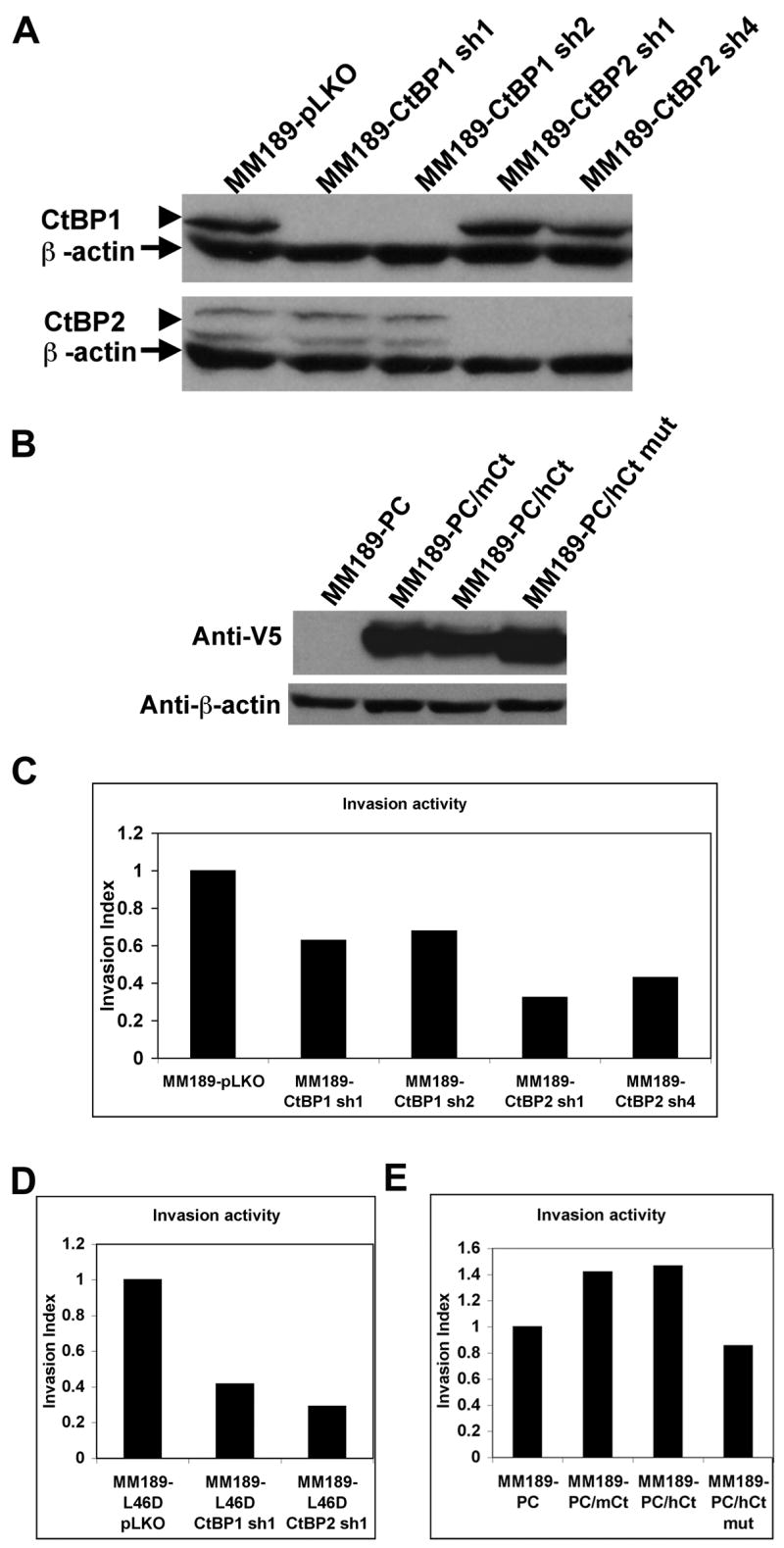
CtBPs regulate tumor cell invasion. (A) Detection of CtBP1 and CtBP2 by immunoblot after introduction of lentiviral vectors encoding shRNAs specific for either family member. Empty pLKO vector was introduced as a control. Protein bands corresponding to CtBP1 or CtBP2 are indicated by arrowheads. β-actin serves a a loading control (indicated by arrows). (B) Detection of ectopic V5 epitope-tagged CtBP2 by immunoblot with anti-V5 antibodies in MM189 cells transfected with pcDNA3-CtBP2 (PC/Ct) or empty vector control (PC). β-actin serves as a loading control. The band shown indicated by the arrow is non-specific. (C) Invasion activity, represented by invasion index (see methods) for MM189 cells infected with either control lentivirus or lentiviruses encoding shRNAs targeting either CtBP1 or CtBP2. (D) Invasion activity of MM189 cells expressing the p19Arf L46D mutant and infected with either control lentivirus or lentiviruses encoding shRNAs specific for either CtBP1 or CtBP2. (E) Invasion activity of MM189 cells transfected with pcDNA3-CtBP2 or empty vector.
To determine whether ectopic expression of CtBP could enhance cell invasion, we introduced a V5-tagged CtBP2 expression construct, or vector control, into MM189 cells. Detection of the introduced CtBP was confirmed by immunoblotting with anti-V5 antibodies (Figure 4B). Interestingly, the levels of the ectopic CtBP appeared to be less than the endogenous levels of CtBP2 as determined by immunoblotting with CtBP2-specific antibodies (data not shown). In cell invasion assays, exogenous CtBP2 expression led to a 1.5-fold increase in invasion activity relative to vector controls, supporting a role for CtBP2 in tumor cell invasion (Figure 4E). Importantly, introduction of a NADH-binding deficient CtBP2 mutant (G189A) (28) failed to stimulate cell invasion (Figure 4E) even though it was expressed at levels similar to wild-type CtBP2 (Figure 4B). Interestingly, in MM189 cells with enforced p19Arf expression, ectopic CtBP2 failed to significantly enhance tumor cell migration and invasion suggesting a dominant effect of p19Arf on the invasion phenotype (data not shown).
Discussion
The Ink4a/Arf locus is regarded as one of the most important anti-tumoral defenses in mammalian systems, and the two protein-coding genes at this locus are frequently independently inactivated in HCC. Studies of human tumors and mouse models have suggested a role for the Arf tumor suppressor in constraining tumor initiation (22, 29, 30). We have recently shown that deletion of the Ink4a/Arf locus enhances the progression of liver tumors induced by polyoma middle T antigen and Trp53 deletion, consistent with findings in a skin carcinogenesis mouse model that indicated that Arf may additionally play a role in tumor progression (31). Further, we showed that mouse HCC cell lines that lacked both Trp53 and Ink4a/Arf displayed enhanced migration and invasion capabilities in cell culture assays compared to a cell line that lacks Trp53 alone (25). We have now shown that p19Arf specifically impairs tumor cell invasion without affecting either proliferation or soft agar colony formation, pointing to a potential role for Arf in regulating tumor metastasis.
p19Arf induces cell cycle arrest in a p53-dependent manner, and relays signals to p53 after oncogene-induced stress that stimulates the onset of cellular senescence, a potent anti-tumorigenic checkpoint. However, accumulating data indicate that Arf may additionally have p53-independent tumor suppressor functions (32). Our data show that Arf-mediated inhibition of tumor cell invasion is independent of its interaction with Mdm2, suggesting that regulation of tumor invasion may be another p53-independent tumor suppressor function of Arf. Importantly, this function is also independent of Arf’s ability to bind to other interacting proteins such as E2F1, c-Myc, and Foxm1b, as deletion mutants that interfere with the ability of Arf to bind to these proteins still effectively block invasion.
Instead, our data demonstrate that the effect of p19Arf on tumor cell invasion occurs via a p53-independent mechanism involving CtBP. Mutant Arf proteins that fail to bind to CtBP do not inhibit cell invasion, and shRNA-mediated ablation of CtBP reduces invasion in HCC cells expressing CtBP-binding-deficient Arf proteins. Guo et al. have previously shown that genetic disruption of Arf in mouse embryonic fibroblasts (MEFs) enhances cell motility in a p53-dependent manner through the stimulation of Rac1 activity (33). Our data indicate that Arf expression in HCC cells does not alter cell morphology or the actin cytoskeleton, as assessed by phalloidin staining. This suggests that the activity of Rac1, or other Rho family GTPases involved in cytoskeletal remodeling, may not be affected by Arf expression, although this has not be formally tested.
Additional studies demonstrated that activated Rho GTPases could stimulate invasion by Trp53 null, but not Arf null, MEFs suggesting that p19Arf regulates cell migration but not invasion (34). Thus, our experiments are the first to show that Arf inhibits tumor cell invasion via a p53-indpendent pathway. The differences between the previous studies and ours may reflect the different cell types utilized - epithelial-derived tumor cell lines versus transformed mouse fibroblasts. Importantly, in a related study, we have also shown that p14Arf inhibits the hypoxia-induced migration of H1299 human lung carcinoma cells in a p53-independent and CtBP-dependent manner (35), and previous studies have indicated that CtBP can regulate the migration of this cell line, although cell invasion was not assayed in these studies (19). Significantly, the leucine 46 residue that is critical for the Arf-CtBP interaction is one of six invariant residues conserved across several species including human, mouse, chicken, pig and opossum (32). Thus, the Arf-CtBP interaction likely mediates a highly conserved tumor suppressor function of Arf, and may have implications for the dissemination of human tumors.
CtBPs play a critical role in cellular regulation by binding to a variety of transcriptional repressors important for development and tumorigenesis (36). CtBPs have also been suggested to play a role in inhibiting anoikis (18). However, the mechanisms by which Arf inhibits CtBP function remain unclear. Our findings suggest that Arf may impede CtBP function, yet the mechanisms by which this might occur remain unclear, although potential links are beginning to emerge. While we have observed that Arf expression does not influence the steady state levels of CtBP, Arf, but not a mutant defective for CtBP binding, stimulates the degradation of CtBP in response to cellular stresses such as UV irradiation [Supplemental Figure 2 and (16)]. Therefore, it is possible that Arf stimulates the degradation of CtBP under stress conditions relevant during cell migration and invasion, such as detachment from the basement membrane. In addition, recent findings from the Grossman lab indicate that Arf can inhibit CtBP-mediated transcriptional repression of a target promoter (R. Kovi and S.R.G., unpublished observations).
CtBP has also been previously shown to induce an epithelial to mesenchymal transition (EMT) (17, 18). EMT, first identified as a critical process during normal embryonic development, involves the downregulation of epithelial cell markers such as E-cadherin and the induction of mesenchymal markers such as vimentin, and is postulated to be involved in tumor cell invasion and metastasis (27). However, our data indicate that our HCC cell lines invade without undergoing EMT, and E-cadherin protein levels are not significantly altered in cells by reintroduction of p19Arf. A similar phenomenon has been recently described by Christofori and colleagues in connection to tumor cell invasion stimulated by the mucin-like protein podoplanin (37). EMT has been postulated to a transient process. Thus, an alternate possibility is that Arf may prevent the transient induction of CtBP, and the consequent transient reduction of E-cadherin levels, in response to pro-invasion stimuli that would allow a tumor cell to initiate invasion. A similar model has been postulated for metastasis of colorectal carcinomas (38).
Thus, our findings indicate a potential new role for Arf in tumor invasion. Further exploration of the pathways involved in this process may yield promising new targets for therapeutic intervention.
Supplementary Material
Acknowledgments
The authors thank Nabeel Bardeesy for mutant p19Arf retroviral constructs, Scott Lowe for the p19Arf-specific shRNA construct, and members of the Lewis and Grossman labs for thoughtful discussion. B.C.L. is the recipient of a Career Development Award in the Biomedical Sciences from the Burroughs Wellcome Fund, and is a Liver Scholar of the American Liver Foundation. Supported by NIH grants CA121171 (B.C.L.) and CA89548 (S.R.G.), and a grant from Our Danny Fund (S.P.).
Financial Support: Career Development Award in the Biomedical Sciences from the Burroughs Wellcome Fund (B.C.L.); Liver Scholar Award from the American Liver Foundation (B.C.L.); NIH grants CA121171 (B.C.L.) and CA89548 (S.R.G.), and Our Danny Fund (S.P.).
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Song TJ, Ip EW, Fong Y. Hepatocellular carcinoma: current surgical management. Gastroenterology. 2004;127:S248–60. doi: 10.1053/j.gastro.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 3.Bosch FX, Ribes J, Borras J. Epidemiology of primary liver cancer. Semin Liver Dis. 1999;19:271–85. doi: 10.1055/s-2007-1007117. [DOI] [PubMed] [Google Scholar]
- 4.Buendia MA. Genetic alterations in hepatoblastoma and hepatocellular carcinoma: common and distinctive aspects. Med Pediatr Oncol. 2002;39:530–5. doi: 10.1002/mpo.10180. [DOI] [PubMed] [Google Scholar]
- 5.Tang ZY, Sun FX, Tian J, et al. Metastatic human hepatocellular carcinoma models in nude mice and cell line with metastatic potential. World J Gastroenterol. 2001;7:597–601. doi: 10.3748/wjg.v7.i5.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Tang Y, Ye L, et al. Establishment of a hepatocellular carcinoma cell line with unique metastatic characteristics through in vivo selection and screening for metastasis-related genes through cDNA microarray. J Cancer Res Clin Oncol. 2003;129:43–51. doi: 10.1007/s00432-002-0396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boissan M, Wendum D, Arnaud-Dabernat S, et al. Increased lung metastasis in transgenic NM23-Null/SV40 mice with hepatocellular carcinoma. J Natl Cancer Inst. 2005;97:836–45. doi: 10.1093/jnci/dji143. [DOI] [PubMed] [Google Scholar]
- 8.Lewis BC, Klimstra DS, Socci ND, Xu S, Koutcher JA, Varmus HE. The absence of p53 promotes metastasis in a novel somatic mouse model for hepatocellular carcinoma. Mol Cell Biol. 2005;25:1228–37. doi: 10.1128/MCB.25.4.1228-1237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Budhu A, Forgues M, Ye QH, et al. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10:99–111. doi: 10.1016/j.ccr.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 10.Ye QH, Qin LX, Forgues M, et al. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med. 2003;9:416–23. doi: 10.1038/nm843. [DOI] [PubMed] [Google Scholar]
- 11.Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 12.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–7. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 13.Pomerantz J, Schreiber-Agus N, Liegeois NJ, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–23. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 14.Martelli F, Hamilton T, Silver DP, et al. p19ARF targets certain E2F species for degradation. Proc Natl Acad Sci U S A. 2001;98:4455–60. doi: 10.1073/pnas.081061398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi Y, Gregory MA, Li Z, Brousal JP, West K, Hann SR. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature. 2004;431:712–7. doi: 10.1038/nature02958. [DOI] [PubMed] [Google Scholar]
- 16.Paliwal S, Pande S, Kovi RC, Sharpless NE, Bardeesy N, Grossman SR. Targeting of C-terminal binding protein (CtBP) by ARF results in p53-independent apoptosis. Mol Cell Biol. 2006;26:2360–72. doi: 10.1128/MCB.26.6.2360-2372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grooteclaes M, Deveraux Q, Hildebrand J, Zhang Q, Goodman RH, Frisch SM. C-terminal-binding protein corepresses epithelial and proapoptotic gene expression programs. Proc Natl Acad Sci U S A. 2003;100:4568–73. doi: 10.1073/pnas.0830998100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grooteclaes ML, Frisch SM. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene. 2000;19:3823–8. doi: 10.1038/sj.onc.1203721. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, Wang SY, Nottke AC, Rocheleau JV, Piston DW, Goodman RH. Redox sensor CtBP mediates hypoxia-induced tumor cell migration. Proc Natl Acad Sci U S A. 2006;103:9029–33. doi: 10.1073/pnas.0603269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liew CT, Li HM, Lo KW, et al. High frequency of p16INK4A gene alterations in hepatocellular carcinoma. Oncogene. 1999;18:789–95. doi: 10.1038/sj.onc.1202359. [DOI] [PubMed] [Google Scholar]
- 21.Matsuda Y, Ichida T, Matsuzawa J, Sugimura K, Asakura H. p16(INK4) is inactivated by extensive CpG methylation in human hepatocellular carcinoma. Gastroenterology. 1999;116:394–400. doi: 10.1016/s0016-5085(99)70137-x. [DOI] [PubMed] [Google Scholar]
- 22.Tannapfel A, Busse C, Weinans L, et al. INK4a-ARF alterations and p53 mutations in hepatocellular carcinomas. Oncogene. 2001;20:7104–9. doi: 10.1038/sj.onc.1204902. [DOI] [PubMed] [Google Scholar]
- 23.Bagchi A, Papazoglu C, Wu Y, et al. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128:459–75. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 24.Lewis BC, Shim H, Li Q, et al. Identification of putative c-Myc-responsive genes: characterization of rcl, a novel growth-related gene. Mol Cell Biol. 1997;17:4967–78. doi: 10.1128/mcb.17.9.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen YW, Klimstra DS, Mongeau ME, Tatem JL, Boyartchuk V, Lewis BC. Loss of p53 and Ink4a/Arf Cooperate in a Cell Autonomous Fashion to Induce Metastasis of Hepatocellular Carcinoma Cells. Cancer Res. 2007;67:7589–96. doi: 10.1158/0008-5472.CAN-07-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitz AA, Govek EE, Bottner B, Van Aelst L. Rho GTPases: signaling, migration, and invasion. Exp Cell Res. 2000;261:1–12. doi: 10.1006/excr.2000.5049. [DOI] [PubMed] [Google Scholar]
- 27.Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740–6. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Mirnezami AH, Campbell SJ, Darley M, Primrose JN, Johnson PW, Blaydes JP. Hdm2 recruits a hypoxia-sensitive corepressor to negatively regulate p53-dependent transcription. Curr Biol. 2003;13:1234–9. doi: 10.1016/s0960-9822(03)00454-8. [DOI] [PubMed] [Google Scholar]
- 29.Ito T, Nishida N, Fukuda Y, Nishimura T, Komeda T, Nakao K. Alteration of the p14(ARF) gene and p53 status in human hepatocellular carcinomas. J Gastroenterol. 2004;39:355–61. doi: 10.1007/s00535-003-1302-9. [DOI] [PubMed] [Google Scholar]
- 30.Kalinichenko VV, Major ML, Wang X, et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–50. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelly-Spratt KS, Gurley KE, Yasui Y, Kemp CJ. p19Arf suppresses growth, progression, and metastasis of Hras-driven carcinomas through p53-dependent and -independent pathways. PLoS Biol. 2004;2:E242. doi: 10.1371/journal.pbio.0020242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6:663–73. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 33.Guo F, Gao Y, Wang L, Zheng Y. p19Arf-p53 tumor suppressor pathway regulates cell motility by suppression of phosphoinositide 3-kinase and Rac1 GTPase activities. J Biol Chem. 2003;278:14414–9. doi: 10.1074/jbc.M300341200. [DOI] [PubMed] [Google Scholar]
- 34.Guo F, Zheng Y. Rho family GTPases cooperate with p53 deletion to promote primary mouse embryonic fibroblast cell invasion. Oncogene. 2004;23:5577–85. doi: 10.1038/sj.onc.1207752. [DOI] [PubMed] [Google Scholar]
- 35.Paliwal S, Kovi RC, Nath B, Chen YW, Lewis BC, Grossman SR. Arf antagonizes hypoxia-induced cancer cell migration via interaction with the CtBP corepressor. Cancer Res. 2007 doi: 10.1158/0008-5472.CAN-07-1743. In Press. [DOI] [PubMed] [Google Scholar]
- 36.Chinnadurai G. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol Cell. 2002;9:213–24. doi: 10.1016/s1097-2765(02)00443-4. [DOI] [PubMed] [Google Scholar]
- 37.Wicki A, Lehembre F, Wick N, Hantusch B, Kerjaschki D, Christofori G. Tumor invasion in the absence of epithelial-mesenchymal transition: podoplanin-mediated remodeling of the actin cytoskeleton. Cancer Cell. 2006;9:261–72. doi: 10.1016/j.ccr.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 38.Spaderna S, Schmalhofer O, Hlubek F, et al. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology. 2006;131:830–40. doi: 10.1053/j.gastro.2006.06.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


