Abstract
The RNA degradosome of Escherichia coli is a multiprotein complex that plays an essential role in normal RNA processing and decay. It was recently shown that the major degradosome constituents are organized in a coiled cytoskeletal-like structure that extends along the length of the cell. Here we show that the endoribonuclease E (RNaseE) and RNA helicase B (RhlB) components of the degradosome can each independently form coiled structures in the absence of the other degradosome proteins. In contrast, the cytoskeletal organization of the other degradosome proteins required the presence of the RNaseE or RhlB coiled elements. Although the RNaseE and RhlB structures were equally competent to support the helical organization of polynucleotide phosphorylase, the cytoskeletal-like organization of enolase occurred only in the presence of the RNaseE coiled structure. The results indicate that the RNA degradosome proteins are components of the bacterial cytoskeleton rather than existing as randomly distributed multiprotein complexes within the cell and suggest a model for the cellular organization of the components within the helical degradosomal structure.
The bacterial cytoskeleton consists of structures that impart long-range order to the cell. These include structures formed by homologs of the major eukaryotic cytoskeletal proteins: tubulin, actin, and intermediate filament proteins. In addition, there are prokaryotic proteins that form independent cytoskeletal structures but have no apparent homology to eukaryotic cytoskeletal proteins (reviewed in Ref. 1).
Several of the bacterial cytoskeletal elements are organized as helical filamentous structures that wind around the cell from pole to pole, associated with the inner surface of the cytoplasmic membrane. These often appear to serve as lattices for the assembly of other proteins into the cytoskeletal framework. For example, the MreBCD structures appear to provide a scaffold for the organization of certain murein biosynthetic enzymes (2, 3) and the MinD helical structure provides a track for the assembly of the MinCDE machinery required for division site selection (4). The cytoskeletal organization presumably provides a mechanism to compartmentalize and coordinate these and other important cellular functions.
We have recently shown that the major protein components of the RNA degradosome are also localized as helical cytoskeletal-like structures within the cell (5). The degradosome is required for the normal maturation of transfer and ribosomal RNA and for the degradation of most messenger RNAs (6–8). The degradosome includes RNaseE,2 an essential endoribonuclease of 1061 amino acids (9) that contains binding sites for the other degradosomal proteins, RNA helicase B (RhlB), polynucleotide phosphorylase (PNPase), and enolase (Fig. 1A) (10–13). The degradosome components have been copurified from cell extracts as complexes of heterogeneous size (10, 12). In degradosome-dependent mRNA decay the RNA helicase activity of RhlB facilitates the degradation of structured RNAs and RNaseE provides the catalytic activity that cuts the RNA into fragments that are then degraded to nucleotides by the 3′ → 5′ exoribonuclease activity of PNPase (reviewed in Ref. 14). The enolase plays a regulatory role in the degradation of specific RNAs such as the glucose transporter mRNA and possibly, by inference, other mRNAs (8, 15, 16).
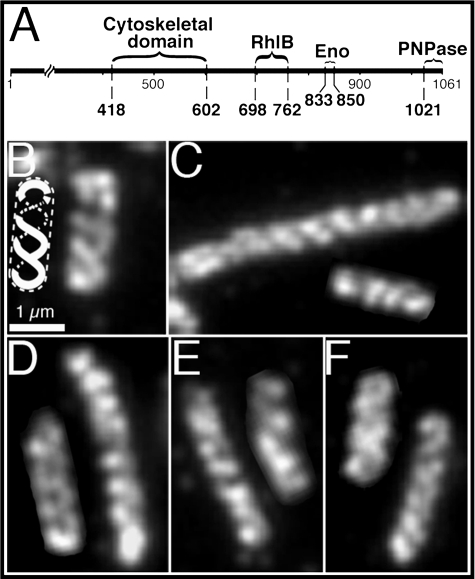
FIGURE 1.
Cellular organization of RNaseE. A, schematic representation of the domains of the 1061-amino acid RNaseE protein. The diagram shows regions containing the binding sites for RhlB, enolase (Eno), and PNPase (25) and the domain required for the helical organization of RNaseE (Cytoskeletal domain) (5). B–F, immunofluorescence micrographs of HA-tagged RNaseE (rne::HA) using monoclonal anti-HA antibody. B, strain AT38 (rne::HA ΔrhlB). The insert is a diagrammatic representation of the structure in the adjacent cell. C, strain AT51 (rne::HA Δpnp). D, strain AT46 (rne::HA Δeno). E, strain AT53 (rne::HA Δpnp,Δeno). F, strain AT52 (rne::HA Δeno,Δpnp,ΔrhlB). Scale bar, 1 μm.
In this study we investigated the roles of the different degradosome proteins in the helical organization of the degradosome. We show that RNaseE and RhlB can form helical cytoskeletal-like structures independently of each other and of the other degradosome constituents. In contrast, the cytoskeletal-like organization of the other degradosome proteins required the presence of the RNaseE or RhlB coiled elements. The RNaseE and RhlB structures were both able to support the helical organization of PNPase, whereas the helical organization of enolase occurred only in the presence of the RNaseE coiled structure. The results suggest a revised model for the cytoskeletal-like organization of the degradosome within the bacterial cell.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, Media, and Growth Condition—Escherichia coli strains were grown in Luria Bertani medium (17) to which 100 μg/ml ampicillin, 25 μg/ml kanamycin, 30 μg/ml chloramphenicol, or 0.4% (w/v) glucose were added when indicated. Δeno strains were grown in M9 medium supplemented with 0.2% tryptone/0.2% glycerol/1 mm MgSO4/0.0001% thiamine and 40 mm succinate. E. coli cells containing plasmids coding for Yfp-MreB were grown in the presence of 10 μm isopropyl β-d-thiogalactoside as described previously (18). For A22 treatment, 10 μg/ml A22 was added to cells that were grown at 37 °C to A600 ∼0.2. The cells were then shaken at the same temperature for 60 or 90 min and fixed with 2% formaldehyde and 0.02% glutaraldehyde. Plasmids and strains are listed in Table 1, and the details of their construction are available upon request. Gene knock-outs were constructed by linear DNA recombination (19). Hemagglutinin (HA) epitope tagging was done as previously described (20), and the associated antibiotic cassettes were eliminated, when indicated, by use of the FLP recombinase expressing plasmid pCP20 (19). In all cases, the chromosomal gene deletions and HA fusions were verified by sequencing. P1-mediated transduction was used to move mutations to different strains (21).
TABLE 1.
Plasmids and strains
| Relevant genotype or description | Reference or source | |
|---|---|---|
| Plasmids | ||
| pCP20 | FLP recombinase expression plasmid | (36) |
| PEJ01 | Plac-pnp::(His)6 | J. García-Mena |
| pHM542 | (His)5::rhlB in pET15-b | M. Cashel |
| pKD46 | Red recombinase expression plasmid | (19) |
| PLE7 | Plac-yfp::mreB | (4) |
| pSU314 | HA epitope tagging | (20) |
| pSU315 | HA epitope tagging | (20) |
| Relevant genotype or descriptiona | Reference or source | |
| Strains | ||
| AT8 | MC1000 rne1–417-catb | (5) |
| AT14 | MC1000 rne1–659-catb | (5) |
| AT20 | MC1000 pnp::HA-kanc | This study |
| AT23 | MC1000 eno::HAc | This study |
| AT24 | MC1000 pnp::HAc | This study |
| AT27 | MC1000 rne1–659::HA-kanb,c | (5) |
| AT29 | MC1000 rne1–417-cat eno::HAb,c | This study |
| AT30 | MC1000 rne1–659-cat eno::HAb,c | This study |
| AT31 | MC1000 rne1–417-cat pnp::HAb,c | This study |
| AT33 | MC1000 rne::HAc | (5) |
| AT34 | MC1000 rne1–659::HAb,c | (5) |
| AT35 | MC1000 rne1–417::HAb,c | (5) |
| AT36 | MC1000 ΔrhlB-cat | This study |
| AT38 | MC1000 rne::HA ΔrhlB-catc | This study |
| AT40 | MC1000 rne1–417::HA ΔrhlB-catb,c | This study |
| AT45 | MC1000 rne1–417::HA Δpnp-catb,c | This study |
| AT46 | MC1000 rne::HA Δeno-kanc | This study |
| AT51 | MC1000 rne::HA Δpnpc | This study |
| AT52 | MC1000 rne::HA Δpnp,ΔrhlB-cat,Δeno-kanc | This study |
| AT53 | MC1000 rne::HA Δpnp,Δeno-kanc | This study |
| AT54 | MC1000 ΔrhlB-cat pnp::HAc | This study |
| AT57 | MC1000 rne1–417-cat ΔrhlB pnp::HA-kanb,c | This study |
| AT58 | MC1000 ΔrhlB-cat eno::HAc | This study |
| AT60 | MC1000 rne1–417-cat ΔrhlB eno::HAb,c | This study |
| AT61 | MC1000 Δmin-kan rne1–659::HAb,c | This study |
| AT63 | MC1000 Δpnp-cat ΔrhlB eno::HAc | This study |
| CF5961 | BL21(DE3)/pHM542 | M. Cashel |
| MC1000 | Wild type strain | (35) |
| PB114 | PB103 ΔminB | (35) |
| TM388 | W3110 mlc Δpnp-cat | (15) |
| TM390 | W3110 mlc ΔrhlB-cat | (15) |
| YLS3 | MC1000 ΔmreB | (18) |
To allow selection during construction of HA fusions, rne truncations, and deletion strains, an antibiotic resistance cassette (kan or cat) was inserted by linear DNA recombination. When necessary, the antibiotic resistance cassette was removed as described under “Experimental Procedures”
The indicated rne truncations replaced the chromosomal copy of rne and therefore were expressed from the native rne promoter
The HA fusions replaced the chromosomal copy of the indicated gene and therefore were expressed from the native promoters
Microscopy—Yfp-labeled cells were examined by fluorescence microscopy as previously described (22). Immunofluorescence experiments were done as previously described (5). Images were not subjected to deconvolution. Monoclonal mouse anti-HA tag (Sigma) was used to detect HA-tagged RNaseE, enolase, and PNPase. Rabbit antisera directed against RhlB and against PNPase, kindly provided by Dr. M. Cashel and Dr. Gianni Deho, respectively, was purified by absorption to purified His-RhlB or PNPase-His bound to polyvinylidene fluoride membrane, followed by elution with 0.2 m glycine (pH 2) and renaturation with 1.5 m Tris base (pH 8.8). Alexa Fluor 488- and Alexa Fluor 594-conjugated goat anti-rabbit and Alexa Fluor 488-conjugated goat anti-mouse secondary antibodies were used (Molecular Probes). Images were collected using the Openlabs image acquisition program (Improvision). 300–350 cells were analyzed for each strain, and the described localization pattern was present in 95–98% of cells.
Immunoblotting Analysis—Quantitative immunoblotting was done on 5, 10, and 30 μg of protein of total cell extracts as previously described (22), using 12% SDS-PAGE.
RESULTS
Formation of the RNaseE Helical Structure—We previously showed that RNaseE is organized in a helical structure that extends between the two cell poles and that the formation of this structure is independent of the known helical MreB and MinD cytoskeletal elements. The other degradosome constituents are also organized as helical elements similar to the RNaseE structure (5). This suggested that the four proteins are probably present in a common coiled structure within the cell. The structure is likely associated with the inner surface of the cytoplasmic membrane (23, 24).
To determine which of the degradosomal proteins provided the primary element(s) of the coiled filamentous array, we first studied the localization pattern of RNaseE in the absence of one, two, or all three other degradosomal proteins. RNaseE was identified by anti-HA immunofluorescence in cells that expressed RNaseE-HA from the native rne promoter. As shown in Fig. 1, the RNaseE coiled structure was present in cells that lacked RhlB, PNPase, or enolase (Fig. 1, B–D), as well as in Δpnp,Δeno cells (Fig. 1E) and Δpnp,Δeno,ΔrhlB cells (Fig. 1F) (see Table 1 for description of genotypes). A similar localization pattern was present in 95% of the cells of the culture (350 cells examined). Thus, the RNaseE coiled filamentous structure can be formed independently of any of the other degradosome constituents. This raised the possibility that the RNaseE structure might be responsible for the helical organization of the other degradosome proteins.
RhlB Forms Helical Structures in the Absence of the RNaseE Helical Elements—RhlB forms a helical cytoskeletal-like structure in wild type cells (5). We therefore asked whether the helical organization pattern of RhlB is autonomous or is due to secondary association with the RNaseE coiled structure. We previously showed that the RNaseE helical structure is absent in cells in which full-length RNaseE is replaced by RNaseE-(1–417). RNaseE-(1–417) does not form coiled arrays (5) and also lacks the known RhlB-, enolase- and PNPase-binding sites of the full-length RNaseE protein (Fig. 1A) (25). To ask whether the RNaseE cytoskeletal structure was required for the helical organization of RhlB, we determined the localization of RhlB by immunofluorescence microscopy in a strain that expresses RNaseE-(1–417) from the native rne promoter. When full-length RNaseE was replaced by the truncated RNaseE-(1–417) protein in AT31 cells (rne1–417 pnp::HA), RhlB was distributed as a regular pattern of diagonal bands along the length of the cell (Fig. 2A), similar to the helical distribution pattern of RNaseE (Fig. 1B). Thus, RhlB can form cytoskeletal-like coiled structures independently of the RNaseE helical structures. The helical localization pattern of RhlB was also unaffected by deletion of PNPase, or enolase, or enolase and PNPase (Fig. 2, B–D).
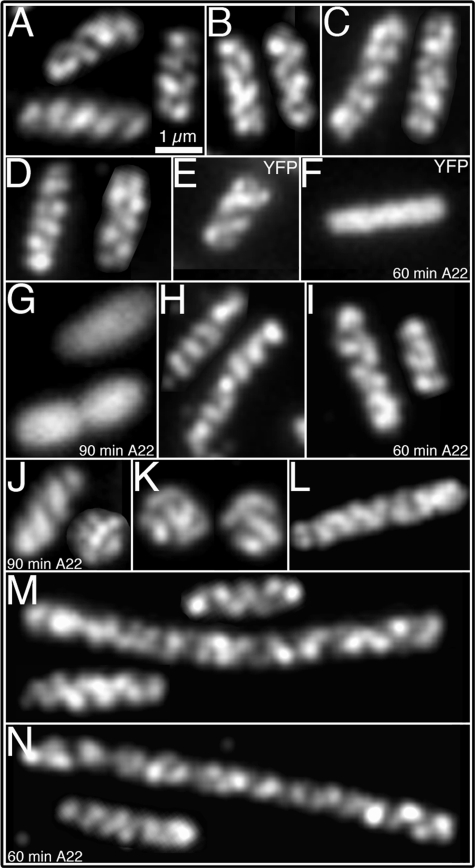
FIGURE 2.
Determinants of the cytoskeletal-like organization of RhlB. Localization of RhlB by immunofluorescence microscopy using purified anti-RhlB antibody (A–D, H–K, M, and N); localization of MreB by fluorescence from Yfp-labeled MreB (E–G); and immunofluorescence localization of HA-tagged RNaseE-(1–659) using anti-HA antibody (L). A, strain AT31 (rne1–417 pnp::HA). B, strain AT45 (rne1–417::HA Δpnp). C, strain AT46 (rne::HA Δeno). D, strain AT53 (rne::HA Δpnp,Δeno). E, MC1000/Plac-yfp::mreB cells grown in the absence of A22. F and G, MC1000/Plac-yfp::mreB A22-treated cells for 60 min (F) or 90 min (G). H, strain AT27 (rne1–659::HA) grown in the absence of A22. I and J, strain AT27 (rne1–659::HA), grown in the presence of A22 for 60 min (I) or 90 min (J). K, strain YLS3 (ΔmreB). L, strain AT61 (rne1–659::HA ΔminCDE) showing the coiled structure of RNaseE-(1–659). M, RhlB localization in strain AT61 (rne1–659::HA ΔminCDE) grown in the absence of A22. N, RhlB localization in strain AT61 (rne1–659::HA ΔminCDE) treated for 60 min with A22. Because of the Min- phenotype the cells grow as a mixture of short filaments of varying cell length (35) (M, N). As previously reported, ΔmreB and cells treated with A22 for 90 min grow as spheroids of different sizes (G, J, K) (18, 26). The described localization pattern was present in 95–97% of cells (see “Experimental Procedures”). Scale bar, 1 μm.
To determine whether the RhlB helical organization required its association with the helical cellular structures that are formed by the cytoskeletal MreB and MinD proteins, we examined the localization pattern of RhlB in the absence of the MreB and/or MinD cytoskeletal elements. The helical structure of MreB can be disrupted by treatment with low concentrations of A22 (S-(3,4-dichlorobenzyl) isothiourea) without change in the rod shape of the cell (26). Studies with a yellow fluorescent protein-MreB derivative (Yfp-MreB) showed that MreB coiled structures disappeared when cells were exposed to 10 μg/ml A22 for 60–90 min (Fig. 2, F and G). At 60 min the cells retained their rod shape (Fig. 2, F and I), and at 90 min the cells formed a mixed population of rods and spheroids (Fig. 2, G and J). This permitted us to examine the localization pattern of RhlB in E. coli cells in which the MreB helical structure was disrupted by A22 treatment. Immunofluorescence studies showed that the RhlB coiled structure was preserved under these conditions (Fig. 2, H and J). This showed that maintenance of the helical cytoskeletal-like organization of RhlB does not require the helical MreB cytoskeleton.
To exclude the possibility that the MreB helical arrays may play a role in formation of the RhlB cytoskeletal structure, although not required for its maintenance (above), we also determined the localization pattern of RhlB in a ΔmreB strain that permanently lacks the MreB protein and therefore grows as spheres of varying sizes as previously described (18). As shown in Fig. 2K, this did not prevent the formation of RhlB filamentous structures that coiled around the spherical cell. The RhlB filaments appeared to cross each other several times (Fig. 2K), reminiscent of the double helical structure of MreB. This demonstrates that formation and maintenance of the RhlB cytoskeletal-like elements are both independent of the MreB cytoskeleton.
The MinCDE proteins also form a helical cytoskeletal structure (4) that is not required for assembly of the RNaseE helical structure (Fig. 2L) (5). To ask whether the RhlB coiled structure might arise from an association of RhlB with the MinD helical cytoskeleton, we determined the localization pattern of RhlB in a ΔminCDE strain. As shown in Fig. 2M, the RhlB helical distribution pattern was retained in the absence of the MinCDE proteins. The RhlB helical pattern was also retained in A22-treated cells of strain AT61 (Δmin rne1–659::HA), in which the MreB and MinD cytoskeletal structures are both absent (Fig. 2N). We conclude that formation of the RhlB cytoskeletal-like structure does not require the presence of the RNaseE, MinD, and/or MreB cytoskeletal elements.
Helical Organization of PNPase—The PNPase component of the degradosome did not form organized cellular structures when expressed in cells that lacked both the RNaseE and RhlB helical elements. Thus, PNPase helical structures were absent in immunostaining studies of strains AT40 (ΔrhlB rne1–417::HA) and AT57 (ΔrhlB rne1–417 pnp::HA), which lack the RNaseE and RhlB cytoskeletal-like structures (Fig. 3, A and B). Quantitative immunoblot analysis showed that the loss of PNPase helical organization was not due to a change in PNPase concentration in these strains (data not shown).
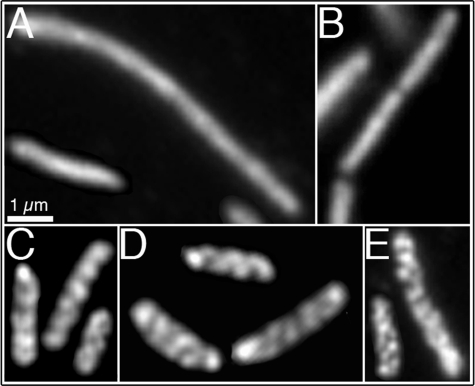
FIGURE 3.
Cellular organization of polynucleotide phosphorylase. Immunofluorescence micrographs using purified anti-PNPase (A, E) or monoclonal anti-HA antibodies (B–D). When indicated, the pnp chromosomal copy was tagged with the HA epitope tag. A, strain AT40 (rne1–417::HA ΔrhlB). B, strain AT57 (rne1–417 ΔrhlB pnp::HA). C, strain AT54 (ΔrhlB pnp::HA). D, strain AT31 (rne1–417 pnp::HA). E, strain AT46 (rne::HA Δeno). Strains in which rne was replaced by rne1–417 grew as mixed populations of filaments and short cells (5). The described localization pattern was present in 95–98% of cells (see “Experimental Procedures”). Scale bar, 1 μm.
Significantly, the ability to form the PNPase coiled structures was restored when PNPase was expressed in the presence of either the RNaseE or RhlB helical array. This was shown in AT54 (pnp::HA ΔrhlB) cells (Fig. 3C), which contained the RNaseE cytoskeletal-like structure but lacked the RhlB protein, and in AT 31 (rne1–417 pnp::HA) cells (Fig. 3D), which contained the RhlB helical structure but lacked the RNaseE structure (Fig. 2A). In both cases, the PNPase coiled arrays were similar to those seen in wild type cells. The helical organization of PNPase was also not perturbed in cells that lacked enolase but contained RNaseE and RhlB (Fig. 3E). The ability of both the RNaseE and RhlB coiled structures to induce the helical distribution pattern of PNPase suggests that both of the coiled structures can act as templates for the helical organization of PNPase. This is consistent with previous reports that showed direct interactions between PNPase and these proteins (25, 27).
The Helical Organization of Enolase—Evidence that the RNaseE cytoskeletal-like structure was required for the helical organization of enolase came from studies of cells that lacked the RNaseE and/or RhlB coiled structures. The helical organization of enolase was retained in AT58 cells (ΔrhlB eno::HA), which contain the RNaseE helical structure but lack the RhlB coiled structure (Fig. 4E). This is consistent with in vivo and in vitro evidence for interaction between enolase and RNaseE (25). The helical localization pattern of enolase was also unaffected when RhlB and PNPase were both deleted (Fig. 4D). In contrast, the helical organization of enolase was lost in AT29 cells (rne1–417 eno::HA), which contain the RhlB helical array but lack the RNaseE coiled structures (Fig. 4A). As expected, the enolase helical arrays were also absent from AT60 cells (rne1–417 ΔrhlB eno::HA) that lack both the RNaseE and RhlB structures (Fig. 4B). Loss of the helical organization was not due to change in the cellular concentration of enolase in quantitative immunoblot experiments (data not shown).
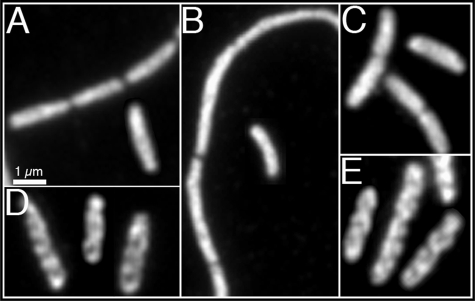
FIGURE 4.
Cellular organization of enolase. Immunofluorescence micrographs using monoclonal anti-HA antibody. The chromosomal copy of eno is tagged with the HA epitope tag. A, strain AT29 (rne1–417 eno::HA). B, AT60 (rne1–417 ΔrhlB eno::HA). C, AT30 (rne1–659 eno::HA). D, AT63 (Δpnp,ΔrhlB eno::HA). E, strain AT58 (ΔrhlB eno::HA). Where rne was replaced by rne1–417 the strains grew as mixed populations of filaments and short cells (5). The described localization pattern was present in 95–98% of cells (see “Experimental Procedures”). Scale bar, 1 μm.
The loss of the helical organization of enolase in AT29 (rne1–417 eno::HA) cells presumably was secondary to loss of the RNaseE helical structures and/or loss of other determinants in the truncated region of the RNaseE-(1–417) protein, such as the enolase-binding site. To investigate the role of the enolase-binding site we studied the enolase localization pattern in AT30 cells (rne1–659 eno::HA). These cells express RNaseE-(1–659), which lacks the enolase-binding site but retains the cytoskeletal determinant (Fig. 1A) and thus maintains the helical organization of RNaseE (Fig. 2L) (5). The diffuse distribution of enolase in the cytoplasm of this strain (Fig. 4C) suggests that the helical organization of enolase is mediated by its binding to the enolase-binding site of RNaseE molecules within the RNaseE helical structures.
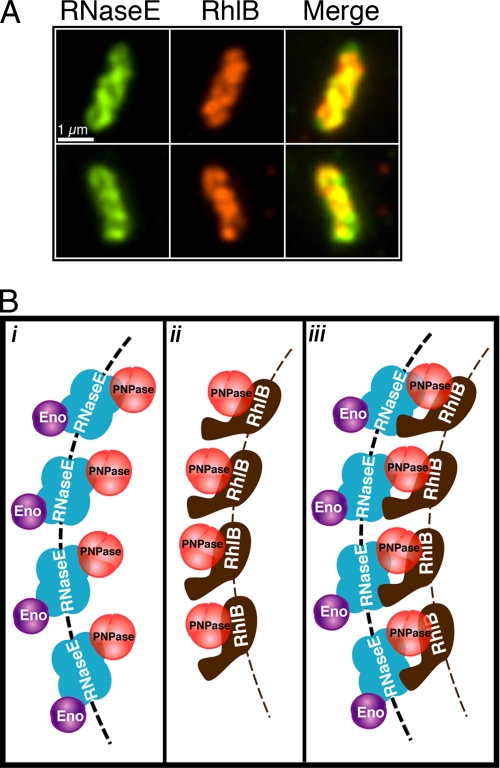
Colocalization of the RNaseE and RhlB Helical Structures—It is likely that the helical structures formed by RNaseE and RhlB (shown above) are both present within the composite helical degradosome structure. Consistent with this assumption, when AT33 (rne::HA) cells were doubly immunostained with mouse anti-HA and rabbit anti-RhlB antibodies the helical loops of RNaseE and RhlB largely colocalized (Fig. 5A).
FIGURE 5.
Organization of the RhlB and RNaseE helical structures. A, double label immunofluorescence microscopy of RNaseE and RhlB. RNaseE (green) and RhlB (red) are shown in the left and middle columns, respectively; overlays are shown in right panels. Yellow and orange regions represent regions of colocalization in the merged images. RNaseE and RhlB images are in slightly different planes of focus. Scale bar, 1 μm. B, proposed model for the cytoskeletal-like organization of the RNA degradosome. i, in the absence of RhlB. ii, in the absence of the RNaseE helical structure (e.g. in RNaseE-(1–417) cells). iii, in wild type cells. RNaseE, RhlB, PNPase, and enolase are shown in blue, brown, red, and purple, respectively. Dashed arcs depict the RNaseE (black) and RhlB (brown) helical strands. It is not known whether the helical strands are formed by RNaseE and RhlB polymerization or by the association of the proteins with an unknown underlying cytoskeletal structure (see “Discussion”). To simplify the figure the molecular dimensions and stoichiometry of the proteins are arbitrary.
DISCUSSION
It is well established that the RNA degradosome is a multiprotein complex that is required for normal RNA processing and decay. The major degradosome constituents have been isolated, and the interactions between the proteins have been studied (reviewed in Ref. 28). Nevertheless, the detailed structure and organization of the degradosome within the cell has remained unclear.
Until recently, it had been assumed that the degradosome exists as individual multiprotein complexes within the cell. Because RNaseE contains binding sites for the other degradosome proteins, RNaseE is believed to form the core of the multiprotein complex, providing a scaffold for the assembly of the mature degradosome (25). The present results suggest a new picture in which the RNA degradosome is organized as a higher order filamentous helical structure within the cell, containing RNaseE and RhlB helical strands that are both likely to participate in organizing the overall structure (Fig. 5B, iii).
It was striking that the RNaseE and RhlB components of the degradosome could each independently assemble into extended helical structures. This supports a model in which the RNaseE and RhlB helices comprise parallel strands within the structure of the complete degradosome. The strands are likely to be held together by direct interaction of RNaseE with RhlB and by interactions of PNPase with both RhlB and RNaseE, with PNPase acting as a bridge between the RNaseE and RhlB helical structures (Fig. 5B, iii). This is consistent with the observation that the RhlB and RNaseE helical elements were each capable of inducing the helical organization of PNPase and with the previous demonstration that direct PNPase-RNaseE and PNPase-RhlB interactions can be detected using protein-protein reconstitution techniques and other methods (10, 25, 27, 29, 30).
In contrast to PNPase, which can assume a helical conformation in the presence of either of the RhlB or RNaseE helical structures, the helical organization of enolase occurred only in the presence of the RNaseE coiled structure. This implies that within the degradosome structure enolase is associated only with the RNaseE helical strand (Fig. 5). This is consistent with the previously reported interaction between RNaseE and enolase (25).
We consider two possible models to explain the helical organization of the primary RhlB and RNaseE structures. (i) The helical strands may be composed of RNaseE and RhlB filamentous polymers similar to those of other cytoskeletal elements, such as MreB and MinD (31–33). Yeast two-hybrid studies have identified two separate RNaseE-RNaseE interaction domains that could participate in polymer assembly (25). Similarly, RhlB-RhlB self-interactions that could participate in RhlB polymer assembly have been demonstrated in bacterial two-hybrid and Biacore surface plasmon resonance studies (27), although this question is still open (34). Because the interacting sites might also be involved in formation of lower order structures such as dimers or tetramers, the idea that the helical cellular structures are composed of long RNaseE and RhlB polymers must remain conjectural until it is directly shown that these proteins can polymerize into extended filaments. (ii) Alternatively, the organization of the RNaseE and RhlB elements could reflect their association with another underlying helical system. Although the RhlB and RNaseE helical structures did not require the known MinD and MreB helical cytoskeletal systems, the existence of as yet unidentified cellular elements that could provide tracks for assembly of the RhlB and RNaseE structures cannot be excluded.
The exonuclease activity of PNPase is believed to complete the process of RNA degradation within the degradosome, acting after the RNaseE endonuclease cuts single-stranded regions of RNA into shorter fragments and the RhlB helicase melts double-stranded RNA domains. The positioning of PNPase as a bridge between the helical strands of RNaseE and RhlB would permit RNA fragments to be presented to the PNPase exonuclease to complete their degradation regardless of the order of action of the RNaseE endonuclease and the RhlB RNA helicase on any individual RNA substrate molecule.
The present work suggests a model for the cytoskeletal-like organization of the E. coli RNA degradosome that includes RhlB and RNaseE helical filamentous elements that interact with each other and with the other degradosome components (Fig. 5B, iii). Further work will be needed to fully define the molecular architecture and functional significance of this unique cytoskeletal-like system.
Acknowledgments
We thank Mary Osborn and Asis Das for helpful discussions and M. Cashel, G. Deho, J. García-Mena, H. Aiba, S. Uzzau, and M. Wachi for providing antibodies, strains, plasmids, and reagents.
This work was supported, in whole or in part, by National Institutes of Health Grant GM R37-06032. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: RNaseE, endoribonuclease E; RhlB, RNA helicase B; PNPase, polynucleotide phosphorylase; HA, hemagglutinin; Yfp, yellow fluorescent protein.
References
- 1.Shih, Y. L., and Rothfield, L. (2006) Microbiol. Mol. Biol. Rev. 70 729-754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dye, N. A., Pincus, Z., Theriot, J. A., Shapiro, L., and Gitai, Z. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 18608-18613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Divakaruni, A. V., Loo, R. R., Xie, Y., Loo, J. A., and Gober, J. W. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 18602-18607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shih, Y.-L., Le, T., and Rothfield, L. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 7865-7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taghbalout, A., and Rothfield, L. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 1667-1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghora, B. K., and Apirion, D. (1978) Cell 15 1055-1066 [DOI] [PubMed] [Google Scholar]
- 7.Ow, M. C., and Kushner, S. R. (2002) Genes Dev. 16 1102-1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein, J. A., Lin, P. H., Cohen, S. N., and Lin-Chao, S. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2758-2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mackie, G. A. (1998) Nature 395 720-723 [DOI] [PubMed] [Google Scholar]
- 10.Carpousis, A. J., Van Houwe, G., Ehretsmann, C., and Krisch, H. M. (1994) Cell 76 889-900 [DOI] [PubMed] [Google Scholar]
- 11.Py, B., Causton, H., Mudd, E. A., and Higgins, C. F. (1994) Mol. Microbiol. 14 717-729 [DOI] [PubMed] [Google Scholar]
- 12.Py, B., Higgins, C. F., Krisch, H. M., and Carpousis, A. J. (1996) Nature 381 169-172 [DOI] [PubMed] [Google Scholar]
- 13.Miczak, A., Kaberdin, V. R., Wei, C. L., and Lin-Chao, S. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 3865-3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carpousis, A. J. (2002) Biochem. Soc. Trans. 30 150-155 [PubMed] [Google Scholar]
- 15.Morita, T., Kawamoto, H., Mizota, T., Inada, T., and Aiba, H. (2004) Mol. Microbiol. 54 1063-1075 [DOI] [PubMed] [Google Scholar]
- 16.Morita, T., Maki, K., and Aiba, H. (2005) Genes Dev. 19 2176-2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maniatis, T., Fritsch, E. F., and Sambrook, J. (1982) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 18.Shih, Y. L., Kawagishi, I., and Rothfield, L. (2005) Mol. Microbiol. 58 917-928 [DOI] [PubMed] [Google Scholar]
- 19.Datsenko, K. A., and Wanner, B. L. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 6640-6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uzzau, S., Figueroa-Bossi, N., Rubino, S., and Bossi, L. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 15264-15269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, J. H. (1992) A Short Course on Bacterial Genetics, pp. 263-274, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 22.Shih, Y.-L., Fu, X., King, G. F., Le, T., and Rothfield, L. I. (2002) EMBO J. 21 3347-3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liou, G. G., Jane, W. N., Cohen, S. N., Lin, N. S., and Lin-Chao, S. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 63-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miczak, A., Srivastava, R. A., and Apirion, D. (1991) Mol. Microbiol. 5 1801-1810 [DOI] [PubMed] [Google Scholar]
- 25.Vanzo, N. F., Li, Y. S., Py, B., Blum, E., Higgins, C. F., Raynal, L. C., Krisch, H. M., and Carpousis, A. J. (1998) Genes Dev. 12 2770-2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gitai, Z., Dye, N. A., Reisenauer, A., Wachi, M., and Shapiro, L. (2005) Cell 120 329-341 [DOI] [PubMed] [Google Scholar]
- 27.Liou, G. G., Chang, H. Y., Lin, C. S., and Lin-Chao, S. (2002) J. Biol. Chem. 277 41157-41162 [DOI] [PubMed] [Google Scholar]
- 28.Carpousis, A. J. (2007) Annu. Rev. Microbiol. 61 71-87 [DOI] [PubMed] [Google Scholar]
- 29.Lin, P. H., and Lin-Chao, S. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 16590-16595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coburn, G. A., Miao, X., Briant, D. J., and Mackie, G. A. (1999) Genes Dev. 13 2594-2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu, Z., Gogol, E., and Lutkenhaus, J. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 6761-6766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suefuji, K., Valluzzi, R., and RayChaudhuri, D. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 16776-16781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esue, O., Cordero, M., Wirtz, D., and Tseng, Y. (2005) J. Biol. Chem. 280 2628-2635 [DOI] [PubMed] [Google Scholar]
- 34.Callaghan, A. J., Aurikko, J. P., Ilag, L. L., Gunter Grossmann, J., Chandran, V., Kuhnel, K., Poljak, L., Carpousis, A. J., Robinson, C. V., Symmons, M. F., and Luisi, B. F. (2004) J. Mol. Biol. 340 965-979 [DOI] [PubMed] [Google Scholar]
- 35.de Boer, P. A. J., Crossley, R. E., and Rothfield, L. I. (1989) Cell 56 641-649 [DOI] [PubMed] [Google Scholar]
- 36.Cherepanov, P. P., and Wackernagel, W. (1995) Gene 158 9-14 [DOI] [PubMed] [Google Scholar]