Abstract
Specialized chromatin exists at centromeres and must be precisely transmitted during DNA replication. The mechanisms involved in the propagation of these structures remain elusive. Fission yeast centromeres are composed of two chromatin domains: the central CENP-ACnp1 kinetochore domain and flanking heterochromatin domains. Here we show that fission yeast Mcl1, a DNA polymerase α (Polα) accessory protein, is critical for maintenance of centromeric chromatin. In a screen for mutants that alleviate both central domain and outer repeat silencing, we isolated several cos mutants, of which cos1 is allelic to mcl1. The mcl1-101 mutation causes reduced CENP-ACnp1 in the central domain and an aberrant increase in histone acetylation in both domains. These phenotypes are also observed in a mutant of swi7+, which encodes a catalytic subunit of Polα. Mcl1 forms S-phase-specific nuclear foci, which colocalize with those of PCNA and Polα. These results suggest that Mcl1 and Polα are required for propagation of centromere chromatin structures during DNA replication.
Introduction
The kinetochore is a multi-protein complex that assembles at the centromere and mediates attachment of chromosomes to spindle microtubules to ensure accurate chromosome segregation at mitosis. The kinetochore is assembled on specialized chromatin containing the histone H3 variant CENP-A and this domain is flanked by pericentric heterochromatin. Pericentric heterochromatin–typified by underacetylation of histone tails and methylation of lysine 9 of histone H3–is bound by heterochromatin protein 1 (HP1) and is required for the recruitment of a high density of cohesin and for the cohesion of sister centromeres.
The DNA sequences of centromeres are not conserved between eukaryotes and, indeed, primary DNA sequence appears not to be an absolute determinant of kinetochore position in most organisms [1]–[3]. Thus, propagation of a particular epigenetic mark could play a pivotal role in propagation of the site of kinetochore assembly. A likely candidate for this mark is CENP-A. To precisely transmit genomic information to daughter cells, these chromatin structures must be maintained during the cell cycle. This is especially the case upon DNA replication when these chromatin structures must be disassembled and reassembled (for review, see [4]). One possibility is that parental CENP-A is redistributed to daughter strands during DNA replication, and the gap is immediately replenished by newly synthesized CENP-A, to reestablish a functional kinetochore. In this case, tight coupling of CENP-A deposition to replication of centromeric DNA would be required to avoid misincorporation of histone H3. However, in both S. pombe and mammalian cells there is evidence that CENP-A can be incorporated at centromeres outside S phase [5]–[9]. In S. pombe, expression of Cnp1 peaks in G1-phase prior to canonical histone H3 and centromeres are replicated in early S-phase [10], [11]. In humans, expression of CENP-A peaks in G2 phase [5] and it has been demonstrated that newly synthesized CENP-A is incorporated at centromeres in a discrete window in G1 [12]
Fission yeast centromeres resemble those of vertebrates in that the kinetochore domain is found embedded in pericentric heterochromatin. More precisely, fission yeast centromeres are composed of two domains: the central core region (cnt), where the kinetochore assembles and the outer repeat region (otr), which is packaged as heterochromatin and bound by the HP1 equivalent Swi6 [13], [14]. S. pombe CENP-A (Cnp1) and some histone H3 containing nucleosomes that are dimethylated on lysine 4 of histone H3 are associated with the central core domain. Central domain chromatin structure is distinct; partial digestion with micrococcal nuclease produces a smear pattern when hybridized with a central core probe in contrast to the canonical ladder pattern observed for the rest of the genome. This unusual chromatin structure correlates with a functional state of fission yeast centromeres [11], [15]–[17]. Marker genes inserted into fission yeast centromeres are transcriptionally silenced [18], [19] and alleviation of silencing at either the central core or outer repeat heterochromatin has been correlated with loss of centromere function [19]–[22]. In addition, it has been observed that mutants affecting silencing at the central core have no effect on silencing at the outer repeats. Moreover, most mutants affecting silencing at the outer repeats have little or no effect on silencing at the central core. For example, cells lacking Swi6 display alleviation of silencing at the outer repeats of the centromere with no effect on the central core [19], whereas mutant alleles of the kinetochore component mis6 alleviate silencing at the central core of the centromere but silencing at outer repeat heterochromatin is unperturbed [23]. Alleviation of central core silencing has been used to identify kinetochore proteins and factors that affect Cnp1 incorporation in the central domain [9], [22]; indeed, mutations in cnp1 itself lead to alleviation of central core silencing [24].
Both central core and heterochromatin domains and their associated proteins are required for full centromere activity [25]. The majority of mutants that alleviate silencing (and concomitantly function) of the centromere are specific for only one of the domains. In an effort to probe the functional relationships between the two domains and the two types of chromatin we implemented a screen to identify cos mutants (central core and outer repeat silencing) which alleviate both central core and outer repeat silencing. From this screen, several mcl1 mutants were identified. We have further analyzed the role of Mcl1 and its binding partner, Swi7 that is a catalytic subunit of DNA polymerase α (Polα), in centromere chromatin structures. The mcl1 and swi7 mutants show defective chromatin structure and impaired CENP-A association at the kinetochore domain. Importantly, acetylation of N-terminus tail of histone H4 was aberrantly increased in mcl1 and swi7 mutants, and some of mcl1 phenotypes were partially suppressed by overexpression of some histone deacetylase genes. Finally, Mcl1 and Swi7 formed S-phase foci in the nucleus that overlapped with the replication processivity clamp PCNA. These results suggest that Mcl1 and Polα maintain the hypoacetylated state of kinetochore domain leading to the efficient kinetochore reassembly during DNA replication. We also observed alleviation of transcriptional gene silencing and increased acetylation of histone H4 at the heterochromatin domain, providing an important role of DNA replication machinery in epigenetic inheritance of centromere structures.
Results
Isolation of central core and outer repeat silencing (cos) mutants
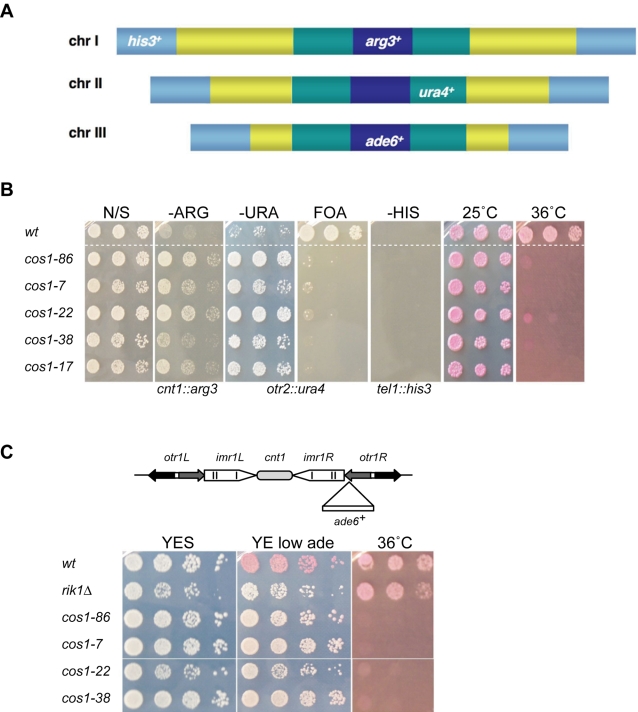
In order to identify candidate factors required for the propagation of chromatin states at centromeres, a screen was performed to isolate fission yeast mutants that alleviate silencing of both central core (kinetochore) and outer repeat (heterochromatin) domains of the centromere. UV-mutagenesis was performed on a strain containing different marker genes inserted at various silent chromatin domains: the central core of centromere 1 (cnt1:arg3+), the outer repeat of centromere 2 (otr2:ura4+), respectively, and a telomere (tel1L:his3+) (Figure 1A) [22]. Mutants were identified that grew faster than wild-type on both -Arg plates and -Ura plates, but did not grow on -His plates, indicating that central core and outer repeat silencing was alleviated, but telomeric silencing remained intact. Thirteen cos (central core and outer repeat silencing) mutants were isolated which fell into 5 complementation groups, cos1 to cos5 (data not shown). Further phenotypic characterization of the cos1 mutants was performed. All cos1 mutants are temperature sensitive and fail to form colonies at 36oC (Figure 1B). cos1 mutants alleviate silencing of the ura4+ marker in the centromere outer repeat of cen2 as evidenced by increased growth on -Ura plates; reciprocally, they fail to grow well on the counter-selective drug on 5-fluoroorotic acid (5-FOA). Outer repeat silencing was also assessed in strains in which ade6+ was inserted in centromere 1 (otr1R(SphI):ade6+); wild-type strains form red colonies on media containing limiting adenine due to transcriptional silencing, whilst heterochromatin mutants such as rik1Δ strongly alleviate outer repeat silencing and form white colonies [14], [26]. All cos1 alleles produced a pale pink colour in this assay suggesting that heterochromatin may not be completely dismantled in these mutants (Figure 1C).
Figure 1. cos1 mutants alleviate central core and outer repeat silencing.
(A) Schematic representation of FY3027 used to isolate mutants defective in central core and outer repeat silencing. FY3027 contains arg3 + at the central core of centromere 1 (dark blue) to monitor alleviation of central core silencing [cnt1:arg3 +], ura4 + at the outer repeat of centromere 2 (green) to monitor alleviation of outer repeat silencing [otr2:ura4 +], his3 + at telomere 1 (light blue) to monitor alleviation of silencing at telomeres [tel1L:his3 +] and ade6 + at the central core of cen3 [cnt3:ade6 +] [22]. Note that the ade6 + marker gene was not used for analysis. (B) Tenfold dilutions were plated onto non-selective (N/S) EMM or appropriate plates. cos1 mutant alleles alleviate silencing at the central core and outer repeats but do not affect silencing at the telomere. cos1 mutants are temperature sensitive for growth and fail to form colonies on Phloxin B at 36°C. (C) cos1 mutants alleviate silencing of ade6 + gene inserted at otr1R. (upper panel) Schematic diagram of the ade6 + gene inserted at the outer repeats of centromere 1. (lower panel) Strains were plated on low adenine supplemented YES plates. In wild-type cells (FY1180), ade6 + is silenced and colonies are red in colour. cos1-86, cos1-7, cos1-22 and cos1-38 were found to partially alleviate silencing at the outer repeats and were pink in colour.
cos1 mutants display chromosome segregation defects including lagging chromosomes
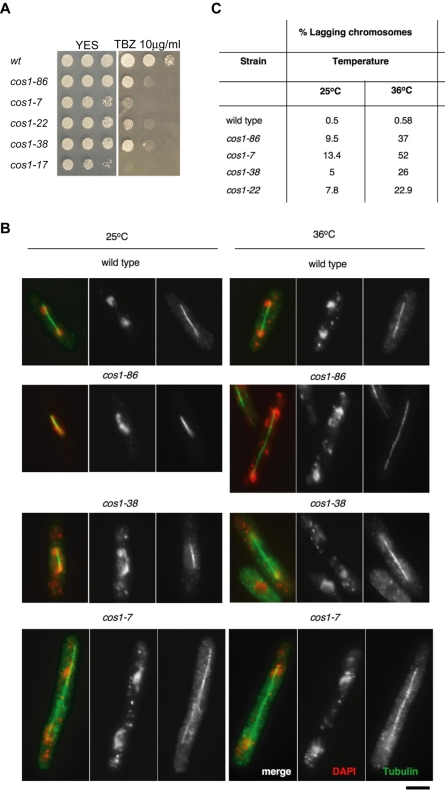
cos1 mutants were found to show sensitivity to the microtubule-disrupting drug Thiabendazole (TBZ) (Figure 2A). Mutants in centromere function are often impaired in their ability to interact with microtubules and the presence of TBZ exacerbates this defect, reducing cell viability, but has only a very mild effect on wild type growth. Mutant strains that alleviate central core silencing such as sim4 or outer repeat silencing, such as swi6Δ, display defective chromosome segregation, with high rates of lagging chromosomes on late anaphase spindles [19]–[22], [27]. To investigate whether cos1 mutations result in chromosome segregation defects or abnormal cell morphology, cos1 mutants and wild-type strain were harvested from log phase cultures grown at permissive and restrictive temperatures (6 hours at 36oC). Cells were fixed and stained with anti-α-tubulin antibody to decorate microtubules and 4-,6-diamidino-2-phenylindole (DAPI) to stain for DNA. Cells were then viewed and analyzed for the presence of segregation defects (n = 200). Segregation defects were rarely seen in wild-type strain (approximately 0.5% at 25°C and 36°C). cos1 mutants displayed multiple forms of chromosome missegregation, including lagging chromosomes at anaphase and unequal segregation of DNA (Figure 2B). The frequency of lagging chromosomes was determined for each cos1 mutant allele, with cos1-7 showing the highest degree of segregation defects (13.4% at 25°C and 52% at 36°C) (Figure 2C). Lagging chromosomes and unequal segregation may suggest a role for the cos1+ gene product in establishing correct microtubule-kinetochore attachments or biorientation of the centromere.
Figure 2. Chromosome segregation defects in cos1/mcl1 mutants.
(A) cos1 mutants are sensitive to the microtubule destabilizing drug TBZ. Tenfold serial dilutions were spotted onto YES media containing 10 µg/ml TBZ. (B) Wild-type and cos1/mcl1 mutant cells were grown at 25°C or at 36°C for 6 hours before fixing and staining with anti-α-tubulin antibody (green) and DAPI stained DNA (red). Bar, 3 µm. (C) Frequency of lagging chromosomes in cos1/mcl1 mutants. Lagging chromosomes were counted in cells with late anaphase spindles and are given as a percentage of total number of cells in anaphase (n = 200). Lagging chromosomes were very rare in wild-type (0.5–0.58%) at both temperatures.
Cloning of cos1+ and identification of cos1 mutations
cos1+ was cloned by complementation of the temperature sensitivity of cos1-86 using an S. pombe genomic library. The complementing plasmid also reimposed silencing at both centromeric domains, and was found to contain two ORFs: one encoding a putative DNA-directed RNA polymerase III complex subunit, rpc82+ (SPAPB1E7.03) and a 2,548 bp ORF (SPAPB1E7.02) encoding mcl1+ [28]. The entire region covering the rpc82 and mcl1 ORFs were sequenced in wild-type and cos1 mutants. cos1 mutants were found to have no mutation in the rpc82 ORF but the following mutations in the mcl 1 ORF: cos1-7, G242R (G to A mutation); cos1-17, V233E (T to A mutation); cos1-22, A574D (G to A mutation); cos1-38, G622E (G to A); cos1-86, Q765STOP (C to T), indicating that cos1 is allelic to mcl1.
Mcl1 was previously identified in a screen for mutants that exhibit frequent mini-chromosome loss. Mcl1 is a homologue of budding yeast Ctf4 and is conserved from yeast to human and members of this family contain WD40 repeats in their N-terminus that might provide a protein-protein interaction surface. The mcl1-1 mutant shows aberrant mitosis such as unequal segregation and lagging chromosomes, and exhibits a defect in sister chromatid cohesion at centromeres that holds sister kinetochores together until anaphase [28]. cos1 mutants isolated here showed similar phenotypes (Figure 2).
In parallel, we performed a screen for mutants that are synthetically lethal with the null mutation of the rad2 + gene (slr). Rad2 is the S. pombe homologue of Rad27/FEN1 nucleases involved in Okazaki fragment processing during lagging strand synthesis. Among these slr mutants, the slr3-1 was found to be a mutation in the mcl1 gene (mcl1-101) and this mutant showed sensitivity to DNA damaging agents such as MMS and HU. In addition, the mcl1+ gene displayed genetic and physical interactions with the swi7 + gene that encodes the largest subunit of Polα, suggesting Mcl1 is involved in lagging strand DNA synthesis [29].
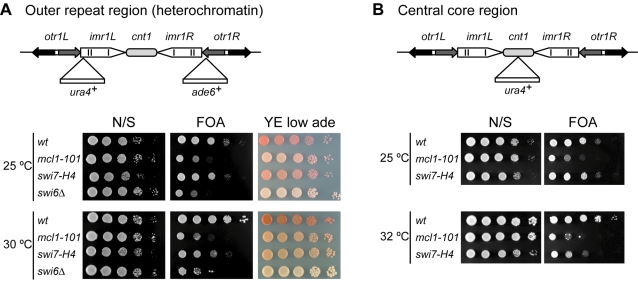
Silencing is alleviated at the two distinct centromeric domains in mcl1-101 and swi7 mutants
To determine whether the mcl1-101 has defects in centromeric silencing like the other mcl1 mutant alleles, we introduced the mcl1-101 mutation into indicator strains. Strain FY1193 harbors the ura4 + and ade6 + genes inserted in the outer repeat imr and otr elements of cen1, respectively [19]. The mcl1-101 mutant harboring maker genes at the centromere was more sensitive to 5-FOA and was pink on YE low adenine plate compared to the wild-type strain which gave a red color (Figure 3A). The level of alleviation in mcl1-101 cells was comparable to that of swi6Δ cells at imr (FOA), but lower at otr (low ade). Previously we have revealed strong interaction between mcl1-101 and swi7-H4, a temperature sensitive allele of swi7+ [29]. Therefore we tested transcriptional gene silencing at heterochromatic regions in this mutant. As reported previously [30], swi7-H4 cells showed alleviation of transcriptional gene silencing at heterochromatic regions (Figure 3A). No dramatic reduction of the heterochromatic marks Swi6 or H3K9me2 on centromeric sequences was apparent in mcl1 mutants (T.N., E.D., A.P., Y.T., and R.A., unpublished data). Therefore, Mcl1 might function at this domain either independently or downstream of Swi6. Recently, Mamnun et al. isolated Mcl1 as an interacting partner of the F-box protein, Pof3, which is a substrate adapter of the SCF ubiquitin ligase and is required for genome integrity. They reported that only a slight change in Swi6-GFP localization could be detected in an mcl1Δ mutant [31], consistent with our observations. As the mcl1-101 mutation genetically interact with rad2Δ or dna2-C2 mutations [29], we examined transcriptional gene silencing in these mutants. However, these mutants did not show any defect in heterochromatin silencing (data not shown), suggesting that Mcl1 and Swi7 might have a specific role in centromeric chromatin structures.
Figure 3. The mcl1-101 and swi7-H4 mutants display alleviation of transcriptional gene silencing at both central core and outer repeat regions.
(A) (upper panel) The schematic diagram of ura4 + and ade6 + genes inserted into imr1L and otr1R, respectively. Note that the ura4 + gene is inserted into heterochromatic domain, which is defined as outside of tRNA genes (vertical lines) [23]. (lower panel) Tenfold serial dilutions were spotted. (B) (upper panel) The schematic diagram of the ura4 + gene inserted into cnt1. (lower panel) Tenfold serial dilutions were spotted.
To examine whether the mcl1-101 and swi7-H4 mutations affect the maintenance of the chromatin structure at the central core region, each mutation was introduced into cnt1:ura4 + strain in which ura4 + marker gene is inserted into cnt1 [19]. As shown in Figure 3B, these mutants were defective in silencing at the kinetochore domain. Thus, both mcl1-101 and swi7-H4 mutants exhibit the cos phenotype.
mcl1 and swi7 mutants have defects in the specialized chromatin structure at the kinetochore
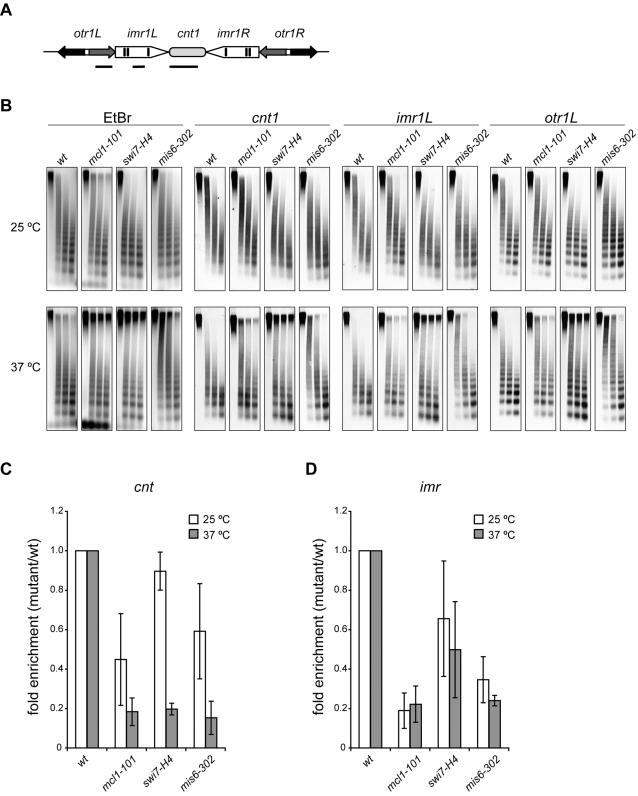
The chromatin structure of the central core domain is known to be unusual since partial micrococcal nuclease (MNase) digestion results in a smeared pattern, rather than the ladder pattern that is indicative of regular nucleosomal packaging [15], [17]. This specialized structure is only associated with a functional centromere context [16]. Importantly, it is reported that integrity of this structure correlates with that of central core silencing [22]. To determine whether Mcl1 and Polα are required for the specialized chromatin structure, chromatin was partially digested with MNase and subjected to Southern hybridization (Figure 4B). In wild-type cells, a typical nucleosomal ladder was observed with a probe corresponding to the heterochromatic domain, outer repeat (otr1L, Figure 4A). In contrast, smeared digestion patterns were detected with probes to central core domain (cnt1 and imr1L, Figure 4A), although wild-type cells shifted to 37°C showed a slightly ladder-like pattern. In mcl1 cells, the smeared patterns of cnt1 and imr1L were partially replaced with a ladder pattern at permissive temperature and the ladder-like pattern of imr1L was significantly enhanced at 37°C compared to wild-type cells. Similar results were obtained in swi7 cells, but only at 37°C. Complete replacement of the smeared pattern was observed in mis6-302 cells at 37°C as reported previously [32]. These data suggest that Mcl1 and Polα are necessary for assembly of the specialized chromatin structure at the central core domain.
Figure 4. The specialized chromatin structure and association of Cnp1 at central core region is impaired in mcl1-101 and swi7-H4 mutants.
(A) Locations of probes used in Southern hybridization are depicted as horizontal lines. (B) The cells were harvested from cultures incubated at 25°C or 37°C for 6 hrs and permeabilized by enzyme treatment and chromatin was partially digested with MNase. DNA was extracted, separated on agarose gel, and subjected to Southern hybridization using probes shown in (A). Smeared digestion patterns characteristic of central core region (cnt1 and imr1L) were partially replaced with ladder patterns in mcl1-101 and swi7-H4 mutants. Strains were wild-type (JY746), mcl1-101 (NYSPC41), swi7-H4 (TN310) and mis6-302 (NYSPL59). (C) ChIP was performed using the antiserum raised to Cnp1. The cells were incubated at 25°C or 37°C for 6 hrs and then fixed. The ratio of ChIPed DNA to input DNA was calculated as described in Materials and methods and average fold enrichment compared to wild-type strain from three experiments is shown. Localization of Cnp1 was decreased even at permissive temperature in the mcl1-101 mutant, and was further decreased at restrictive temperature. Decreased Cnp1-association was also seen at 37°C in the swi7-H4 mutant. Error bars represent the standard deviation. Strains were same as described in (B).
Genetic interactions with kinetochore mutants
To further investigate the involvement of Mcl1 in the kinetochore function, we examined genetic interactions with kinetochore mutants. S. pombe Cnp1 is essential for faithful chromosome segregation and constitutively associates with the centromere central domain to assemble unique nucleosomal structure [11]. The cnp1-FH allele was obtained by tagging C-terminus of endogenous cnp1+ gene with 3x FLAG and 6x His (FH). The cnp1-FH strain is viable but shows slow growth and sensitivity to TBZ (data not shown). The mcl1-101 mutant was crossed with the cnp1-FH, but the double mutant could not be recovered even at permissive temperature, 25°C (Figure S1A), suggesting that mcl1-101 and cnp1-FH are synthetically lethal.
Next, we examined genetic interactions with mutants deficient in Cnp1 loading to kinetochores. In fission yeast it has been proposed that two distinct pathways mediate Cnp1 loading: a Mis6-dependent pathway and an Ams2-dependent pathway [7]. Mis6 is a homologue of vertebrate CENP-I that constitutively localizes to kinetochores and is required for Cnp1 loading during S and G2 phase [32]. Although the mcl1-101 mutation was viable in combination with a temperature sensitive allele of mis6 +, mis6-302, the double mutant showed a decreased permissive temperature compared to each single mutant (Figure S1B). Ams2 is a cell cycle regulated GATA factor required for Cnp1 loading during S phase [33]. Synthetic lethality was observed between mcl1-101 and a null allele of ams2 (Figure S1C).
Mis16 is a homologue of human RbAp46 and RbAp48, which are found in some histone deacetylase (HDAC) complexes and CAF-1 complex. Mis18 is required for histone hypoacetylation at the kinetochore domain and for the Mis6-dependent Cnp1-loading pathway [34]. mis16 was tagged with myc at the endogenous locus (mis16-myc) and showed no detectable phenotype such as temperature sensitivity or TBZ sensitivity (data not shown). However, construction of a mis16-myc mcl1-101 strain revealed that myc-tagged Mis16 is not able to cover all the functions of the wild-type Mis16 as the strain showed a drastic decrease in the growth permissive temperature (Figure S1D), indicating that mis16 and mcl1 have a synthetic phenotype. Thus, Mcl1 has close functional relationships with Cnp1 and factors involved in its deposition.
Impaired association of Cnp1 to central core region in mcl1 and swi7 mutants
Impaired chromatin structure (Figure 4B) and genetic interactions with Cnp1 and its loading factors (Figure S1) suggested that Mcl1 might be required for Cnp1 association with the central core region. To determine whether Cnp1 association is impaired in mcl1 cells, chromatin immunoprecipitation (ChIP) was performed using antiserum raised to Cnp1 (Figure 4C). The fraction of cnt DNA found in Cnp1 immunoprecipitates was 2-fold lower, even at permissive temperature, in the mcl1 mutant compared to wild-type, and was further decreased at restrictive temperature. Decrease in cnt DNA in Cnp1 immunoprecipitates was also observed in the swi7 mutant only at restrictive temperature. The fraction of imr DNA was severely decreased regardless of growth temperature in the mcl1 mutant and, to a lesser extent, in the swi7 mutant.
Loss of Cnp1 from the central core is accompanied by increase of histone H3 in the ams2Δ mutant [33], indicating that histone H3 is misincorporated instead of Cnp1. ChIP was also performed using antibody against histone H3. cnt and imr sequences in histone H3 immunoprecipitates were increased in the mcl1 and swi7 mutants indicating that histone H3 is misincorporated into central core region in these mutants (Figure S2). These observations suggest that Mcl1 and Polα are important for efficient Cnp1 incorporation into chromatin of the central core domain. Previous observations that an mcl1Δ mutation affected formation of Cnp1-GFP foci at centromeres also support our conclusion [31].
Aberrant acetylation of histone H4 at centromere in mcl1 and swi7 mutants
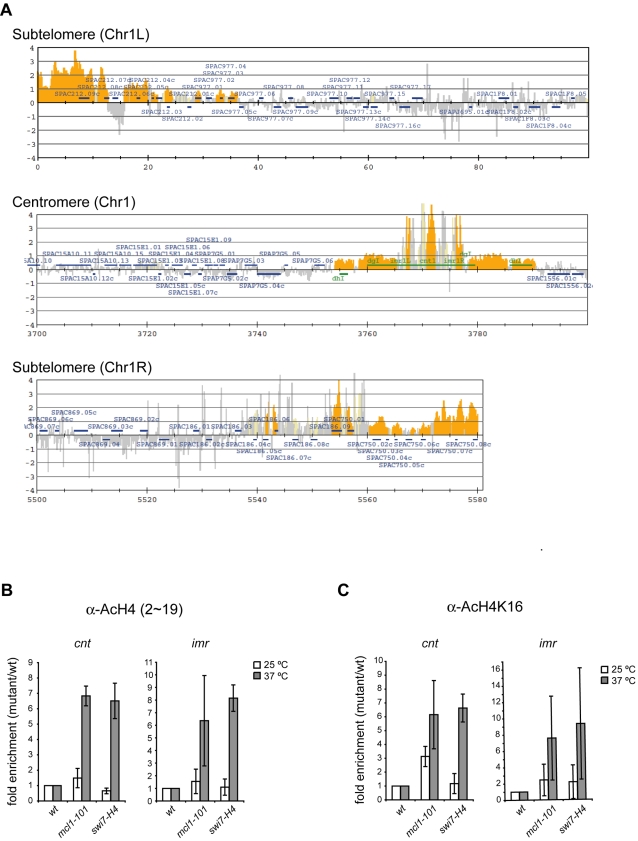
The acetylation of N-terminal tails of histone H3 and H4 is maintained at a lower level in the central core region than in coding regions. Here, mcl1 and, to a lesser extent, swi7 mutants showed sensitivity to the HDAC inhibitor, Trichostatin A (Figure S3), suggesting that Mcl1 and Polα might be required for maintenance of histone hypoacetylation. To examine this possibility, we performed ChIP-on-chip assays using antiserum raised to a peptide corresponding to amino acid 2-19 of histone H4 acetylated at K5, K8, K12, and K16 (H4-KN). We determined the acetylation status of histone H4 across the entire genome (Figure S4A). Acetylation of histone H4 was found at the promoter regions of most genes, whereas H4 was hypoacetylated at centromeric regions. Although acetylation peaks similar to those in wild-type were observed at most genes in the mcl1 mutant, histone H4 acetylation was elevated at the centromere region in mcl1 mutant (Figure S4B). Comparison of H4-KN data obtained from mcl1 and wild-type indicates that acetylation level was increased at centromere and sub-telomeric regions in the mcl1 mutant (Figure 5A).
Figure 5. Acetylation of histone H4 is aberrantly increased in the central kinetochore domain in mcl1 and swi7 mutants.
(A) ChIP-on-chip was performed using the antiserum raised to a peptide corresponding to amino acid 2–19 of histone H4 that is acetylated at K5, K8, K12, and K16 (AcH4-KN). The cells were prepared as described in Figure 4C. Values obtained from AcH4-KN in the mcl1 mutant were divided by those in wild-type strain. The orange shading represents the binding ratio of loci that show significant enrichment as described previously [68]. Representative results for subtelomeric and centromeric regions of chromosome I are shown. (B,C) ChIP was performed using AcH4-KN (B) and AcH4-K16 (C), respectively. The cells were prepared as described in Figure 4C. Values obtained in the mutants were normalized to those obtained in wild-type strain. Values are further normalized to euchromatic lys1 locus. Average fold enrichment compared to wild-type are obtained from two repetitions. Strains were wild-type (JY879), mcl1-101 (NYSPC52), swi7-H4 (TN403).
To confirm these results, we performed independent ChIP analyses and quantified the precipitated DNA by quantitative real-time PCR (qPCR). Consistent with the ChIP-on-chip data, the level of H4-KN acetylation at the kinetochore domain was increased in the mcl1 mutant compared to the wild-type strain. A similar increase in acetylation was seen in the swi7 mutant (Figure 5B). When we used the specific antibody against acetylated H4-K16, acetylation at this residue was also increased in both mutants at restrictive temperature and, to a lesser extent, at permissive temperature in the mcl1 mutant (Figure 5C). These results suggest that Mcl1 and Polα are required for the hypoacetylated state of the histone H4 N-terminal tail including K16 in the kinetochore domain of fission yeast centromeres.
Multicopy suppression of mcl1 phenotypes by histone deacetylase genes
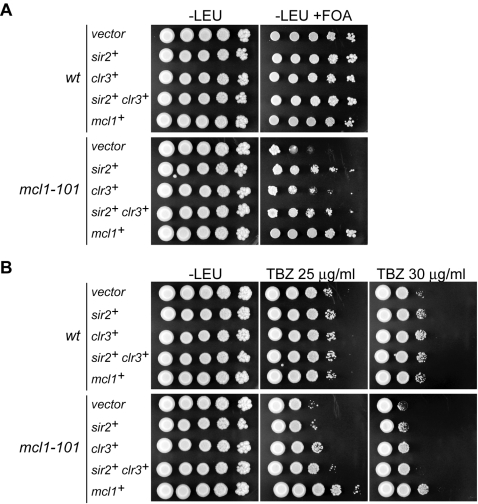
Increased acetylation of histone H4 in the mcl1 mutant raised the possibility that Mcl1 might regulate HDAC(s) to maintain the hypoacetylated state of the kinetochore domain. The S. pombe genome encodes six HDACs that belong to three distinct classes. Among these HDACs, Clr3, Clr6 and Hst4 are required for transcriptional gene silencing at the central core region [35], [36]. In addition, the HDAC Sir2 preferentially localizes to the central core region [37]. To examine whether these HDACs are functionally related to Mcl1, multi-copy plasmids carrying each of the HDAC genes were introduced into cnt1:ura4+ mcl1-101 cells, and sensitivity to 5-FOA and TBZ, were examined. As shown in Figure 6A, alleviation of silencing in the mcl1 mutant was partially suppressed by multi-copy plasmids of sir2+ gene and, to a lesser extent, of the clr3+ gene. Furthermore, TBZ sensitivity was partially suppressed by the clr3+ plasmid and simultaneous introduction of sir2+ and clr3+ genes further relieved the sensitivity (Figure 6B). These results suggest that Mcl1 functionally interacts with Sir2 and Clr3 HDACs in maintaining centromere integrity.
Figure 6. Multi-copy suppression of mcl1-101 phenotypes by sir2 + and clr3 + HDAC genes.
Wild-type and mcl1-101 mutant harboring the ura4 + insertion at the cnt1 were transformed with multi-copy plasmid carrying sir2 +, clr3 +, or both, respectively. Tenfold serial dilutions were spotted onto EMM-Leu and EMM-Leu containing 5-FOA (A) or EMM-Leu containing indicated concentration of TBZ (B). In panel A, sir2 + and, to a lesser extent, clr3 + partially suppressed 5-FOA sensitivity of the mcl1-101 mutant. In panel B, clr3 + partially suppressed TBZ sensitivity of the mcl1-101 mutant, and sir2 + also relieved the sensitivity only in the presence of clr3 +.
Localization of Mcl1 to replication foci
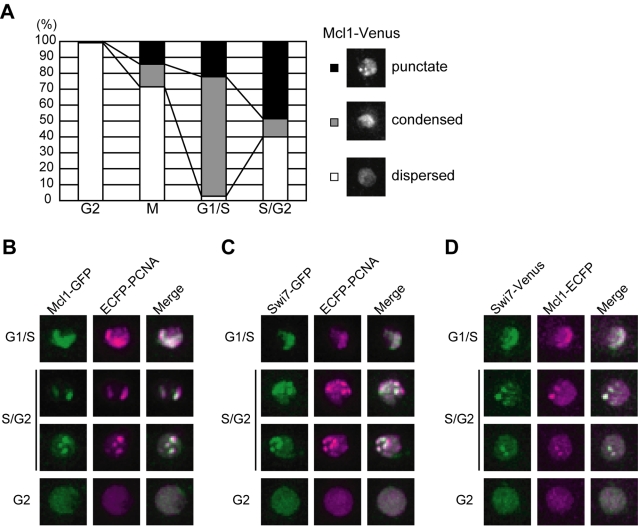
Mcl1 was previously shown to be a constitutive nuclear protein that associates with chromatin from G1 to S phase [28]. To further examine the nuclear localization of Mcl1, the endogenous mcl1+ gene was C-terminally tagged with the fluorescent marker Venus [38]. The mcl1+-venus cell did not show any phenotypes such as HU and MMS sensitivity (data not shown). Mcl1-Venus localized to the nucleus throughout the cell cycle, and a portion of cells showed a punctate (Figure 7A, top) or condensed pattern (Figure 7A, middle) of Mcl1-Venus signal. To determine which stage of the cell cycle shows such localization pattern, mcl1+-venus cells were stained with Hoechst 33432 to visualize nucleus and septum, and roughly classified into four cell cycle stages (Figure 7A). A diffuse Mcl1-Venus signal was observed in the nucleus of most cells with a single nucleus (G2-phase). Condensed localization of Mcl1-Venus peaked in cells with a septum (G1/S-phase) and decreased in separating cells (S/G2-phase). The punctate localization of Mcl1-Venus increased from G1/S-phase and accounted for about half of S/G2 cells. These results indicate that Mcl1 changes its nuclear localization pattern during DNA replication.
Figure 7. Mcl1 localizes to replication foci during S-phase.
(A) Mcl1 changes its nuclear localization during S-phase. Cells expressing Mcl1-Venus from the endogenous locus were classified into 4 cell cycle stages according to their morphology. The percentage of cells showing punctate (closed bar), condensed (shaded bar), and dispersed (open bar) localization of Mcl1-Venus in various cell cycle stages are shown (left panel). Representative photographs of each localization pattern are shown (right panel). (B–D) Mcl1 and Swi7 colocalize with PCNA in the S-phase nucleus. Mcl1-GFP and ECFP-PCNA (B), Swi7-GFP and ECFP-PCNA (C), and Swi7-Venus and Mcl1-ECFP (D) are shown. Merged images are also shown.
In vertebrate cells, DNA polymerases and their accessory proteins such as PCNA cluster together and form nuclear foci called “replication factories” [39]. Similar nuclear structures are also observed in budding and fission yeasts. In S-phase budding yeast cells, POLα, POLε and PCNA tagged with GFP or EYFP forms globular nuclear signals, and more importantly, DNA replication actually occurs in these foci [40]. Fission yeast PCNA-ECFP also forms foci during DNA replication [41]. Since Mcl1 physically interacts with Polα and associates with chromatin in a G1/S-phase specific manner, we tested whether the Mcl1 foci formed during S-phase corresponds to replication factories. PCNA-ECFP was expressed from its native promoter together with Mcl1-GFP. Multiple nuclear foci of ECFP-PCNA were observed in S-phase cells as previously reported, and these PCNA foci overlapped with those of Mcl1-GFP (Figure 7B). We also examined cells expressing Swi7-GFP and ECFP-PCNA (Figure 7C). Consistent with the physical interaction between them, Mcl1 and Swi7 also colocalized in S-phase nuclei (Figure 7D). Together, these observations strongly suggest that Mcl1 localizes to the replication fork, interacting with Polα. We also examined the relationship between kinetochores and Mcl1 foci formed during S-phase. ECFP-Cnp1 was expressed from nmt41 promoter in mcl1 +-GFP cells, but the signals did not colocalize in most cells (data not shown), which might reflect the short time window in which centromeres are replicated.
Discussion
In this study, we identified several cos mutants, which alleviate silencing at both the central kinetochore and outer heterochromatin regions of fission yeast centromeres. cos1 is allelic to mcl1 which was previously show to be important for chromosome replication, segregation and sister chromatid cohesion. In addition, we demonstrate that a mutant in the catalytic subunit of Polα, Swi7, also shows the cos phenotype. In both mcl1 and swi7 mutants the levels of the centromeric histone Cnp1 associated with the central domain are reduced and there is partial loss of the unique chromatin structure of this region. Although outer repeat silencing was alleviated, no reduction of the heterochromatic marks Swi6 or H3K9me2 on centromeric sequences was apparent (T.N., E.D., A.P., Y.T., and R.A., unpublished data). Interestingly, the acetylation of histone H4 N-terminal tails was aberrantly increased at both domains. Finally, we showed that Mcl1 co-localized with both PCNA and Swi7 mainly during S-phase. From these results, we propose that Mcl1 and Swi7 are involved in propagation of centromereic chromatin structures during DNA replication by regulating the level of histone acetylation. Our observation that the overexpression of Sir2 and/or Clr3 partially suppressed some of mcl1 phenotype suggests that Mcl1 might control these HDACs during DNA replication.
The role of Mcl1 in regulation of specific chromatin structures
Here we show that Mcl1 and Polαhave roles in regulation of specific chromatin structures: the central core and outer heterochromatin regions of fission yeast centromeres. Evidence presented here suggests that Mcl1 may act on central core chromatin by ensuring that Cnp1 chromatin is established and maintained at centromeres during replication. In fission yeast, Cnp1 incorporation into centromeres depends on two distinct pathways in S and G2; Mis6, a homologue of CENP-I in vertebrates, is required in both S- and G2-phase loading [7], [11] and a cell cycle-regulated GATA factor, Ams2, is also critical for Cnp1 loading during S-phase [7], [33]. In addition, Mis16 that is homologous to human RbAp46 and RbAp48, which are found in some histone deacetylase (HDAC) complexes and with histone chaperone CAF-1 [42]–[46], is also required for Cnp1 loading [34]. As the mcl1-101 mutation is synthetically lethal with both mis6-302 and ams2Δ and also shows genetic interactions with mis16, it may be that Mcl1 performs a function in Cnp1 loading that is independent of both S and G2 loading pathways.
Mcl1 may also regulate chromatin at centromeres by influencing the levels of histone acetylation at central core and heterochromatin domains. In fission yeast, a global function in chromatin regulation has also been reported for the CHD (chromo-ATPase/helicase-DNA binding) domain-containing chromatin remodeling factor, Hrp1 [47]. Interestingly, human CHD proteins, CHD3 and CHD4, were co-purified with HDAC1/2 to form a protein complex called NuRD (nucleosomal remodeling histone deactylase), and ATP-dependent remodeling activity is required for deacetylation of oligonucleosomal histones [43]–[45]. The S. pombe hrp1Δ mutant, like mcl1-101, shows alleviation of silencing at both domains of the centromere and the mat locus, and is defective in Cnp1 incorporation into the central core region, concomitant with aberrant acetylation of histone H3 and H4. Notably, Hrp1 localizes to the kinetochore domain in early S-phase when centromeres are replicated [10], suggesting that Hrp1 acts during DNA replication [47]. One possibility is that Hrp1 may enhance histone deacetylation by modulating chromatin structure at the central core region. It is also possible that Mcl1 might control chromatin remodeling in concert with Hrp1, or the HDACs Sir2 and Clr3, to enhance subsequent histone deacetylation.
Histone deacetylation during DNA replication
Does the removal of histone acetylation during or soon after replication have any importance for the epigenetic inheritance of chromatin structures? This is the case in human cells. In mammalian cells, DNA synthesis occurs at discrete sites in the nucleus called replication foci or replication factories [39]. In early S-phase, numerous foci of pulse labeled BrdU are distributed throughout the nucleus except for nucleoli, and these foci correspond to transcriptionally active chromatin. In late S-phase, replication foci can be found along the nuclear periphery and perinucleolar regions, and finally at a few larger sites, which represent the replication of heterochromatic regions. Interestingly, acetylation at K5 and K12 in histone H4, which is a hallmark of newly synthesized histones, is removed within 20 min after DNA synthesis at late replicating foci, but not early replicating foci, and this immediate deacetylation is inhibited by Trichostatin A treatment [48]. These findings suggest that certain HDAC(s) act(s) only on late replicating foci to couple the incorporation of highly-acetylated histones to subsequent deacetylation events.
Furthermore, in human cells it has been demonstrated that late replication foci contain a HDAC and also that PCNA interacts with the DNA methyltransferase DNMT1, which localizes to both early and late replication foci [49]. Intriguingly, DNMT1 also interacts with HDAC2, and they colocalize only in late replicating foci, suggesting that HDAC activity is preferentially recruited to late replication foci by the replication machinery. We have shown here that Mcl1 colocalizes to S-phase nuclear foci of PCNA, which might correspond to mammalian replication foci. Thus, Mcl1 might regulate the recruitment of a HDAC to the kinetochore domain in certain replication foci, though fission yeast centromeres are known to be replicated early in S-phase. Since the level of histone acetylation is determined by a balance between HDAC and HAT activities, it remains possible that Mcl1 is essential to prevent HAT activity from acting on kinetochore domain.
Mcl1 and the DNA replication fork
We demonstrate that the Mcl1 nuclear foci formed during S-phase overlap with those of PCNA and Swi7. Although we currently have no direct evidence, we suppose that Mcl1 localizes to the progressing DNA replication fork for the following reasons. Firstly, Mcl1 interacts physically with Swi7 in vivo and in vitro and binds to chromatin tightly during S-phase [28], [29], [50]. Secondly, the budding yeast homologue, Ctf4, moves along chromosomes with the replication fork as revealed by ChIP-on-chip experiments [51]. Finally Ctf4 is a component of replication progressive complex (RPC) including other replication proteins such as the MCM proteins and the GINS complex [52]. Because we were not able to detect stable colocalization of Mcl1-GFP foci with ECFP-Cnp1 foci in asynchronous culture (data not shown), Mcl1 might localize to centromere chromatin only in a discrete window when centromeric DNA is replicated.
We have previously shown that the mcl1 mutant shows genetic interactions with mutations defective in Okazaki fragment maturation such as rad2 (RAD27/FEN1) and dna2, and it accumulates double strand breaks after completion of bulk DNA synthesis, suggesting that Mcl1 is involved in lagging strand DNA synthesis [29]. It is possible that defective centromere chromatin structures observed in mcl1 and swi7 mutants might be a secondary effect of an unstable DNA replication fork. However, the rad2Δ mutant did not show any defect in kinetochore and heterochromatin silencing (data not shown), suggesting that Mcl1 and Swi7 might have a specific role in maintaining centromeric chromatin structures.
An understanding of the roles of Mcl1 and Swi7 in the maintenance of centromeric chromatin structures and how this relates to their roles in DNA replication remains to be uncovered. The swi7-H4 allele used in this study (G889D) is located within homology box VI, which is in the conserved nucleotide binding domain found in all DNA polymerases [30]. Another swi7 mutation, swi7-1, maps to the non-conserved C-terminal region and shows a defect in heterochromatic silencing [53]. This mutation also showed alleviation of kinetochore domain silencing (data not shown), suggesting that Swi7 has a role in centromeric chromatin assembly independent of its catalytic activity. All six alleles of mcl1 show defects in centromeric silencing and we have shown that mcl1-101 has defects in both DNA replication and maintenance of chromatin structures (this study and [29], [31]). To understand the role of Mcl1 in kinetochore assembly, further experiments such as isolation of separation of function mutations, which show a defect in only maintenance of chromatin structures or DNA replication, might be required. Alternatively, all phenotypes observed in the mcl1-101 mutant could be attributable to loss of the sole function of Mcl1 (for example, regulation of histone deacetylation) and thus be inseparable.
Mcl1 and budding yeast Ctf4 have conserved functions in chromosome metabolism
The budding yeast homologue of Mcl1, Ctf4, is required for chromosome stability [54], [55] It physically interacts with Pol1, a catalytic subunit of Polα in budding yeast [56], [57]. Ctf4 travels along chromosomes with the replication fork [51]; deletion of CTF4 results in premature separation of sister chromatids [58]. These findings suggest that Ctf4 is required for establishment of sister chromatid cohesion during S-phase. Interestingly, deletion of CTF4 also shows defects in transcriptional silencing at the cryptic mating type locus, HMR, and at telomeric regions [59], suggesting that Ctf4 is involved in the regulation of chromatin structures as Mcl1 is. The molecular function of Ctf4, however, remains unclear, and it is intriguing to address whether Ctf4 is implicated in modulating the level of histone acetylation. In addition, Ctf4 competes for binding to Pol1 with the Spt16 subunit of FACT complex, which removes H2A-H2B dimers during transcript elongation and is also required for DNA replication [60]. Deletion of the S. pombe FACT subunit Pob3 causes alleviation of silencing at otr, although the effects at cnt are less clear-cut [61]. Therefore, our observations reported here strengthen the importance of Polα and its accessory proteins, such as FACT and Mcl1/Ctf4, in faithful propagation of chromatin structures during DNA replication.
Materials and Methods
Strains, plasmids, and growth conditions
S. pombe strains used in this study is listed in Table S1. S. pombe cells were routinely grown in YES, EMM or PMG supplemented with appropriate nutrients. Standard genetic manipulations were used as previously described [62]. A LEU2 + S. pombe genomic library was kindly provided by Chikashi Shimoda. The mis16+, cnp1+, mcl1+, and swi7+ were tagged at their endogenous loci by PCR-based tagging modules as described previously [63]. For C-terminal tagging with the Venus gene, The PacI-AscI fragment containing the gfp gene of pFA6a-GFP-kanMX6 was replaced with the Venus gene (a gift from Atsushi Miyawaki) to give pFA6a-Venus-kanMX6. The pFA6a-Flag His (FH)-kanMX6 was kindly provided by Takashi Morishita. The nucleotide sequences of primers used here are listed in Table S2.
For deletion of the ams2+ gene, the genomic SacI-HindIII fragment containing the ams2+ gene was amplified from 972h- with Ams2-F0 and Ams2-R0, and cloned into SacI and HindIII sites of pUC19 to give pUC19-Ams2. The 1.4-kb fragment containing was amplified from pFA6a-kanMX6 [63] with KanMX6-F0 and KanMX6-R0, and digested with SmaI. The BstXI fragment containing most ORF of ams2+ gene in pUC19-ams2 was replaced with the 1.4-kb PTEF-kanr-TTEF fragment by blunt-end ligation to give pUC19-ams2::kanr. To introduce multi-copy of HDAC genes, the genomic fragments containing sir2+ and clr3+ genes were amplified from 972 h − strain with Sir2-F0 and Sir2-R0, Clr3-F0 and Clr3-R0, respectively. These fragment were digested with PstI (sir2+) and BamHI (clr3+), and cloned into PstI and BamHI sites of pSP102 (ars2004, LEU2) [64] to give pSP102-Sir2 and pSP102-Clr3, respectively. The 3.5-kb BamHI fragment containing the clr3+ gene was excised from pSP102-Clr3, blunt-ended with T4 DNA polymerase, and cloned into the SmaI site of pSP102-Sir2 to give pSP102-Sir2-Clr3. The nucleotide sequences of primers used here are listed in Table S2.
Isolation and cloning of cos mutants
Wild-type strain (FY3027 h+) was plated either onto medium lacking arginine (-Arg) and then onto medium lacking uracil (-Ura) or directly on to double selection medium lacking both arginine and uracil (-Arg, -Ura) and mutagenised by UV (3–5 mJ, 50–80% killing). Plates were incubated at 25oC for 5–20 days and fast growing colonies (184 colonies from –Arg plates, 54 colonies from -Arg, -Ura plates) were picked and streaked onto –Arg plates to retest for alleviation of silencing. Fast growing colonies were subsequently picked from these –Arg plates and streaked onto –Ura plates to assay alleviation of silencing at outer repeats (otr). In total, 13 cos mutants that alleviate silencing at both the central core and the outer repeat domains were isolated.
In order to clone the cos1+ gene, a LEU2 + S. pombe genomic library (gift from Chikashi Shimoda) was transformed into cos1 mutants and cells were screened for rescue of their ts phenotype. Transformants were plated on minimal medium lacking leucine and containing phloxin (0.02% v/v). After 5 days growth at 25°C, plates were shifted to 36°C for 1–2 days, to select for colonies that were now capable of growing at this temperature. At 36°C, 1 single colony out of 18,000 cos1-86 colonies analysed was pale pink in color and could grow at the restrictive temperature.
Micrococcal nuclease assay
Micrococcal nuclease assay was performed as described previously [65]. MNase-digested samples were separated on 1.5% agarose gel and analyzed by Southern hybridization using AlkPhos Direct Labelling and Detection System (Amersham). Since the efficiency of MNase digestion varied among strains, samples that show a similar digestion efficiency judged by EtBr staining were used for analysis. The nucleotide sequences of primers used for amplifying probe DNA are listed in Table S2.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as described previously [66]. Antibodies used were antiserum raised to N-terminal peptide of Cnp1, α-acetyl-histone H4 antiserum (Upstate), α-acetyl-histone H4 (Lys16) antiserum (Upstate), and α-histone H4 pan antibody (Upstate), respectively. Quantification of ChIPed DNA was performed by real-time PCR. The nucleotide sequences of primers used in real-time PCR are listed in Table S2.
ChIP-on-chip was carried out using IVT amplification method as described previously [67]. All the array data are available at GEO database (http://www.ncbi.nlm.nih.gov/projects/geo/) through the accession number of GSE11102.
Fluorescence microscopy
Cells were grown to mid-log phase and washed with DW and adhered to glass bottom dish coated with lectin. Z-stack images were captured at intervals of 0.3 µm using DeltaVison (Applied Precision). After 3D deconvolution, projected images were used for judging colocalization of two differently tagged proteins. To stain nucleus and septum, cells were treated with 1 µg/ml of Hoechst 33432 for 30 min.
Supporting Information
The mcl1-101 mutant interacts with kinetochore mutants genetically. (A) The mcl1-101 mutation is synthetically lethal with cnp1-FH. The mcl1-101 mutant (NYSPC40) was crossed with cnp1-FH (TN705). Resultant asci were dissected and incubated at 25 °C. Open squares indicate locations of double mutant spores. (B) Permissive growth temperature of mcl1-101 mutant is decreased by the mis6-302 mutation. Cells were grown at 25 °C and ten-fold serial dilutions were plated onto YES plates. Plates were incubated at indicated temperature for 3 days. Strains were derived from a cross between mcl1-101 (NYSPC41) and mis6-302 (NYSPL58). (C) The mcl1-101 mutation is synthetically lethal with ams2Δ mutation. The mcl1-101 mutant (NYSPC40) was crossed with ams2Δ (NYSPK66) as described in A. (D) Permissive growth temperature of mcl1-101 mutant is decreased by the mis16-13myc allele. Strains were wild-type (TN212), mis16-13myc (TN968), mcl1-101 (NYSPC41), mis16-13myc mcl1-101 (TN1035).
(0.70 MB TIF)
Histone H3 is aberrantly incorporated into kinetochore domain. ChIP was performed using antibody against C-terminal part of human histone H3. The ratio of immunoprecipitated DNA to input DNA was calculated and normalized to that of euchromatic lys1 locus. Fold enrichment compared to wild-type is shown. Open bars and shaded bars indicate results of 25 °C and 37 °C, respectively. Strains were wild-type (JY879), mcl1-101 (NYSPC52), swi7-H4 (TN403).
(0.10 MB TIF)
The mcl1 and swi7 mutants are sensitive to an HDAC inhibitor, Trichostatin. A(TSA)Ten-fold serial dilutions of wild-type (JY746), mcl1-101 (NYSPC41), and swi7-H4 (TN310) were plated onto YES containing 0 or 30 µg/ml of TSA and incubated for 4 days at the permissive (25 °C) or semi-permissive (33 °C) temperature.
(0.51 MB TIF)
Comparison of acetylation level between wild-type strain and mcl1 mutant. ChIP-on-chip was performed as described in Materials and Methods. Values obtained from AcH4-KN were normalized to those obtained from the antibody that recognizes amino acid 25-28 of histone H4 in wild-type strain (A) or mcl1-101 mutant (B), respectively. Similar results were observed in all chromosomes and the representative results in subtelomeric and centromeric regions of chromosome I were shown.
(1.63 MB TIF)
(0.12 MB DOC)
(0.05 MB DOC)
Acknowledgments
We thank Hironori Niki for giving useful comments and advice on fluorescence microscopy; Hironori Araki, Tetsuji Kakutani, Yasushi Hiromi, Tetsuro Kokubo, Junko Kanoh for helpful discussion; Giuseppe Baldacci, Takashi Morishita, Osami Niwa, Hiroto Okayama, Chikashi Shimoda, Hideo Shinagawa, Masayuki Yamamoto, Yeast Genetic Resource Center (Japan) for S. pombe strains and regents; Atsushi Miyawaki for Venus vector; Emi Fukuchi and Terumi Nishimura for technical help.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Grants-in Aids from the Ministry of Education, Culture, Sports, Science, and Technology of Japan Wellcome Trust Program Grant 065061/Z and 069286/AICR: Association for International Cancer Research Grant: 00-0297
References
- 1.Mythreye K, Bloom KS. Differential kinetochore protein requirements for establishment versus propagation of centromere activity in Saccharomyces cerevisiae. J Cell Biol. 2003;160:833–843. doi: 10.1083/jcb.200211116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karpen GH, Allshire RC. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- 3.Baum M, Sanyal K, Mishra PK, Thaler N, Carbon J. Formation of functional centromeric chromatin is specified epigenetically in Candida albicans. Proc Natl Acad Sci U S A. 2006;103:14877–14882. doi: 10.1073/pnas.0606958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris CA, Moazed D. Centromere assembly and propagation. Cell. 2007;128:647–650. doi: 10.1016/j.cell.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Shelby RD, Monier K, Sullivan KF. Chromatin assembly at kinetochores is uncoupled from DNA replication. J Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sullivan KF. A solid foundation: functional specialization of centromeric chromatin. Curr Opin Genet Dev. 2001;11:182–188. doi: 10.1016/s0959-437x(00)00177-5. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi K, Takayama Y, Masuda F, Kobayashi Y, Saitoh S. Two distinct pathways responsible for the loading of CENP-A to centromeres in the fission yeast cell cycle. Philos Trans R Soc Lond B Biol Sci. 2005;360:595–606. doi: 10.1098/rstb.2004.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furuyama T, Dalal Y, Henikoff S. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc Natl Acad Sci U S A. 2006;103:6172–6177. doi: 10.1073/pnas.0601686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunleavy EM, Pidoux AL, Monet M, Bonilla C, Richardson W, et al. A NASP (N1/N2)-Related Protein, Sim3, Binds CENP-A and Is Required for Its Deposition at Fission Yeast Centromeres. Mol Cell. 2007;28:1029–1044. doi: 10.1016/j.molcel.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim SM, Dubey DD, Huberman JA. Early-replicating heterochromatin. Genes Dev. 2003;17:330–335. doi: 10.1101/gad.1046203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- 12.Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 14.Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 15.Polizzi C, Clarke L. The chromatin structure of centromeres from fission yeast: differentiation of the central core that correlates with function. J Cell Biol. 1991;112:191–201. doi: 10.1083/jcb.112.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marschall LG, Clarke L. A novel cis-acting centromeric DNA element affects S. pombe centromeric chromatin structure at a distance. J Cell Biol. 1995;128:445–454. doi: 10.1083/jcb.128.4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, et al. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allshire RC, Javerzat JP, Redhead NJ, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 19.Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- 20.Ekwall K, Javerzat JP, Lorentz A, Schmidt H, Cranston G, et al. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science. 1995;269:1429–1431. doi: 10.1126/science.7660126. [DOI] [PubMed] [Google Scholar]
- 21.Ekwall K, Nimmo ER, Javerzat JP, Borgstrom B, Egel R, et al. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J Cell Sci. 1996;109 ( Pt 11):2637–2648. doi: 10.1242/jcs.109.11.2637. [DOI] [PubMed] [Google Scholar]
- 22.Pidoux AL, Richardson W, Allshire RC. Sim4: a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J Cell Biol. 2003;161:295–307. doi: 10.1083/jcb.200212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Partridge JF, Borgstrom B, Allshire RC. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- 24.Castillo AG, Mellone BG, Partridge JF, Richardson W, Hamilton GL, et al. Plasticity of fission yeast CENP-A chromatin driven by relative levels of histone H3 and H4. PLoS Genet. 2007;3:e121. doi: 10.1371/journal.pgen.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pidoux AL, Allshire RC. Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res. 2004;12:521–534. doi: 10.1023/B:CHRO.0000036586.81775.8b. [DOI] [PubMed] [Google Scholar]
- 26.Ekwall K, Cranston G, Allshire RC. Fission yeast mutants that alleviate transcriptional silencing in centromeric flanking repeats and disrupt chromosome segregation. Genetics. 1999;153:1153–1169. doi: 10.1093/genetics/153.3.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pidoux AL, Uzawa S, Perry PE, Cande WZ, Allshire RC. Live analysis of lagging chromosomes during anaphase and their effect on spindle elongation rate in fission yeast. J Cell Sci. 2000;113 Pt 23:4177–4191. doi: 10.1242/jcs.113.23.4177. [DOI] [PubMed] [Google Scholar]
- 28.Williams DR, McIntosh JR. mcl1+, the Schizosaccharomyces pombe homologue of CTF4, is important for chromosome replication, cohesion, and segregation. Eukaryot Cell. 2002;1:758–773. doi: 10.1128/EC.1.5.758-773.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsutsui Y, Morishita T, Natsume T, Yamashita K, Iwasaki H, et al. Genetic and physical interactions between Schizosaccharomyces pombe Mcl1 and Rad2, Dna2 and DNA polymerase alpha: evidence for a multifunctional role of Mcl1 in DNA replication and repair. Curr Genet. 2005;48:34–43. doi: 10.1007/s00294-005-0584-2. [DOI] [PubMed] [Google Scholar]
- 30.Ahmed S, Saini S, Arora S, Singh J. Chromodomain protein Swi6-mediated role of DNA polymerase alpha in establishment of silencing in fission Yeast. J Biol Chem. 2001;276:47814–47821. doi: 10.1074/jbc.M109186200. [DOI] [PubMed] [Google Scholar]
- 31.Mamnun YM, Katayama S, Toda T. Fission yeast Mcl1 interacts with SCF(Pof3) and is required for centromere formation. Biochem Biophys Res Commun. 2006;350:125–130. doi: 10.1016/j.bbrc.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 32.Saitoh S, Takahashi K, Yanagida M. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell. 1997;90:131–143. doi: 10.1016/s0092-8674(00)80320-7. [DOI] [PubMed] [Google Scholar]
- 33.Chen ES, Saitoh S, Yanagida M, Takahashi K. A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol Cell. 2003;11:175–187. doi: 10.1016/s1097-2765(03)00011-x. [DOI] [PubMed] [Google Scholar]
- 34.Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, et al. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Freeman-Cook LL, Sherman JM, Brachmann CB, Allshire RC, Boeke JD, et al. The Schizosaccharomyces pombe hst4+ gene is a SIR2 homologue with silencing and centromeric functions. Mol Biol Cell. 1999;10:3171–3186. doi: 10.1091/mbc.10.10.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grewal SI, Bonaduce MJ, Klar AJ. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics. 1998;150:563–576. doi: 10.1093/genetics/150.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freeman-Cook LL, Gomez EB, Spedale EJ, Marlett J, Forsburg SL, et al. Conserved locus-specific silencing functions of Schizosaccharomyces pombe sir2+. Genetics. 2005;169:1243–1260. doi: 10.1534/genetics.104.032714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 39.Berezney R, Dubey DD, Huberman JA. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 2000;108:471–484. doi: 10.1007/s004120050399. [DOI] [PubMed] [Google Scholar]
- 40.Kitamura E, Blow JJ, Tanaka TU. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell. 2006;125:1297–1308. doi: 10.1016/j.cell.2006.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meister P, Poidevin M, Francesconi S, Tratner I, Zarzov P, et al. Nuclear factories for signalling and repairing DNA double strand breaks in living fission yeast. Nucleic Acids Res. 2003;31:5064–5073. doi: 10.1093/nar/gkg719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 44.Xue Y, Wong J, Moreno GT, Young MK, Cote J, et al. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 45.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 46.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 47.Walfridsson J, Bjerling P, Thalen M, Yoo EJ, Park SD, et al. The CHD remodeling factor Hrp1 stimulates CENP-A loading to centromeres. Nucleic Acids Res. 2005;33:2868–2879. doi: 10.1093/nar/gki579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taddei A, Roche D, Sibarita JB, Turner BM, Almouzni G. Duplication and maintenance of heterochromatin domains. J Cell Biol. 1999;147:1153–1166. doi: 10.1083/jcb.147.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 50.Williams DR, McIntosh JR. Mcl1p is a polymerase alpha replication accessory factor important for S-phase DNA damage survival. Eukaryot Cell. 2005;4:166–177. doi: 10.1128/EC.4.1.166-177.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, et al. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell. 2006;23:787–799. doi: 10.1016/j.molcel.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 52.Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 53.Nakayama J, Allshire RC, Klar AJ, Grewal SI. A role for DNA polymerase alpha in epigenetic control of transcriptional silencing in fission yeast. EMBO J. 2001;20:2857–2866. doi: 10.1093/emboj/20.11.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kouprina N, Kroll E, Bannikov V, Bliskovsky V, Gizatullin R, et al. CTF4 (CHL15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5736–5747. doi: 10.1128/mcb.12.12.5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kouprina N, Tsouladze A, Koryabin M, Hieter P, Spencer F, et al. Identification and genetic mapping of CHL genes controlling mitotic chromosome transmission in yeast. Yeast. 1993;9:11–19. doi: 10.1002/yea.320090103. [DOI] [PubMed] [Google Scholar]
- 56.Miles J, Formosa T. Protein affinity chromatography with purified yeast DNA polymerase alpha detects proteins that bind to DNA polymerase. Proc Natl Acad Sci U S A. 1992;89:1276–1280. doi: 10.1073/pnas.89.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miles J, Formosa T. Evidence that POB1, a Saccharomyces cerevisiae protein that binds to DNA polymerase alpha, acts in DNA metabolism in vivo. Mol Cell Biol. 1992;12:5724–5735. doi: 10.1128/mcb.12.12.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hanna JS, Kroll ES, Lundblad V, Spencer FA. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol. 2001;21:3144–3158. doi: 10.1128/MCB.21.9.3144-3158.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suter B, Tong A, Chang M, Yu L, Brown GW, et al. The origin recognition complex links replication, sister chromatid cohesion and transcriptional silencing in Saccharomyces cerevisiae. Genetics. 2004;167:579–591. doi: 10.1534/genetics.103.024851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou Y, Wang TS. A coordinated temporal interplay of nucleosome reorganization factor, sister chromatin cohesion factor, and DNA polymerase alpha facilitates DNA replication. Mol Cell Biol. 2004;24:9568–9579. doi: 10.1128/MCB.24.21.9568-9579.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lejeune E, Bortfeld M, White SA, Pidoux AL, Ekwall K, et al. The chromatin-remodeling factor FACT contributes to centromeric heterochromatin independently of RNAi. Curr Biol. 2007;17:1219–1224. doi: 10.1016/j.cub.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 63.Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 64.Okuno Y, Okazaki T, Masukata H. Identification of a predominant replication origin in fission yeast. Nucleic Acids Res. 1997;25:530–537. doi: 10.1093/nar/25.3.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pidoux A, Mellone B, Allshire R. Analysis of chromatin in fission yeast. Methods. 2004;33:252–259. doi: 10.1016/j.ymeth.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 66.Ekwall K, Partridge JF. Fission yeast chromosomal analysis: fluorescent in situ hybridisation (FISH) and chromatin immunoprecipitation (CHIP). W A Bickmore (ed), Chromsome structural analysis: a practical approach Oxford University Press, Oxford, United Kingdom. 1999:39–57. [Google Scholar]
- 67.Gregan J, Riedel CG, Pidoux AL, Katou Y, Rumpf C, et al. The kinetochore proteins Pcs1 and Mde4 and heterochromatin are required to prevent merotelic orientation. Curr Biol. 2007;17:1190–1200. doi: 10.1016/j.cub.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, et al. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The mcl1-101 mutant interacts with kinetochore mutants genetically. (A) The mcl1-101 mutation is synthetically lethal with cnp1-FH. The mcl1-101 mutant (NYSPC40) was crossed with cnp1-FH (TN705). Resultant asci were dissected and incubated at 25 °C. Open squares indicate locations of double mutant spores. (B) Permissive growth temperature of mcl1-101 mutant is decreased by the mis6-302 mutation. Cells were grown at 25 °C and ten-fold serial dilutions were plated onto YES plates. Plates were incubated at indicated temperature for 3 days. Strains were derived from a cross between mcl1-101 (NYSPC41) and mis6-302 (NYSPL58). (C) The mcl1-101 mutation is synthetically lethal with ams2Δ mutation. The mcl1-101 mutant (NYSPC40) was crossed with ams2Δ (NYSPK66) as described in A. (D) Permissive growth temperature of mcl1-101 mutant is decreased by the mis16-13myc allele. Strains were wild-type (TN212), mis16-13myc (TN968), mcl1-101 (NYSPC41), mis16-13myc mcl1-101 (TN1035).
(0.70 MB TIF)
Histone H3 is aberrantly incorporated into kinetochore domain. ChIP was performed using antibody against C-terminal part of human histone H3. The ratio of immunoprecipitated DNA to input DNA was calculated and normalized to that of euchromatic lys1 locus. Fold enrichment compared to wild-type is shown. Open bars and shaded bars indicate results of 25 °C and 37 °C, respectively. Strains were wild-type (JY879), mcl1-101 (NYSPC52), swi7-H4 (TN403).
(0.10 MB TIF)
The mcl1 and swi7 mutants are sensitive to an HDAC inhibitor, Trichostatin. A(TSA)Ten-fold serial dilutions of wild-type (JY746), mcl1-101 (NYSPC41), and swi7-H4 (TN310) were plated onto YES containing 0 or 30 µg/ml of TSA and incubated for 4 days at the permissive (25 °C) or semi-permissive (33 °C) temperature.
(0.51 MB TIF)
Comparison of acetylation level between wild-type strain and mcl1 mutant. ChIP-on-chip was performed as described in Materials and Methods. Values obtained from AcH4-KN were normalized to those obtained from the antibody that recognizes amino acid 25-28 of histone H4 in wild-type strain (A) or mcl1-101 mutant (B), respectively. Similar results were observed in all chromosomes and the representative results in subtelomeric and centromeric regions of chromosome I were shown.
(1.63 MB TIF)
(0.12 MB DOC)
(0.05 MB DOC)