Abstract
Background
Non-small cell lung cancer (NSCLC) is the most common cause of cancer-related death in Western countries. Developing more effective NSCLC therapeutics will require the elucidation of the genetic and biochemical bases for this disease. Bronchioalveolar stem cells (BASCs) are a putative cancer stem cell population in mouse models of oncogenic K-ras-induced lung adenocarcinoma, an histologic subtype of NSCLC. The signals activated by oncogenic K-ras that mediate BASC expansion have not been fully defined.
Methodology/Principal Findings
We used genetic and pharmacologic approaches to modulate the activity of phosphatidylinositol 3-kinase (PI3K), a key mediator of oncogenic K-ras, in two genetic mouse models of lung adenocarcinoma. Oncogenic K-ras-induced BASC accumulation and tumor growth were blocked by treatment with a small molecule PI3K inhibitor and enhanced by inactivation of phosphatase and tensin homologue deleted from chromosome 10, a negative regulator of PI3K.
Conclusions/Significance
We conclude that PI3K is a critical regulator of BASC expansion, supporting treatment strategies to target PI3K in NSCLC patients.
Introduction
Non-small cell lung cancer (NSCLC) is the leading cause of cancer-related death in Western countries and its incidence is rising in Asia. Once it has metastasized, there are no curative therapies for NSCLC. A better understanding of the pathogenesis and risk factors for this disease will contribute not only to improvements in early detection and prevention but also to the development of more effective therapies for it.
Approximately 10% of NSCLC specimens carry activating mutations in K-ras [1]. The histologic subtype of NSCLC that most frequently has K-ras mutations is adenocarcinoma; 30% of adenocarcinomas have these mutations. K-ras mutations have also been identified in atypical adenomatous hyperplasia (AAH) lesions, which are thought to precede the development of lung adenocarcinoma [2]. A growing body of evidence indicates that K-ras mutations are important in the initiation of lung adenocarcinoma development. Mouse models have been developed that express mutant K-ras conditionally, somatically, or inducibly [3]–[7]. These mice develop AAH lesions rapidly and with high penetrance, and a subset of these lesions progress to adenocarcinomas.
A candidate lung cancer progenitor cell (bronchioalveolar stem cell or BASC) has been identified in murine models of K-ras-induced lung cancer. BASCs have a specific pattern of stem cell marker expression (Sca-1pos CD34pos CD45neg Pecamneg), are located at the terminal bronchi, and have the capacity to self-renew and undergo multi-linage differentiation [8]. These cells may be the same as the variant Clara (or ClaraV) cells, which are located adjacent to clusters of neuroendocrine bodies in bronchi and bronchioles. Similar to BASCs, ClaraV cells are capable of restoring the Clara cell population after naphthalene injury [9]. After intra-nasal installation of adenoviral Cre into KrasLSL mice, which develop lung adenocarcinoma due to activation of a conditional oncogenic K-ras allele [5], BASCs rapidly proliferate prior to the development of histologic abnormalities [8]. The BASCs in these mice contain K-ras mutations identical to those found in tumors [8]. On the basis of this evidence, BASCs are postulated to be progenitors of lung adenocarcinoma in mice. The biochemical signals that initiate the expansion of this cellular population have not been fully elucidated. This deficiency is important given that the elucidation of these signals might lead to better treatment for NSCLC patients.
Premalignant lesions typically undergo a transient expansion followed by programmed senescence, and only a minority of early lesions progresses to a fully transformed state. In the case of Ras-dependent cancer mouse models, the programmed senescence of premalignant lesions is activated by mitogen-activated protein kinase kinase/extracellular signal-regulated kinase signaling, which inhibits Ras through feed-back pathways involving increased expression of SPROUTY proteins, Ras GTPase activating proteins, and mitogen-activated protein kinase phosphatase-3 [10]. The senescence program in premalignant lesions can be blocked by phosphatidylinositol 3-kinase (PI3K) activation [10], indicating that PI3K is a crucial determinant of the malignant progression of these lesions. In fact, PI3K signaling is constitutively activated in NSCLC cells through inactivating somatic mutations in, or epigenetic silencing of, phosphatase and tensin homologue deleted from chromosome 10 (Pten), a lipid phosphatase that negatively regulates the PI3K signaling cascade [11]–[15].
In this study, we evaluated the importance of PI3K in regulating the expansion of BASCs in two genetic mouse models of oncogenic K-ras-induced lung cancer. The oncogenic K-ras alleles in these models are activated somatically in one (KrasLA1) and conditionally in the other (KrasLSL) (5, 7). We found increased numbers of BASCs in these mice relative to those of wild-type littermates. BASC expansion and malignant progression in the lung were attenuated by pharmacologic inhibition of PI3K and enhanced by genetic inactivation of Pten. We speculate on the basis of these findings that pharmacologic strategies to inhibit PI3K will be useful in the prevention and treatment of NSCLC.
Results
PX-866 inhibits lung tumorigenesis in KrasLA1 mice
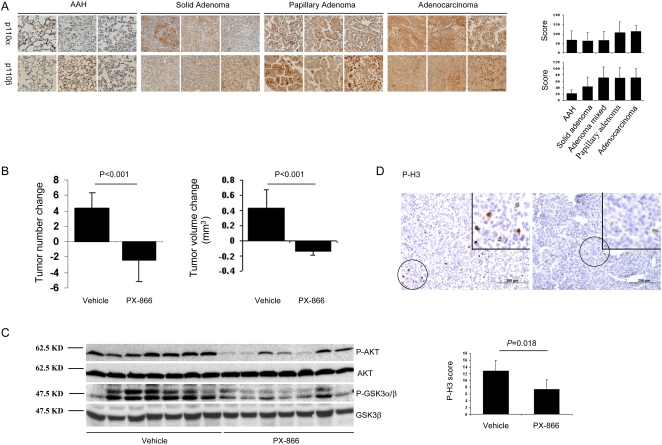
We postulated that PI3K is required for malignant progression in lung cancer and tested this hypothesis by using KrasLA1 mice as a model of lung tumorigenesis. Several weeks after birth, KrasLA1 mice exhibit multifocal AAH lesions that, by 2–3 months of age, coalesce into solid or papillary adenomas, which enlarge and undergo histologic transformation into adenocarcinomas by 6–8 months [7]. We first evaluated the expression of PI3K (p110α and β isoforms) in lung tissues at different stages of tumorigenesis (AAH, adenoma, or adenocarcinoma). These PI3K isoforms were detected and their abundance increased with malignant progression (AAH lesions versus adenocarcinomas: P = 0.006 for p110α; P<0.001 for p110β) (Figure 1A).
Figure 1. PI3K promotes tumorigenesis in KrasLA1 mice.
(A) PI3K expression increases with malignant progression. Representative immunohistochemical staining of lesions in the tissue microarray and their quantification in adjacent bar graphs. Calibration bar in lower right panel represents 100 μm. Scores of mixed (solid/papillary) adenomas are included in bar graphs but not in PI3K staining images. (B) PI3K is required for tumor growth. Mean changes in tumor multiplicity and volume determined by micro-computed tomography performed before and after treatment. (* P<0.05 compared to vehicle). (C) PX-866 inhibits PI3K in lung tissues. Western blotting of Ser473-phosphorylated AKT (P-AKT) and Ser21/9-phosphorylated GSK3 (P-GSK3α/β) in whole lung lysates from mice (n = 7 in each treatment cohort). Positions of molecular weight markers are indicated on the left. (D) PX-866 decreases tumor cell proliferation. Representative immunohistochemical staining of Ser28-phosphorylated histone H3 (P-H3) in adenomas in the tissue microarray (x 20 magnification, areas with positive cells encircled) and quantification of staining in 16 lesions from vehicle-treated mice and 4 lesions from PX-866-treated mice (bar graph, * P<0.05 compared to vehicle). Inset illustrates encircled cells at ×40 magnification. Calibration bars represent 200 μm.
Based on its increased abundance in fully transformed cells, we next sought to determine whether PI3K was required for tumor growth. Treatment of K-rasLA1 mice with PX-866, a PI3K inhibitor [16], decreased lung lesion volume and multiplicity (Figure 1B). This was accompanied by reductions in PI3K activity as shown by western blotting of whole-lung lysates for phosphorylated AKT and glycogen synthase kinase-3 (Figure 1C). Tumor cells underwent a reduction in proliferation caused by PX-866 based on intra-lesional phosphorylated histone H3 (Figure 1D) but exhibited no biochemical evidence of apoptosis as shown by western blotting for cleaved caspase-3 and Poly (ADP-ribose) polymerase (data not shown). Thus, treatment with a PI3K antagonist profoundly inhibited the growth of these early lesions.
BASCs are highly sensitive to PX-866 in vivo and in vitro
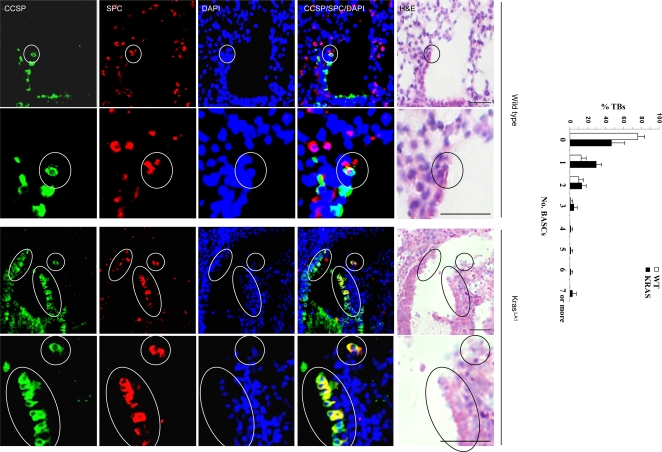
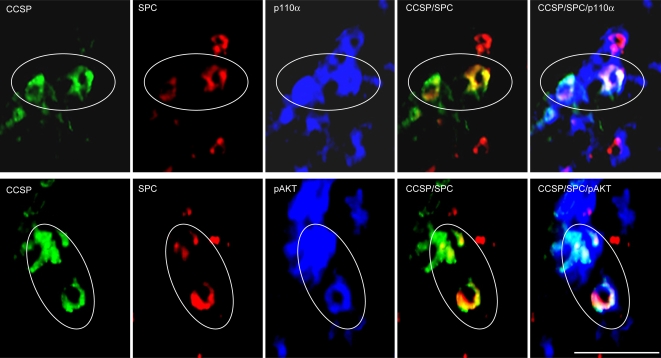
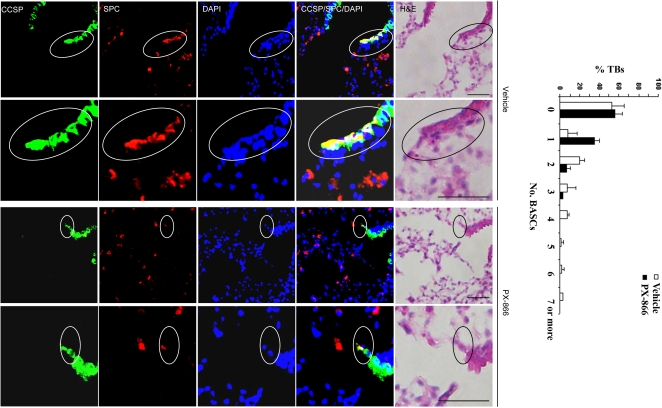
We next determined whether the anti-tumor effect of PX-866 in KrasLA1 mice was partly due to inhibition of BASC expansion. At the terminal bronchi, a subset of epithelial cells expressed both clara cell specific protein (CCSP) and surfactant protein-C (SPC) (Figure 2), which is a hallmark of BASCs [8]. Quantification of dual-positive cells in wild-type mice (with 83 bronchi) and K-rasLA1 mice (with 66 bronchi) revealed that no wild-type mice had more than 3 BASCs per terminal bronchus, whereas a subset of terminal bronchi in K-rasLA1 mice had 4 to 7 (P = 0.042, wild-type versus KrasLA1) (Figure 2). Thus, a subset of terminal bronchi in KrasLA1 mice had BASC expansion. Furthermore, triple immunofluorescence studies revealed that BASCs expressed p110α and had detectable phosphorylation of AKT (Figure 3), a downstream mediator of PI3K, providing evidence of PI3K activation in these cells. Using tissue sections from the KrasLA1 mice treated with PX-866 (with 88 terminal bronchi) or vehicle (with 81 terminal bronchi), we determined the number of BASCs per terminal bronchus in the treatment groups. The vehicle-treated mice had a subset of terminal bronchi with 4 to 7 BASCs per terminal bronchus, whereas none of the PX-866-treated mice had more than three BASCs per terminal bronchus (P = 0.025, vehicle versus PX-866-treated) (Figure 4).
Figure 2. KrasLA1 mice have an accumulation of BASCs at terminal bronchi.
Immunofluorescent staining to detect cells at terminal bronchi that co-express CCSP and SPC. Encircled BASCs in top panels (x10 magnification) illustrated in lower panels at higher magnification (x40). Bar graph indicates percentages of terminal bronchi with the indicated numbers of BASCs. Calibration bars in images of H&E stained tissues represent 50 μm.
Figure 3. BASCs express p110α and have Ser473-phosphorylated AKT.
Immunofluorescent staining of tissue sections to detect triply stained (CCSP/SPC/p110α or pAKT) cells at terminal bronchi. BASCs encircled (x40 magnification). Calibration bar in lower right panel represents 50 μm.
Figure 4. PX-866 treatment decreases BASC numbers in KrasLA1 mice.
Immunofluorescent staining to detect cells at terminal bronchi that co-express CCSP and SPC. Encircled BASCs in top panels (×10 magnification) illustrated in lower panels at higher magnification (×40). Bar graph indicates percentages of terminal bronchi with the indicated numbers of BASCS. Calibration bars in images of H&E stained tissues represent 50 μm.
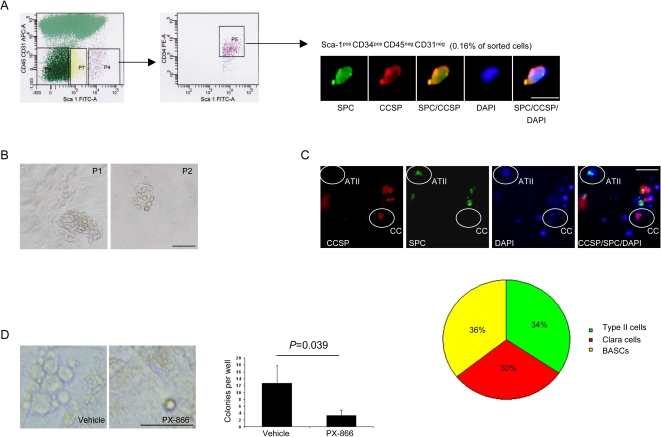
To determine whether PX-866 had direct effects on KrasLA1–derived BASCs, we used primary BASC cultures, which were isolated from the lungs of KrasLA1 mice by sorting for cells that were Sca-1pos, CD34pos, CD45neg, and CD31neg. These sorted cells co-expressed CCSP and SPC (Figure 5A), formed colonies when plated on feeder cultures (Figure 5B), and exhibited multi-potential differentiation capacity when plated in matrigel (Figure 5C), which are hallmarks of BASCs [8]. PX-866 treatment led to a prominent decrease in BASC colony forming activity (Figure 5D), indicating that PX-866 had a direct effect on BASCs.
Figure 5. PX-866 inhibits BASC expansion in vitro.
(A) Sorted cells co-express SPC and CCSP. KrasLA1 whole lung tissues were dissociated and sorted to isolate Sca-1pos CD45neg CD31neg CD34pos cells (P4 and P5 in left and middle panels, respectively), which were subjected to immunofluorescent staining (x40 magnification, right panel). Calibration bar in far right panel represents 10 μm. (B) BASCs form colonies when plated on feeder cultures. Photograph of colony (x10 magnification) formed upon initial plating (P1) and passage 2 (P2). Calibration bar in panel on right represents 50 μm. (C) BASCs differentiate when plated in matrigel. Cells were stained after 7 d in matrigel (x10 magnification) and the numbers of cells with features of alveolar type II cells (CCSPneg SPCpos), clara cells (CCSPpos SPCneg), or BASCs (CCSPpos SPCpos) were counted and expressed as the percentages of the total 158 counted cells in a pie chart. Examples of cells with features of ATII cells (ATII) and clara cells (CC) are indicated. Calibration bar in far right panel represents 50 μm. (D) PX-866 inhibits BASC colony formation. Photographs (x10 magnification) and quantification of colonies per well (5 wells per condition) formed after 6 d in the presence or absence of PX-866. Calibration bar in panel on right represents 50 μm.
Pten deficiency enhances oncogenic K-ras-induced BASC accumulation
As another approach to evaluating the role of PI3K in BASC expansion, we inactivated Pten, a negative regulator of PI3K, in CCSP-expressing cells. Pten was genetically inactivated by using Pten flox/+ mice, which have a Pten allele that contains LoxP sites surrounding the PTEN phosphatase domain encoded by exon 5 [17]. Pten was recombined specifically in the lung by interbreeding these mice with CCSPCre/+ mice, which express Cre under the control of the CCSP gene [18]. In addition, we evaluated whether Pten inactivation cooperated with oncogenic K-ras to promote BASC expansion. Pten-deficient mice were interbred with K-rasLSL mice, in which the expression of oncogenic K-rasG12D is controlled by a removable transcription termination STOP element [5]. Using this approach, mice were generated with bronchi that are Pten-deficient (PtenΔ5/Δ5; CCSPCre/+), express oncogenic K-ras (KrasLox/+; CCSPCre/+), have Pten deficiency and oncogenic K-ras expression (PtenΔ5/Δ5; KrasLox/+; CCSPCre/+), or have neither (Ptenflox/flox; CCSP+/+). Mice were sacrificed at 4 weeks or 12 weeks, and their lung tissues were subjected to immunofluorescence studies to quantify BASCs.
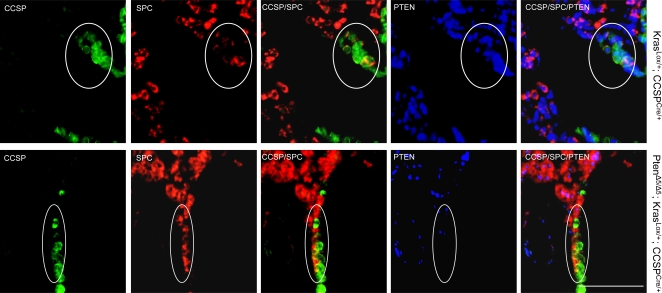
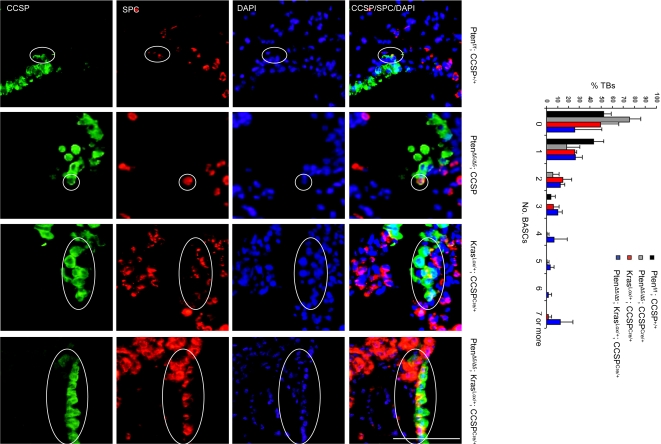
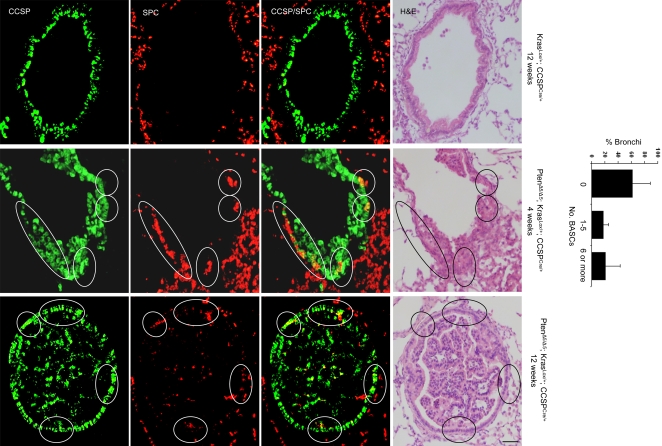
As expected, triple immunofluorescence staining (CCSP/SPC/PTEN) revealed loss of PTEN expression in BASCs from mice that were both Pten-deficient and K-ras-transformed (PtenΔ5/Δ5; KrasLox/+; CCSPCre/+) but not in mice that were only K-ras-transformed (KrasLox/+; CCSPCre/+) (Figure 6). Quantification of BASCs per terminal bronchus at 4 weeks revealed that Pten-deficiency alone (PtenΔ5/Δ5; CCSPCre/+) was not sufficient to change BASC numbers relative to that of control mice (Ptenflox/flox; CCSP+/+) (Figure 7). However, comparison of BASC numbers in PtenΔ5/Δ5; KrasLox/+; CCSPCre/+ mice to that of KrasLox/+; CCSPCre/+ mice demonstrated that Pten deficiency strikingly enhanced BASC expansion by oncogenic K-ras (P = 0.006, PtenΔ5/Δ5; KrasLox/+; CCSPCre/+ versus KrasLox/+; CCSPCre/+) (Figure 7). In addition to these findings at the terminal bronchi, clusters of CCSPpos SPCpos cells were observed in the bronchioles and small bronchi of PtenΔ5/Δ5; KrasLox/+; CCSPCre/+ mice; these cell clusters were evident as early as 4 weeks of age and were not present in the bronchi or bronchioles of KrasLox/+; CCSPCre/+ mice (Figure 8). We conclude that Pten inactivation cooperated with oncogenic K-ras to enhance BASC accumulation.
Figure 6. Loss of PTEN expression in BASCs from PtenΔ5/Δ5; KrasLox/+; CCSPCre/+ mice.
Immunofluorescent staining of tissue sections to detect triply stained (CCSP/SPC/PTEN) cells at terminal bronchi. BASCs (encircled) illustrated at ×10 magnification. Calibration bar in lower right panel represents 100 μm.
Figure 7. BASC expansion at terminal bronchi of PtenΔ5/Δ5; KrasLox/+; CCSPCre/+ mice.
Immunofluorescent staining to detect cells at terminal bronchi that co-express CCSP and SPC. BASCs (encircled) illustrated at ×40 magnification. Bar graph indicates percentages of terminal bronchi with the indicated numbers of BASCS. Calibration bar in lower right panel represents 100 μm.
Figure 8. BASCs identified in small bronchi/bronchioles of PtenΔ5/Δ5; KrasLox/+; CCSPCre/+ mice but not KrasLox/+; CCSPCre/+ mice.
Immunofluorescent staining to detect cells that co-express CCSP and SPC. BASCs (encircled) illustrated at ×10 magnification. Bar graph illustrates percentages of bronchi with the indicated numbers of BASCS. Calibration bar in lower right panel represents 100 μm.
Discussion
Here we report that PI3K was required for BASC expansion initiated by oncogenic K-ras and, when constitutively activated by Pten inactivation, PI3K cooperated with oncogenic K-ras in this process. This conclusion was based on findings from experiments that incorporated pharmacologic and genetic approaches to target PI3K in two different genetic mouse models and an in vitro model. We have previously reported that Pten inactivation cooperates with oncogenic K-ras to accelerate lung tumorigenesis, causing lung tumors that are more histologically advanced and more rapidly obstruct bronchial lumina and replace alveolar spaces than those of mice with oncogenic K-ras alone [18]. Together, these findings raise the possibility that PI3K promoted lung tumorigenesis through its effects on BASCs. However, conclusive evidence in this regard will require proof that adenocarcinomas arise from BASCs. Transplantation studies to determine whether BASC injections are sufficient to recapitulate lung adenocarcinoma development in mice have thus far not been reported. A stem cell population has been identified in the lungs of mice and in tumors from NSCLC patients that expresses CD133, exhibits stem cell characteristics in vitro (forms spheres and can be induced to undergo terminal differentiation), and is tumorigenic in nude mice [19]. Although certain features of that stem cell population are similar to those of BASCs (i.e. response to naphthalene-induced lung injury), their relationship remains to be elucidated.
When initiated during embryogenesis, conditional Pten inactivation in the lung leads to hypoxia and perinatal death, indicating that Pten expression is required for normal lung development [20]. In this study, Cre expression in CCSPCre mice began at postnatal day 4, allowing us to evaluate Pten's role in maintaining lung function after birth. Although we can not exclude the possibility that subtle changes in lung function occurred, the Pten-deficient mice in this study had no obvious alterations in their lung structure and exhibited no morbidity at up to 12 months of age, suggesting that, after birth, lung tissues can function normally without Pten. This apparent resilience of lung tissues to Pten deficiency stands in contrast to adult hematopoietic stem cells, which are depleted within 40 days after inactivation of Pten [21], indicating that the self-renewal capabilities of hematopoietic stem cells can not be maintained in the absence of PTEN's negative growth regulatory signals. Although the reasons for this difference are not clear, it may be related in part to the relative infrequency with which lung stem cells normally divide [9].
PI3K has been reported to play an important role in tumorigenesis initiated by oncogenic K-ras. KrasLA2 mice that have a knock-in PIK3CA mutation that blocks the interaction of PI3K with K-ras develop far fewer lung tumors than do KrasLA2 mice with wild-type PIK3CA [22], indicating a key role for PI3K as a downstream mediator of K-ras. Corroborating this point, we found that PX-866 treatment inhibited lung tumor growth in KrasLA1 mice. However, based on other findings reported here, the importance of PI3K in K-ras-induced lung tumorigenesis may not be restricted to its role as a downstream mediator of K-ras. First, PI3K expression increased as KrasLA1 lung tissues underwent malignant progression from AAH to adenocarcinoma, suggesting that PI3K was activated in these histologically advanced lesions in part through K-ras-independent mechanisms. Second, findings reported here and elsewhere [18] indicate that Pten inactivation enhanced oncogenic K-ras–induced lung tumorigenesis in mice. Extrapolating from these findings, somatic mutations in NSCLC cells involving genes that regulate PI3K-dependent signaling (i.e. Pten, Egfr, and PIK3CA, among others [ref. 23]) may transform these cells in part by cooperating with oncogenic K-ras. The potential importance of secondary somatic mutations in promoting oncogenic K-ras-induced lung tumorigenesis is illustrated by the long latencies of adenocarcinomas that arise in mice engineered to express mutant K-ras, which develop lung lesions rapidly and with high penetrance, but only a fraction of the lesions progress into adenocarcinomas and require several months to do so [3]–[7].
Findings reported here differed from those reported previously in a different strain of mice that undergo conditional Pten deletion induced by SPC-driven Cre expression [20]. In that report, Pten inactivation was sufficient to increase BASC numbers and induce lung tumors, which stands in contrast to findings presented here and elsewhere [18] that Pten deficiency is not sufficient to increase BASC numbers or induce lung tumors. That previous report also differed with respect to the genetic events that cooperated with Pten deficiency. In that study, Pten inactivation accelerated lung tumorigenesis induced by urethane, a carcinogen that frequently induces K-ras mutations, but only 2 of 6 tumors evaluated had K-ras mutations, which leaves open the possibility that urethane cooperates with Pten loss by inducing somatic mutations of genes other than K-ras. This stands in contrast to our model in which the conditional K-ras and Pten alleles recombined in the same (CCSP-expressing) cells. Hence, the disparate outcomes of these studies might be related to secondary genetic events in the urethane-treated mice or differences in experimental design, such as the genetic backgrounds of the mice or the specific promoter elements driving Cre expression.
Methods
Reagents
Antibodies purchased for these studies include rabbit anti-p110α (sc-7174), p110β (sc-7175), SPC (sc-13979), goat anti-CC10 (sc-9772) and PTEN (sc-6818) (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-total AKT (9272), Ser473-phosphorylated AKT (9271), Ser21/9-phosphorylated GSK3α/β (9331), total GSK3 (9332), cleaved caspase-3 (9661), Poly (ADP-ribose) polymerase (9542), phosphorylated-histone H3 (9701), Ser28-phosphorylated histone H3 (9713P) (Cell Signaling Technologies, Danvers, MA), rabbit anti-SPC (WRAB-SPC, Seven Hills Bioreagents, Cincinnati, OH), rabbit anti-CCSP (07-623, Upstate Biotechnologies, Lake Placid, NY), FITC-conjugated anti-Sca-1 (557405), biotin-conjugated anti-CD45 (553771) biotin-conjugated-CD31 (558737), PE-conjugated anti-CD34 (12-0341-33, eBioscience), anti-rabbit IgG Alexa Flour 488 (A11008), and anti-goat IgG Alexa Flour 594 (A11058) (Invitrogen, Carlsbad, CA). Other purchased reagents include streptavidin-APC (554067, BD Pharmigen), TSA kits 22 (T20932), 25 (T20935), and 27(T20937), Avidin/Biotin blocking kit (00-4303) (Invitrogen), Dispase (354235, BD Biosciences), collagenase/dispase (10269638001, Roche Biosciences), and 7-aminoactinomycin D (A1310, Invitrogen).
Mouse studies
To test whether PI3K promotes tumor development, 9 K-rasLA1 mice were treated for 4 weeks with 2 mg/kg/d intra-gastric PX-866, a small molecule inhibitor of PI3K [12], and 12 were given vehicle beginning at 4 months, an age at which lung lesions (AAH and adenomas) are measurable by micro-computed tomography scans. Mice were subjected to these scans at the beginning and completion of treatment to count lesion numbers and evaluate changes in lesion volumes over time as previously reported [24].
Prior to their initiation, all mouse studies were submitted to and approved by the Institutional Animal Care and Use Committee at the University of Texas–M. D. Anderson Cancer Center. Mice received standards of care and were euthanized according to the standards set forth by the IACUC. Mice and cells and were obtained through collaborations with Dr. Tyler Jacks, MIT (KrasLA1 mice, and KrasLSL mice), Dr. Hong Wu, UCLA (Ptenflox mice), and Dr. Francesco Demayo, Baylor College of Medicine (CCSPCre mice).
Tissue microarrays and immunohistochemical analyses
We performed immunohistochemical analysis of PI3K on a tissue microarray containing AAH (n = 17), solid adenomas (n = 31), mixed solid/papillary adenomas (n = 36), papillary adenomas (n = 45), and adenocarcinomas (n = 14) to determine whether the levels of PI3K changed during malignant progression. We analyzed the expression of PI3K isoforms (p110α and β).
For immunohistochemical staining, 4 μm paraffin-embedded sections were baked at 60°C for 30 min and then cooled to ambient temperature. Sections were sequentially incubated in Xylene (5 min twice), 100% ethyl alcohol (5 min twice), 95% ethyl alcohol (5 min twice), and 80% ethyl alcohol (5 min). After washing with water, the sections were antigen-retrieved using citrate buffer (pH 6.0, DAKO) in a steamer for 20 min and cooled to ambient temperature. Sections were then washed with TBS-T and quenched with 3% hydrogen peroxide in TBS for 10 min, blocked for Avidin/Biotin activity, and incubated with primary antibody as follows: For p110α and p110β staining, sections were blocked with 5% goat serum for 1 h, incubated with primary antibodies for 1 h at ambient temperature, and then washed with TBS-T; for Ser28-phosphorylated histone H3 staining, sections were blocked with 10% goat serum at 4°C overnight, incubated with primary antibody for 30 min at ambient temperature, and washed with TBS-T. The staining was developed using DAB substrate system. Negative controls included omission of the primary antibody and pre-incubation of primary antibody with blocking peptide. Staining was quantified by one investigator (M.G.R.) blinded as to treatment group. Intra-lesional expression was scored based on staining intensity and extension as described previously [24].
Detection of BASCs in lung tissues
Antigens were retrieved from mouse lung tissues with 0.01 mol/L citrate buffer (pH 6; DakoCytomation, Glostrup, Denmark) for 25 min in a steamer. The slides were quenched in 1.5% hydrogen peroxide in Tris-buffered saline for 10 minutes and blocked in DAKO serum-free protein block (Dako) for 1 h at ambient temperature. The slides were then incubated with anti-SPC (sc-7705, Santa Cruz Biotechnology; WRAB-SPC, Seven Hills Bioreagents) and anti-CCSP (07-623, Upstate Biochemicals) antibodies at 4°C overnight. The immunofluorescence was developed using TSA kits 25 and 22 (Invitrogen) on the basis of the manufacturer's instructions. We used blocking peptides and omitted the primary antibodies as negative controls to determine the specificity of the immunostaining results. Immunofluorescence was visualized using a microscope with a reflected fluorescence system (Olympus Model IX71SIF-2). Color acquisition and background fluorescence were optimized by using the DPController and DPManager software (Olympus).
BASC isolation and culture
The mice were killed after 12 weeks, and the lungs were perfused with 10 mL of Hank's balanced salt solution through the right ventricle until they were cleared of blood. The tracheas were injected with Dispase (undiluted liquid, BD Biosciences) followed by 0.5–1 mL 1% LMP agarose in water. The lungs were placed on ice, chopped into pieces, incubated with 0.001% DNAse (Sigma D-4527) and 2 mg/mL of collagenase/dispase (100 mg/mL, Roche) in phosphate-buffered saline at 37° C for 45 min, filtered (100 μm followed by 40 μm pore-sized filter), and centrifuged. The resulting pellets were resuspended in 2 mL of PF10 (10% fetal bovine serum in phosphate-buffered saline), which typically yields 1–2 million total cells per mouse. The red cells were lysed. BASCs were isolated by sorting of the Sca-1pos CD45neg Pecamneg CD34pos cell population on a three-laser 13-color FACS Aria fluorescence-activated cell sorter (BD Biosciences) that is capable of sorting four sub-populations at a low pressure with a maximum rate of 20 million cells/hour. BASCs represented approximately 0.1% of the total sorted cell population.
The uniformity or purity of the sorted cells was not routinely checked because the cells of interest would have been consumed in the process. That being said, the cells were sort-purified to the highest possible purity by gating on size (by forward and side scatter), excluding doublets (by forward scatter width versus area and side scatter width versus area), and isolating cells with the markers of interest. In addition, the uniformity of the sorted cell populations used in these assays was reflected by the reproducibility of our findings; the differentiation and colony formation assays were performed each time using cells derived from three different mice, which yielded similar results.
For colony formation assays, the sorted cells were plated in DMEM/HEPES/10% FBS/Pen-Strep/L-glutamine on 96-well plates with feeder cells (irradiated DR4 mouse embryo fibroblasts, SCRC-1045.1, American Type Culture Collection, Manassas, VA) to maintain the undifferentiated state (1000 cells per well). PX-866 was added to the cultures the day following plating. For differentiation assays, BASCs were plated for 7 d on 96-well plates (10,000 cells per well) coated with 100 μL growth factor-reduced Matrigel (356231, BD Biosciences) and subjected to immunofluorescent staining for SPC and CCSP as described [8]. The yield of BASCs (Sca-1pos CD45neg Pecamneg CD34pos) is typically 0.1% of sorted cells from the lung.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: R01 CA117965, R01 CA105155, U01 CA105352
References
- 1.Aviel-Ronen S, Blackhall FH, Shepherd FA, Tsao MS. K-ras mutations in non-small-cell lung carcinoma: a review. Clin Lung Cancer. 2006;8:30–38. doi: 10.3816/CLC.2006.n.030. [DOI] [PubMed] [Google Scholar]
- 2.Mori M, Tezuka F, Chiba R, Funae Y, Watanabe M, et al. Atypical adenomatous hyperplasia and adenocarcinoma of the human lung: their heterology in form and analogy in immunohistochemical characteristics. Cancer. 1996;77:665–674. [PubMed] [Google Scholar]
- 3.Fisher GH, Wellen SL, Klimstra D, Lenczowski JM, Tichelaar JW, et al. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes & Dev. 2001;15:3249–3262. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerra C, Mijimolle N, Dhawahir A, Dubus P, Barradas M, et al. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4:111–120. doi: 10.1016/s1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 5.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji H, Houghton AM, Mariani TJ, Perera S, Kim CB, et al. K-ras activation generates an inflammatory response in lung tumors. Oncogene. 2006;25:2105–2112. doi: 10.1038/sj.onc.1209237. [DOI] [PubMed] [Google Scholar]
- 7.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 8.Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 9.Rawlins EL, Hogan BL. Epithelial stem cells of the lung: privileged few or opportunities for many? Development. 2006;133:2455–2465. doi: 10.1242/dev.02407. [DOI] [PubMed] [Google Scholar]
- 10.Courtois-Cox S, Genther Williams SM, Reczek EE, Johnson BW, McGillicuddy LT, et al. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell. 2006;10:459–472. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eng C. PTEN: one gene, many syndromes. Human Mutat. 2003;22:183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- 12.Marsit CJ, Zheng S, Aldape K, et al. PTEN expression in non-small-cell lung cancer: evaluating its relation to tumor characteristics, allelic loss, and epigenetic alteration. Hum Pathol. 2005;36(7):768–76. doi: 10.1016/j.humpath.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Soria JC, Lee HY, Lee JI, et al. Lack of PTEN expression in non-small cell lung cancer could be related to promoter methylation. Clin Cancer Res. 2002;8(5):1178–84. [PubMed] [Google Scholar]
- 14.Xia D, Srinivas H, Ahn YH, et al. Mitogen-activated protein kinase kinase-4 promotes cell survival by decreasing PTEN expression through an NF kappa B-dependent pathway. J Biol Chem. 2007;282(6):3507–19. doi: 10.1074/jbc.M610141200. [DOI] [PubMed] [Google Scholar]
- 15.Forgacs E, Biesterveld EJ, Sekido Y, et al. Mutation analysis of the PTEN/MMAC1 gene in lung cancer. Oncogene. 1998;17(12):1557–65. doi: 10.1038/sj.onc.1202070. [DOI] [PubMed] [Google Scholar]
- 16.Ihle NT, Williams R, Chow S, Chew W, Berggren MI, et al. Molecular pharmacology and antitumor activity of PX-866, a novel inhibitor of phosphoinositide-3-kinase signaling. Mol Cancer Ther. 2004;3:763–772. [PubMed] [Google Scholar]
- 17.Lesche R, Groszer M, Gao J, Wang Y, Messing A, et al. Cre/loxP-mediated inactivation of the murine Pten tumor suppressor gene. Genesis. 2002;32:148–149. doi: 10.1002/gene.10036. [DOI] [PubMed] [Google Scholar]
- 18.Iwanaga K, Yang Y, Raso MG, Ma L, Hanna AE, et al. Pten inactivation accelerates oncogenic K-ras–initiated tumorigenesis in a mouse model of lung cancer. Cancer Res. 2008;68:1119–1127. doi: 10.1158/0008-5472.CAN-07-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504–514. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 20.Yanagi S, Kishimoto H, Kawahara K, Sasaki T, Sasaki M, et al. Pten controls lung morphogenesis, bronchioalveolar stem cells, and onset of lung adenocarcinomas in mice. J Clin Investig. 2007;117:2929–2940. doi: 10.1172/JCI31854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 22.Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 23.Angulo B, Suarez-Gauthier A, Lopez-Rios F, Medina PP, Conde E, et al. Expression signatures in lung cancer reveal a profile for EGFR-mutant tumours and identify selective PIK3CA overexpression by gene amplification. J Pathol. 2008;214:347–356. doi: 10.1002/path.2267. [DOI] [PubMed] [Google Scholar]
- 24.Wislez M, Spencer ML, Izzo JG, Juroske DM, Balhara K, et al. Inhibition of mammalian target of rapamycin reverses alveolar epithelial neoplasia induced by oncogenic K-ras. Cancer Res. 2005;65:3226–3235. doi: 10.1158/0008-5472.CAN-04-4420. [DOI] [PubMed] [Google Scholar]