Abstract
Both latent transforming growth factor-β (TGF-β)-binding proteins fibrillins are components of microfibril networks, and both interact with members of the TGF-β family of growth factors. Interactions between latent TGF-β-binding protein-1 and TGF-β and between fibrillin-1 and bone morphogenetic protein-7 (BMP-7) are mediated by the prodomain of growth factor complexes. To extend this information, investigations were performed to test whether stable complexes are formed by additional selected TGF-β family members. Using velocity sedimentation in sucrose gradients as an assay, complex formation was demonstrated for BMP-7 and growth and differentiation factor-8 (GDF-8), which are known to exist in prodomain/growth factor complexes. Comparison of these results with complex formation by BMP-2, BMP-4 (full-length and shortened propeptides), BMP-10, and GDF-5 allowed us to conclude that all, except for BMP-2 and the short BMP-4 propeptides, formed complexes with their growth factors. Using surface plasmon resonance, binding affinities between fibrillin and all propeptides were determined. Binding studies revealed that the N-terminal end of fibrillin-1 serves as a universal high affinity docking site for the propeptides of BMP-2, -4, -7, and -10 and GDF-5, but not GDF-8, and located the BMP/GDF binding site within the N-terminal domain in fibrillin-1. Rotary shadowing electron microscopy of molecules of BMP-7 complex bound to fibrillin-1 confirmed these findings and also showed that prodomain binding targets the growth factor to fibrillin. Immunolocalization of BMP-4 demonstrated fibrillar staining limited to certain tissues, indicating tissue-specific targeting of BMP-4. These data implicate the fibrillin microfibril network in the extracellular control of BMP signaling and demonstrate differences in how prodomains target their growth factors to the extracellular space.
Several different molecular mechanisms are known to regulate transforming growth factor-β (TGF-β)2 signaling in the extracellular space. First, TGF-βs are secreted as latent complexes consisting of a processed growth factor dimer in association with its propeptides. The propeptide of TGF-β1, called LAP, for latency-associated peptide, confers latency either by blocking binding of the receptor to the growth factor domain or by altering the conformation of the growth factor domain such that it cannot bind to its receptors (1). Second, covalent interactions between the propeptides and latent TGF-β-binding proteins (LTBPs) target latent TGF-β complexes to the extracellular matrix (2–3). Third, these large latent TGF-β·LTBP complexes interact with fibrillin-1 (4) in the extracellular matrix. These three molecular interactions are required to properly regulate TGF-β signaling, since mutations in latency-associated peptide in LTBPs or in fibrillin-1 correlate with dysregulated TGF-β signaling in humans and mice (5–8).
LTBPs and fibrillins constitute a family of structurally homologous molecules. These molecules are composed of multiple calcium binding epidermal growth factor-like modules interspersed by domains containing eight cysteines (8-Cys domains) (9). Latency-associated peptide is disulfide-bonded to a specific 8-Cys domain in LTBP (3, 10), so these domains are also called TB (TGF-β binding) modules (11). In the human genome there are 33 8-Cys domains, and these are found only in LTBPs and fibrillins. Because of these structural similarities, we hypothesized that any member of the family of TGF-β-related growth factors may interact with LTBPs or with fibrillins. We first tested this hypothesis by recombinantly expressing full-length bone morphogenetic protein-7 (BMP-7) complex and demonstrating that the BMP-7 prodomain binds to fibrillin and targets BMP-7 growth factor to fibrillin microfibrils (12).
Fibrillins form structures called microfibrils, which are ubiquitous in the connective tissue space and which can be defined at the ultrastructural level as small diameter (10–12 nm) fibrils that display a hollow or beaded appearance. LTBPs are associated with fibrillin microfibrils, but they are not required to form the microfibrils. The fibrillin microfibril network, including associated LTBPs, forms a physical scaffold to which TGF-β-related growth factors are targeted. Changes or disruptions in the microfibril network may affect the appropriate targeting of growth factors and may subtly or unsubtly perturb signaling activities of these growth factors. For example, heterozygous mutations in FBN1 result in poor assembly of fibrillin microfibrils by Marfan fibroblasts and disrupted patterns of fibrillin microfibrils in skin biopsies from individuals with the Marfan syndrome (13). Abnormal TGF-β activation has been implicated in the pathogenesis of the Marfan syndrome (6–7). However, it remains important to determine how structural alterations affect signaling activities of the growth factors targeted to the microfibril network. It is also important to identify which members of the TGF-β superfamily are targeted to tissue-specific microfibril networks and how the microfibril network coordinates the signaling activities of multiple different targeted growth factors.
In this study we have extended our previous investigations to additional TGF-β-related growth factors. We first addressed whether other TGF-β family members form prodomain/growth factor complexes similar to TGF-β and BMP-7. For this purpose, we performed novel in vitro reconstitution experiments using recombinantly expressed growth factor propeptides titrated with their commercially available growth factor dimers. In addition, we tested whether various propeptides interact with fibrillins, and we identified a universal high affinity binding site present in fibrillins. We also tested whether growth factors are targeted to fibrillin microfibrils using commercially available antibodies. Based on these investigations, a general framework of how growth factors of the TGF-β superfamily are targeted to fibrillin microfibrils has emerged.
EXPERIMENTAL PROCEDURES
Recombinant Proteins—Expression and purification of the BMP-7 complex and its separation into the BMP-7 prodomain and growth factor dimer were performed as described (12). Construction, transfection, purification, and characterization of recombinant human fibrillin-1 peptides rF23 (14), rF31 (15), rF37 (15), rF38 (16), rF45 (17), and rF47 (18) have been described. Human growth factors (BMP-2, BMP-4, BMP-5, BMP-6, BMP-7, BMP-10), mouse growth factors (GDF-5, GDF-8), and the mouse GDF-8 propeptide were purchased from R&D Systems, Minneapolis, MN. All purchased R&D products contained 0.1% bovine serum albumin as carrier protein.
Antibodies—The following antibodies were used: polyclonal antibody (pAb) 9543 (fibrillin-1) (19), pAb 0868 (fibrillin-2) (18), monoclonal antibody (mAb) 2 (BMP-7 prodomain) (12), mAb 6 (BMP-7 growth factor) (12), and commercially available mAbs specific for BMP-2 growth factor, BMP-2 prodomain, BMP-4 growth factor, BMP-7 growth factor, GDF-5 growth factor, GDF-8 growth factor, and His6 tag, and polyclonal anti-GDF-8 prodomain (R&D Systems). pAb anti-BMP-4 prodomain was generated in rabbits against bacterially expressed BMP-4 prodomain, which was purified over a Co2+-charged GE Healthcare Hi-Trap column, equilibrated with 50 mm sodium phosphate buffer, pH 7.2, 8 m urea, and 1 m NaCl (buffer A), and eluted in a gradient run starting with buffer A containing 0–100% buffer B (buffer A containing 0.25 m imidazole).
Other Reagents—Escherichia coli BL21 (DE3) and the pET11a vector were obtained from Stratagene (La Jolla, CA); DH5α cells were from Invitrogen. The pCEP-SP (20) and the AC7/pCEP-Pu (21) vectors have been described.
Human Cell Lines—SW1353 human chondrosarcoma cells (HTB-94) and MG63 human osteosarcoma osteoblast cells (CRL-1427) were obtained from American Type Culture Collection (Manassas, VA), and human embryonic kidney cells, 293/EBNA, were from Invitrogen.
Production of BMP/GDF Propeptides—cDNA encoding human BMP-7 propeptide was amplified by PCR using the BMP-7-pcDNA3.1(+) vector containing full-length BMP-7 cDNA as a template and master clone (12). The cDNAs from SW1353 and MG63 cells were generated using the Bio-Rad iScript™ cDNA synthesis kit, after harvesting total RNA using TRIzol reagent (Invitrogen). Coding regions for BMP and GDF prodomains were amplified from these cDNA sources by PCR with the Platinum™ Pfx DNA polymerase system using appropriate 5′- and 3′-primers designed from GenBank™ information (Table 1). The 5′ primers introduced an NdeI restriction site, whereas a BamHI site and six histidine residues in tandem followed by a termination signal were added to the downstream primers. PCR products were cloned into a NdeI/BamHI-digested pET11a vector such that each final construct contained the entire prodomain-coding sequence starting from the predicted endogenous signal peptide cleavage site and ending with the predicted furin cleavage site followed by a C-terminal His6 tag and a stop codon. Each vector construct was transformed into competent cells of E. coli DH5α, and the insert structure was verified by restriction analysis and DNA sequencing. Each BMP/GDF propeptide was overexpressed in E. coli BL21 (DE3) cells and purified using chelating chromatography under the same conditions as described previously (22), with slight changes in the case of the BMP-10 and the GDF-5 propeptides. Overexpressed propeptides of BMP-10 and GDF-5 showed higher solubility and were, therefore, purified out of the soluble fraction of the E. coli lysate under the same buffer conditions but with urea concentrations not higher than 2 m instead of 8 m. Purified propeptides are shown in Fig. 1A.
TABLE 1.
Sequences of 5′- and 3′-primer pairs for PCR amplification of BMP/GDF prodomain and GDF-8 complex cDNAs The design of the primer pairs was based on available GenBank™ information.
| TGF-β family member | GenBank™ accession no. | cDNA source | Primer forward (5′–3′) | Primer reverse (5′–3′) |
|---|---|---|---|---|
| BMP-2 | NM_001200 | MG63 | 5′–ggaattccatatgctcgttccggagctgggc–3′ | 5′–catgggatcctcagtgatggtgatggtgatgacgtttttctcttttgtggagagga–3′ |
| BMP-4 (S1) | M22490 | MG63 | 5′–ggaattccatatgagtttgatacctgagacggg–3′ | 5′–catgggatcctcagtgatggtgatggtgatgacgcttggccctcc–3′ |
| BMP-4 (S2) | M22490 | BMP-4 (S1) | 5′–ggaattccatatgagtttgatacctgagacggg–3′ | 5′–catgggatcctcagtgatggtgatggtgatgtcggctaatcctgacatg–3′ |
| BMP-10 | NM_014482 | MG63 | 5′–ggaattccatatgagccccatcatgaacctagag–3′ | 5′–catgggatcctcagtgatggtgatggtgatgccttctgattcgggcagtg–3′ |
| GDF-5 | NM_000557 | SW1335 | 5′–ggaattccatatggcccctgacttgggccagagac–3′ | 5′–catgggatcctcagtgatggtgatggtgatgccgccgttttcgccgctg–3′ |
| GDF-8 | NM_010834 | I.M.A.G.E. clone #40047208 | 5′–gtcagctagctgatttttaaaatgatgcaaaaactgc–3′ | 5′–gtcactcgagtcatgagcacccacagcggtctac–3′ |
FIGURE 1.
Coomassie Blue-stained quality control gels of the recombinant BMP/GDF pd and fibrillin peptides used in this study. A, non-reducing SDS-PAGE (12.5% acrylamide) of BMP-2, BMP-4 (full-length, S1, and shortened, S2), BMP-7, BMP-10, and GDF-5 pds. B, non-reducing SDS-PAGE of recombinant fibrillin-1 and fibrillin-2 peptides rF36, rF63, rF85, rF86, rF87, rF92, and rF93 in 5–12.5% acrylamide gels. Growth factor pds, expressed in bacteria, and fibrillin peptides, expressed in 293 cells, were purified to apparent homogeneity as described under “Experimental Procedures.”
Construction and Expression of Recombinant Fibrillin Subdomains and the GDF-8 Complex—To assemble expression vectors for recombinant production of fibrillin-1 and fibrillin-2 subdomains, FBN1 or FBN2 clones were PCR-amplified using Platinum™ Pfx DNA polymerase. To subclone the PCR products, the primers were designed to introduce an additional NheI restriction site at the 5′ end and either a NotI or XhoI restriction site at the 3′ end. In addition, the sequences for six histidine residues were introduced into the PCR product at the 3′ end (with the exception of rF63, where the 6×histidine tag sequence was placed at the 5′ end) to facilitate purification of the recombinant peptide using chelating chromatography. Ligation of the PCR products into the episomal expression vectors pCEP4/2III4 or AC7/pCEP-Pu (21), which contain the sequence for the BM40/SPARC signal peptide (23), results in secretion of recombinant peptides with four additional N-terminal amino acid residues (APLA) preceding the authentic fibrillin sequence. Human embryonic kidney cells, 293/EBNA, were transfected and selected with hygromycin B or puromycin (Sigma). These methodologies and protein purification methods have been described in more detail (20).
Expression vector rF86 was constructed from human FBN2 cDNA clones obtained by screening λgt11 unamplified placenta library (Clontech, Palo Alto, CA) with FBN1-specific PCR products, as described previously (9). One clone, UPΦ22-3, was used to amplify sequences for rF86 by PCR using appropriate primers (Table 2). For rF87, rF92, and rF93, the rF23 expression construct (14) was used as a template for PCR. To generate rF85, two cDNA fragments, rF85A and rF85B, were generated by PCR using sequence specific primers and a fibrillin-1 full-length clone, HFBN29 (9), as a template. PCR fragment rF85A was digested with NheI/SpnI, rF85B with KpnI/NotI, and a full-length cDNA clone rF100 with KpnI/SpnI. All three obtained fragments were ligated into a pCEP-SP vector that had been predigested with NheI/NotI. Purified proteins from these newly constructed fibrillin recombinant constructs are shown in Fig. 1B. The expression construct for rF90 was generated as described earlier for rF11 (14) with the addition of a 6×histidine tag sequence at the 3′ end of the sequence coding for rF11.
TABLE 2.
Sequences of 5′- and 3′-primer pairs for PCR amplification of cDNAs coding for fibrillin-1 and fibrillin-2 subdomains
| Fibrillin fragment | Name | Prime forward (5′–3′) | Name | Primer reverse (5′–3′) |
|---|---|---|---|---|
| rF36 | rF36-S | 5′–ctgctagcagatttgcgaatgagctactgttatg–3′ | rF36-AS | 5′–ctctcgagttaatggtgatggtgatggtgtgctccattggtcatgaatcc–3′ |
| rF63 | rF63-S | 5′–taatgctagcacaccatcaccatcaccatggagacaatcgggaagggta–3′ | rF100AS | 5′–tgaaaatccaggttttgcttcactgagcggccgcgtcgacgggcccatag–3′ |
| rF85A | rF82-S | 5′–cgtagctagcagatatcaatgagtgcaagatg–3′ | FBN1–4383AS | 5′–gcaagttccaaagacaca–3′ |
| rF85B | FBN1–5200S | 5′–ggacagtgtcccatcccaa–3′ | rF85SA | 5′–agtttagcggccgctagtgatggtgatggtgatgagtgttactgcatgtgcccag–3′ |
| rF86 | FBN2–84S | 5′–gctagctagcccagcctcagcctcctcc–3′ | DR70 | 5′–atagtttagcggccgctagtgatggtgatggtgatgtatacaatctccatttgcatcctgc—3′ |
| rF87 | pCEP-5′ | 5′–gggcctggatcttctttctcc–3′ | rF87 AS | 5′–gactcgagtcagtgatggtgatggtgatgtatgcatctggtaccatctgg–3′ |
| rF92 | rF92 (1) | 5′–gtcagctagcggacgccaatttgg–3′ | rF92 (2) | 5′–gtcactcgagactagtgatggtgatggtgatgttcacactggggtccag–3′ |
| rF93 | rF93 (1) | 5′–gtcagctagctattgtccccatttgccgg–3′ | rF93 (2) | 5′–ggtttgtccaaactcatcaatg–3′ |
For the expression of GDF-8 complex, a cDNA fragment coding for full-length mouse GDF-8 was generated by PCR using specific primers (Table 1) and a cDNA clone (#40047208) purchased from the I.M.A.G.E consortium as a template. The amplified fragment was digested with NheI/XhoI and ligated into a pCEP-Pu vector. The resulting construct was transfected into 293/EBNA cells for protein expression.
Rotary Shadowing and Electron Microscopy—Purified BMP-7 complex (100 μg/ml) was dialyzed together with rF90 (160, 340, and 680 μg/ml) in 0.2 m NH4HCO3 with or without 2 mm CaCl2. These amounts were equivalent to molar ratios from 1:1 to 1:4 of BMP-7 complex to rF90. The presence of CaCl2 did not result in any noticeable difference. Each sample was diluted to 70% glycerol, then sprayed onto freshly cleaved mica and rotary-shadowed with Pt-C using a Balzers BAE 250 vacuum evaporator. Photomicrographs were taken using a Philips EM410 transmission electron microscope operated at 80 kV.
Velocity Sedimentation—Recombinant propeptides and growth factor dimers were mixed at molar ratios mentioned under “Results” and dialyzed against TBS or TBS containing 1 m urea. Because the GDF-8 and GDF-5 prodomain (pd) were more soluble than the BMP pd, experiments with GDF pd were conducted in TBS without urea. Aliquots (200 μl) were then pipetted onto the top of a 5–20% (w/v) sucrose gradient (3.6 ml total volume), buffered with TBS, and formed in Polyallomer tubes (11×3×60 mm; Beckman, Fullerton, CA). Ultracentrifugation experiments were performed for 22 h 15 min at 42,000 rpm (ω2t: 1.55 · 1012) at 4 °C in a Beckman L8-M ultracentrifuge using a Beckman SW 60Ti rotor. After a small hole was pricked with a pin in the bottom of the tubes, 8-drop fractions were collected. Fractions were trichloroacetic acid-precipitated, separated by non-reducing SDS-PAGE containing 12.5% (w/v) acrylamide, and analyzed by Western blot analysis. Protein loading was checked by Ponceau stain. Nitrocellulose membranes were developed with either SuperSignal™ (Pierce) or the Opti 4-CN™ substrate kit (Bio-Rad) according to the manufacturer's instructions. In some cases membranes were redeveloped after stripping with Restore Western blot Stripping Buffer (Pierce) and additional first and secondary antibody incubations.
Surface Plasmon Resonance—Binding analyses were performed using a BIAcoreX (BIAcore AB, Uppsala, Sweden). Propeptides of BMP-2, -4, -7, and -10 and GDF-5 and -8 (500 response units of each molecule) were covalently coupled to CM5 sensor chips (research grade) using the amine coupling kit following the manufacturer's instructions (BIAcore AB). Binding responses due to analyte interaction with the surface coupled ligand were normalized by subtraction of background binding to control flow cells. Binding assays were performed at 25 °C in 10 mm Hepes buffer, pH 7.4, containing 0.15 m NaCl, 3 mm EDTA, and 0.005% (v/v) P20 surfactant (HBS-EP buffer, BIAcore AB). Fibrillin peptides were diluted in HBS-EP buffer and then injected at several concentrations and different flow rates over immobilized BMP propeptides. For competition assays, rF23 was preincubated at a constant concentration of 20 nm with the competitor BMP propeptide at concentrations of 400-5 nm before injection. To account for variations of the rF23 signal due to buffer changes caused by the addition of different amounts of competitor, we generated for each competition sensorgram a buffer-matched control without competitor, and the maximum response was set in each case as the 100% reference signal. The surface was regenerated with a pulse of 10 mm glycine, pH 1.7. Kinetic constants were calculated by nonlinear fitting (1:1 interaction model with mass transfer) to the association and dissociation curves according to the manufacturer's instructions (BIAevaluation 3.0 software). Apparent equilibrium dissociation constants (KD values) were then calculated as the ratio of kd/ka.
Immunofluorescence Microscopy—Wild type mouse tissues were examined by immunofluorescence microscopy. C57Bl/6 pups were sacrificed at P1-P5, and limbs were sectioned and stained according to routine protocols (18). BMP-4 (mAb 751) and BMP-7 (mAb 3541) antibodies from R&D were diluted 1:50 in phosphate-buffered saline; pAb 9543 and 0868 were diluted 1:200. For coimmunolocalization, P3 limbs were sectioned and stained simultaneously with BMP-4 mAb and fibrillin-1 or fibrillin-2 pAb, detected with anti-mouse Alexa 488 and anti-rabbit Alex 568 (Invitrogen), and coverslipped with Prolong Gold antifade mounting medium with 4′,6-diamidino-2-phenylindole (Invitrogen). Micrographs were taken using a Zeiss Axiovert microscope equipped with Axiovision software.
RESULTS
Velocity Sedimentation Assay for BMP-7 Complex Formation—The BMP-7 complex consists of a mature growth factor dimer (gfd) and a pd (12). Velocity sedimentation was performed to analyze the migration of BMP-7 gfd, pd, and complex through sucrose gradients. Each sample was loaded onto a 5–20% sucrose gradient and subjected to ultracentrifugation. The entire gradient was collected in fractions beginning from the bottom to the top of the tube. Each fraction was trichloroacetic acid-precipitated and analyzed by non-reducing SDS-PAGE followed by immunoblotting using monoclonal antibodies specific for the BMP-7 gfd (mAb6) and pd (mAb2). Control reference runs of the 293-expressed and separated components showed that peak signals for the BMP-7 gfd and pd (Mr for each component is ∼31) appeared in fraction numbers 18–21 (gfd: Fig. 2A, right upper panel) and 21–24 (pd: Fig. 2A, left upper panel). In contrast, the control reference run of the BMP-7 complex (Mr ∼ 94.6) showed antibody signals for both BMP-7 components in the middle of the gradient (peak fractions 12–18, Fig. 2A, lower panels), demonstrating migration of the pd and gfd through the gradient together as a higher molecular weight complex. Trailing of the peak signals for the pd and the gfd in the complex reference runs (signals in fractions 17–20) were not present in reference runs without urea (data not shown), indicating that 1 m urea causes a slight complex dissociation. Bands at around 60–70 kDa in the reference run of the complex represent minor amounts of unprocessed full-length BMP-7 species, which represent up to 5% of the expressed recombinant BMP-7 (12).
FIGURE 2.
Velocity sedimentation analyses of BMP-7 complex formation. The ability of BMP-7 pd (expressed in bacteria) and separated BMP-7 gfd (expressed as a complex in 293 cells and separated as described (12)) to form a complex was tested using velocity sedimentation through sucrose gradients. A, control reference runs of separated BMP-7 pd (top left panel) and separated BMP-7 gfd (top right panel) show a peak around fraction 23 (immunoblotted with anti-BMP-7 pd antibody) and one around fractions 19–20 (immunoblotted with anti-BMP-7 gfd antibody). The control reference run for the BMP-7 complex (expressed in 293 cells) shows a peak around fraction 14 when detected with an anti-BMP-7 pd antibody (lower left panel) or with an anti-BMP-7 gfd antibody, indicating that BMP-7 pd and gfd migrate together as a complex through the gradient. B, BMP-7 complex formation after incubation of BMP-7 pd (expressed in bacteria) with separated BMP-7 gfd at a molar ratio of 2:1 (BMP-7 pd:BMP-7 gfd) was demonstrated by immunoblotting of fractions after velocity sedimentation using an anti-BMP-7 pd antibody (upper left panel) and an anti-BMP-7 gfd antibody (upper right panel). The BMP-7 pd and gfd signals were found in a similar position in the gradient as the native BMP-7 complex in the reference run (A), indicating the ability of the BMP-7 pd expressed in bacteria to form a complex with the BMP-7 gfd. The direction of sedimentation through the sucrose gradient is indicated under the fraction numbers (lower right panel).
To test whether the BMP-7 complex can be reconstituted from the individual components, BMP-7 gfd was incubated with BMP-7 pd, expressed in E. coli, in TBS containing 1 m urea overnight. The addition of 1 m urea was necessary because of the low solubility of the propeptide. To determine whether BMP-7 complex formation occurred, velocity sedimentation was performed. A reference run with E. coli-expressed BMP-7 pd showed peak signals in fractions 19–22 (Fig. 2B, lower left panel), sedimenting somewhat faster than 293-expressed and separated pd. Incubated together in a molar ratio of 2:1 BMP-7 pd:BMP-7 gfd, the sample showed a shift of the BMP-7 pd signal to the middle of the gradient (peak fractions 12–16, Fig. 2B, upper left panel). The BMP-7 gfd signal shifted along with the BMP-7 pd signal, indicating that complex formation with the pd had occurred (Fig. 2B, right panel). Moreover, since the experimentally constituted BMP-7 complex migrated through the sucrose gradient in a manner very similar to the genuine native BMP-7 complex (compare Fig. 2A, lower panels, with Fig. 2B), these data suggest that the BMP-7 complex can be properly constituted from its individual components.
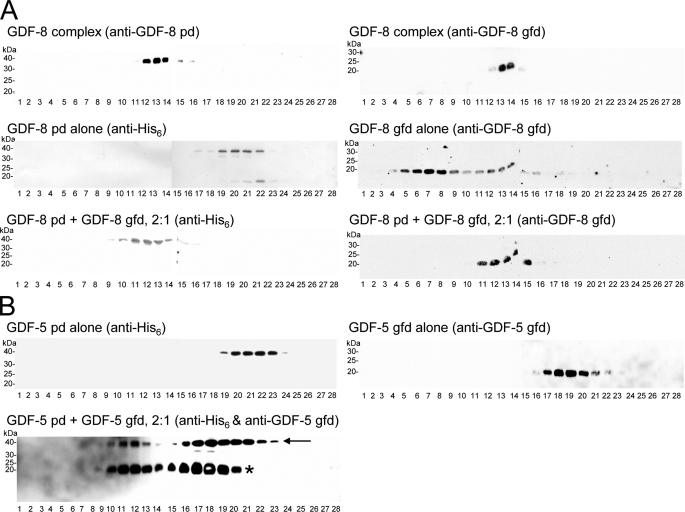
Velocity Sedimentation Assays for Complex Formation between pds and gfds of Other Members of the TGF-β Superfamily—To extend our results for BMP-7 complex formation, we compared recombinant GDF-8 complex expressed in 293 cells with complex formed in vitro from separately expressed components. A reference run for the GDF-8 complex, expressed in 293 cells, showed peak signals for both the GDF-8 pd and gfd in fractions 12–14 (pd: Fig. 3A, left upper panel; gfd: Fig. 3A, right upper panel), demonstrating that both the pd and gfd migrate together in a complex and that the GDF-8 complex and the BMP-7 complex (Fig. 2A, lower panels) sediment similarly.
FIGURE 3.
Complex formation by GDF-8 and the GDF-5 pds and gfds. A, the control reference run for the GDF-8 complex (expressed in 293 cells) shows peak signals in fractions 12–14 when detected with anti-GDF-8 pd (left upper panel) or with anti-GDF-8 gfd (right upper panel), showing that GDF-8 pd and gfd migrate together as a complex through the gradient. GDF-8 pd alone sedimented in fractions 19–22 (left middle panel). The GDF-8 pd was detected using an antibody specific for the 6×histidine tag. After incubation with GDF-8 gfd (pd:gfd ratio was 2:1), the GDF-8 pd shifted farther down in the gradient to fractions 10–15 (left lower panel). Immunoblotting with anti-GDF-8 gfd showed that the GDF-8 gfd co-migrated with the pd, indicating the formation of a complex similar to the BMP-7 complex (right lower panel). The reference run of the GDF-8 gfd alone showed peak signals in fractions 5–9 trailing past fraction 18 (right middle panel), demonstrating that in the absence of its cognate pd the gfd forms higher molecular weight aggregates of variable sizes. B, GDF-5 pd alone sedimented in fractions 19–23 (left upper panel), detected with anti-histidine. Sedimentation profiles of the GDF-8 and GDF-5 pds were similar to the BMP-7 pd. GDF-5 gfd alone sedimented in fractions 17–22 (right upper panel). Incubation of GDF-5 gfd with GDF-5 pd (pd:gfd ratio of 2:1) resulted in signals for the GDF-5 pd in fractions 10–23 (arrow, left lower panel). Detection with anti-GDF-5 gfd showed signals in fractions 10–20 (asterisk, left lower panel).
When sedimented by itself, the GDF-8 pd was detected primarily in fractions 19–22 (Fig. 3A, left middle panel), whereas incubation with the GDF-8 gfd resulted in a shift of the pd toward earlier fractions, fractions 10–15 (Fig. 3A, left lower panel). Using the same stripped blot, reprobing with anti-GDF-8 gfd demonstrated signals in the same fractions only, indicating quantitative complex formation of the GDF-8 gfd with its pd (Fig. 3A, right lower panel) and suggesting that the GDF-8 complex can be properly constituted from its separate components.
To test whether other members of the TGF-β superfamily can form complexes similar to the BMP-7 complex and GDF-8 complex, we incubated purchased BMP-2, BMP-4, BMP-10, and GDF-5 gfd together with their recombinantly expressed pd in a ratio of 2:1 (pd:gfd) and analyzed the samples after velocity sedimentation through sucrose gradients by SDS-PAGE and Western blotting of each fraction.
Analyses of GDF-5 complex formation showed a broad GDF-5 pd distribution in fractions 10–23 (Fig. 3B, arrow, left lower panel) after incubation with GDF-5 gfd. These fractions represented a significant shift farther down in the gradient from the reference run of uncomplexed GDF-5 pd alone (fractions 19–23) (Fig. 3B, left upper panel). GDF-5 gfd signals were also present in fractions 10–20 (Fig. 3B, asterisk, left lower panel), whereas uncomplexed GDF-5 gfd sedimented in fractions 16–22 (Fig. 3B, right upper panel). Comparison of these results with the reference runs for BMP-7 complex (fractions 12–18) and GDF-8 complex (fractions 12–14) and with complex formation by GDF-8 (fractions 10–15) suggests that GDF-5 in fractions 10–14 represents fully assembled complex. GDF-5 in fractions 15–18 may represent a partial complex of only one pd molecule in association with the gfd, and this partial complex was in equilibrium with uncomplexed pd and gfd present in the remaining fractions. Results similar to these with GDF-5 components were obtained with BMP-10 pd and gfd (data not shown).
When BMP-2 pd and gfd were tested, complex formation was poor. Using a 2:1 molar ratio of pd:gfd, BMP-2 pd signals appeared in fractions 17–23 (Fig. 4B, left upper panel) compared with signals in fractions 21–24 for the BMP-2 pd reference run (Fig. 4A, left upper panel). The reference control signals for the BMP-2 gfd appeared in fractions 19–23 (Fig. 4A, right upper panel). These results were consistent with only a weak interaction between the BMP-2 pd and gfd.
FIGURE 4.
Complex formation by BMP-4 and BMP-2 pds and gfds. A, BMP-2 pd alone sedimented in fractions 21–24 (left upper panel); BMP-4 pd alone sedimented in fractions 21–23 (left lower panel). Reference runs for the gfds showed peak signals in fractions 19–23 for BMP-2 gfd (right upper panel) and in fractions 17–22 for BMP-4 gfd (right lower panel). B, after incubation of BMP-4 gfd and pd at various molar ratios of BMP-4 pd:BMP-4 gfd (1:1, 2:1, and 4:1), the BMP-4 pd sedimented farther down in the sucrose gradient, indicating the formation of a BMP-4 complex. In contrast, at a 2:1 molar ratio of BMP-2 pd:BMP-2 gfd, BMP-2 pd signals were found in fractions 17–23, suggesting only very weak complex formation. C, the sequence of the C-terminal end of the BMP-4 prodomain is shown with cleavage sites boxed and marked as S1 or S2. When the pd ending with the S2 sequence (RISR) was incubated with BMP-4 gfd and tested by velocity sedimentation, the BMP-4 pd signal was found in fractions 22–24, similar to the sedimentation of the BMP-4 pd reference run (A, right lower panel) except that the peak fractions were slightly retarded.
Using a 2:1 molar ratio of BMP-4 pd:BMP-4 gfd, the BMP-4 pd shifted from fractions 21–23 (BMP-4 pd reference run, Fig. 4A, left lower panel) to fractions 14–19, seven fractions farther down in the gradient (Fig. 4B, molar ratio 2:1). Comparison of these results with the BMP-7 reference run (fractions 12–18) indicated that the BMP-4 pd and gfd had formed a complex. To further explore this complex formation, titration experiments varying the pd:gfd ratio from 1:1 to 4:1 were performed. With lower than stoichiometric amounts of pd, signals were found in fractions 16–19 (Fig. 4B, molar ratio 1:1), suggesting that species found in these fractions represented primarily one pd associated with one gfd. With higher than stoichiometric amounts of pd, only faint signals appeared in fractions 13 and 14, slightly farther down in the gradient (Fig. 4B, molar ratio 4:1), indicating that excess prodomain could not further influence complex formation.
BMP-4 has a second cleavage site (S2) upstream of the consensus furin motif (S1) (these sites are marked in the sequence shown in Fig. 4C). The shorter form of the BMP-4 pd, ending at the S2 site, was expressed recombinantly in bacteria, purified, and incubated with the BMP-4 gfd at molar ratios of 2:1 and 4:1 of pd:gfd. Velocity sedimentation assays showed that, even with excess pd (4:1), no shift of the pd signal could be induced, suggesting that the sequence between the two cleavage sites is essential for the formation of a complex. Sedimentation of the short BMP-4 pd appeared in fractions 22–24 (Fig. 4C), similar to the BMP-4 pd reference run (Fig. 4A, left lower panel). Consistent with the smaller size of the BMP-4 pd shortened at the S2 site, the peak was in fraction 22–24, slightly retarded in comparison with the reference run where the peak was in fractions 21–23.
Assays for Heterologous Complex Formation—To test whether complex formation between pds and gfds is specific or whether pds and gfds exhibit promiscuous interactions, velocity sedimentation assays were performed in which heterologous complex formation within the same subgroup of BMPs was tested. The BMP-5 or BMP-6 gfd was combined with the BMP-7 pd (Fig. 5, upper panels), the BMP-2 gfd with the BMP-4 pd (Fig. 5, right lower panel), and the BMP-4 gfd with the BMP-2 pd (Fig. 5, left middle panel). These attempts to form heterologous complexes between members of the same BMP subgroup were unsuccessful, indicating that specific interactions occur between pds and gfds.
FIGURE 5.
Formation of heterologous complexes between pds and gfds. Although BMP gfds within the BMP-2/BMP-4 and the BMP-5/-6/-7 subgroup share a high sequence similarity of 71–86% (similarity of pds within the subgroups: 54–69%), heterologous complexes consisting of a BMP-6 or BMP-5 gfd and a BMP-7 pd could not be formed (upper panels). The BMP-7 pd signals showed some trailing farther down in the gradient, but no quantitative shift was induced from its reference peak fractions (fractions 19–22). Similar trailing was found in attempts to assemble BMP-2/BMP-4 heterologous complexes (middle panels), without substantial shifts farther down in the gradient from the reference peak signals in fractions 21–23. Combination of BMP-7 pd with BMP-2 gfd also failed to result in a shift of the pd farther down in the gradient (lower panel).
Although sequence similarities between members belonging to different BMP subgroups are low (51–57% for gfds and 19–27% for pd sequences), it has been speculated that BMP-2/BMP-6 heterodimers may exist in vivo (24). Therefore, formation of heterologous complexes between subgroups was also tested. When BMP-7 pd was combined with BMP-2 gfd, complex formation did not occur (Fig. 5, lower left panel).
Interactions between Growth Factor Propeptides and Fibrillin—Recombinant BMP and GDF propeptides were tested for binding to recombinant fibrillin polypeptides (Figs. 1A and 6A) using surface plasmon resonance technology (Table 3 and Fig. 6B). The propeptides of BMP-2, -4, -7, and -10 and GDF-5 interacted with high affinities (KD = 5–10 nm) with rF23, a fibrillin-1 peptide composed of the N terminus followed by the subsequent 9 domains. In contrast, binding of the GDF-8 propeptide to rF23 was negative (data not shown). To test whether these propeptides also interact with fibrillin-2, a recombinant peptide, rF86, was constructed to resemble rF23. This fibrillin-2 peptide also interacted with the propeptides with comparable high affinities. In contrast, a fibrillin-2 peptide, rF37, which begins with the first epidermal growth factor (EGF)-like domain and ends with the 10th calcium binding EGF-like domain, failed to bind to all tested BMP/GDF propeptides, indicating that the major BMP/GDF propeptide binding site in fibrillin-2 is localized within the first two N-terminal domains.
FIGURE 6.
Surface plasmon resonance interaction experiments with immobilized BMP/GDF propeptides and fibrillin peptides in solution. A, schematic drawings represent the recombinant fibrillin polypeptides used as ligands. B, sensorgrams show binding of rF23 or rF86 at various concentrations to propeptides of BMP-2, -4, -7, -10, and GDF-5 (immobilized on CM5 chip). Numbers assigned to curves refer to the specified rF23 or rF86 concentrations. cb, calcium binding.
TABLE 3.
Dissociation constants (KD) determined by fitting surface plasmon resonance curves obtained for the interaction of different fibrillin peptides (Fig. 6A) and various BMP/GDF propeptides (BMP/GDF propeptides were used as ligands immobilized on CM5 chips and fibrillin peptides were used as analytes in solution) NB, no binding; ND, not determined. Kinetic constants were calculated by nonlinear fitting (1:1 interaction model with mass transfer) to the association and dissociation curves. Immobilized ligands are listed across the top of the table; analytes are listed in the rows.
| BMP-2 | BMP-4 | BMP-7 | BMP-10 | GDF-5 | |
|---|---|---|---|---|---|
| nm | |||||
| rF23 | 7 | 34 | 10 | 33 | 20 |
| rF45 | 50 | 190 | NB | 65 | 312 |
| rF85 | NB | NB | NB | NB | NB |
| rF36 | NB | NB | NB | NB | NB |
| rF63 | 21 · 103 | 3 · 103 | NB | 30 | NB |
| rF86 | 8 | 25 | 15 | 13 | 24 |
| rF37 | NB | NB | NB | NB | NB |
| rF47 | 150 | 20 · 103 | NB | nd | nd |
The BMP-2, -4, -10, and GDF-5 propeptides also interacted with strong to moderate affinities (KD = 50–300 nm) for rF45, which begins with the fourth epidermal growth factor-like domain and ends with the third 8-cysteine domain. A similar fibrillin-2 peptide rF47 also displayed weak to moderate binding (150 nm-20 μm) to BMP-2 and BMP-4 propeptides. Rather weak to strong affinities (KD = 21 μm–30 nm) of BMP-2, -4, and -10 propeptides were found for rF63, a peptide that represents the C-terminal end of fibrillin-1. A comparable fibrillin-2 peptide was not available for testing.
Competition of BMP-2, -4, -7, -10, and GDF-5 Propeptides for the Same Site on rF23—Competition studies were performed by preincubating rF23 (concentration of rF23 was kept constant at 20 nm) with increasing amounts of competitor propeptide (0–100 nm or 0–400 nm) followed by injection of the mixture onto a Biacore CM5 chip immobilized with the selected ligand propeptide (Table 4 and Fig. 7). All propeptides competed with inhibition constants ranging from 69 nm to 5.3 × 103nm (I50), suggesting that all propeptides bind to a common site in rF23.
TABLE 4.
Inhibition constants (I50) for competition of different propeptides against each other for the rF23 binding site BMP/GDF propeptides were immobilized as ligands on CM5 chips The fibrillin-1 peptide rF23 at 20 nm was injected in the presence of varying concentrations (400–100 to 0 nm) of competitor propeptides. The inhibition constants were determined by graphing the decrease in percentage of the rF23 signal at 20 nm without competitor (100%). Immobilized ligands are listed across the top of the table; competitors are listed in the rows.
| BMP-2 | BMP-4 | BMP-7 | BMP-10 | GDF-5 | |
|---|---|---|---|---|---|
| nm | |||||
| BMP-4 | 1.5 · 103 | 1.7 · 103 | 2.7 · 103 | 5.3 · 103 | |
| BMP-7 | 215 | 69 | 420 | 680 | |
| BMP-10 | 130 | 750 | 120 | 94 |
FIGURE 7.
Competition of BMP/GDF propeptides for rF23. Immobilized BMP/GDF propeptides were injected with 20 nm rF23 in the presence of increasing concentrations of different inhibitor propeptides. The signal in resonance units (RU) obtained for rF23 at 20 nm without competitor was set as 100%. The decrease of the 100% rF23 signal was graphed against the inhibitor concentrations to determine the inhibition constant for each competition reaction (Table 4). To account for variations of the rF23 signal due to buffer changes caused by the addition of different amounts of competitor, we generated for each competition sensorgram a buffer-matched control without competitor, which was set in each case as the 100% reference signal.
Localization of the High Affinity Binding Site for the BMP/GDF Propeptides within the Fibrillin-1 N Terminus—To further define the binding site of the BMP/GDF propeptides in rF23, immobilized BMP-7 propeptide was tested with recombinant fibrillin-1 peptides that represent subregions of rF23 (Fig. 8A). Recombinant fibrillin-1 peptides rF38, rF31, and rF93 failed to bind to BMP-7, whereas rF87 and rF92 interacted with dissociation constants of 16–20 nm (Table 5 and Fig. 8B) comparable with KDs obtained with rF23. These results indicated that the universal propeptide binding site may reside in the very N-terminal region of fibrillin-1.
FIGURE 8.
Localization of the BMP/GDF propeptide binding site within the N-terminal end of fibrillin-1. A, schematic drawings represent the recombinant fibrillin-1 subdomains of rF23 used in these experiments. B, surface plasmon resonance sensorgrams demonstrate binding of rF92 to immobilized BMP-7 pd. cb, calcium binding; RU, resonance units.
TABLE 5.
Dissociation constants (KD in nm) for the BMP-7 propeptide interaction with fibrillin-1 peptides spanning the N-terminal end of fibrillin-1 BMP-7 propeptide was used as immobilized ligand, and fibrillin peptides were used as analytes in solution. NB, no binding. Kinetic constants were calculated by nonlinear fitting (1:1 interaction model with mass transfer) to the association and dissociation curves. Fig. 8A shows a schematic representation of the fibrillin peptides.
| BMP-7 | |
|---|---|
| rF87 | 16 |
| rF92 | 20 |
| rF93 | NB |
| rF38 | NB |
| rF31 | NB |
As a second approach to localizing the binding site, BMP-7 complex was incubated with the N-terminal half of fibrillin-1 and visualized by electron microscopy after rotary shadowing (Fig. 9A). The globular BMP-7 complex appeared to interact at one end of the extended wiggly fibrillin molecule. Length measurements of rF90 molecules (shown in Fig. 9B) peaked at 90 nm, whereas the lengths of rF90 mixed with the BMP-7 complex peaked at 100–105 nm (Fig. 9D). Because the circumference of the BMP-7 complex (Fig. 9C) was determined to be 46 nm (12), the diameter of the complex is ∼15 nm. Therefore, the peak lengths of the rF90 molecules, with attached BMP-7 complex, suggested that the BMP-7 complex binds to the very end of the fibrillin-1 N terminus and is not bound within the first 15 nm of the N-terminal end. These data supported and extended the binding data shown in Fig. 8. Moreover, these results demonstrated that the BMP-7 pd, in association with its gfd, binds to fibrillin-1.
FIGURE 9.
Rotary shadowing electron microscopy depicting binding of the BMP-7 complex to the N-terminal half of fibrillin-1 (rF90). A, rotary shadowing electron microscopy showed globular molecules of BMP-7 complex bound to the very end of rF90. Images of individual molecules shadowed alone are shown for rF90 (B) and the BMP-7 complex (C). Scale bar = 25 nm. D, the lengths of rF90 molecules peaked at 90 nm (n = 122), whereas the peak lengths of rF90 + BMP-7 complex composites were 100–105 nm (n = 36).
Immunolocalization of BMPs in Mouse Tissues—We previously showed co-localization of BMP-7 pd and BMP-7 gfd with fibrillin-1 in human fetal skin and kidney using specific mAbs we generated (12). To test whether other BMP or GDF gfds are targeted to specific tissues, we used commercially available antibodies specific for BMP-2, BMP-4, BMP-7, GDF-5, and GDF-8 gfds and performed immunofluorescence microscopy on wild type mouse tissues. Commercial mAbs specific for BMP-7 and for BMP-4 stained tissues in a fibrillar pattern. Results with the R&D Systems mAb specific for BMP-7 (data not shown) were consistent with staining patterns obtained with our mAb 6, which we characterized previously (12). However, available antibodies specific for BMP-2, GDF-5, and GDF-8 were negative on the tissues (early postnatal limbs) that we tested.
Representative micrographs demonstrating fibrillar immunofluorescent staining patterns for fibrillin-1 (Fig. 10A, middle, and B, middle), fibrillin-2 (Fig. 10C, middle), and BMP-4 (Fig. 10A–C, left panels) are shown in sections of blood vessel (Fig. 10A) and dermis (Fig. 10, B and C). Sections were treated simultaneously with BMP-4 mAb and fibrillin-1 or fibrillin-2 pAb. Individual images as well as merged images (right panels) demonstrated colocalization of BMP-4 with fibrillin-1 and with fibrillin-2. Arrows point to selected areas of colocalization. Arrowheads indicate areas where BMP-4 appeared to be fibrillar, but fibrillin-2 localization to these areas was minimal. It is possible that these structures represent fibrils that were composed more of fibrillin-1 than fibrillin-2. In addition, BMP-4 staining was found in peripheral nerves and appeared to be absent in the skeletal muscle and perichondrium (data not shown).
FIGURE 10.
Co-immunolocalization of BMP-4 and fibrillins in wild type mouse tissues. A, P3 blood vessel, stained simultaneously with BMP-4 mAb (left panel) and pAb 9543 (specific for fibrillin-1) (middle panel), showed colocalization to fibrillar structures in the wall of the blood vessel. B, sections of dermis demonstrated colocalization of BMP-4 (left panel) and fibrillin-1 (middle panel) to fibrillar structures. C, fibrils in the dermis were also costained with BMP-4 (left panel) and fibrillin-2 pAb 0868 (middle panel). Arrows point to selected fibrils that are stained with both BMP-4 and fibrillin antibodies. Arrowheads indicate fibrils that are well stained with BMP-4 antibodies but not so well stained with fibrillin-2 antibodies. Panels on the right are merged images that also show 4′,6-diamidino-2-phenylindole staining of nuclei. Scale bar = 20 μm.
DISCUSSION
In this study we investigated whether BMP-2, -4, and -10 and GDF-5 and -8 can form complexes consisting of propeptides and processed growth factors and whether propeptides of these TGF-β family members can interact with fibrillin. We investigated complex formation by testing migration of propeptides alone or in the presence of growth factors through sucrose gradients. These velocity sedimentation experiments allowed us to demonstrate complex formation between the pd and gfd of BMP-7 and GDF-8 because migration of the reconstituted complexes in the sucrose gradient was the same as the migration of native BMP-7 or GDF-8 complexes.
Previously, we showed that the BMP-7 complex can be separated into its components, the BMP-7 gfd and the BMP-7 pd, using harsh denaturing conditions (8 m urea plus 20 mm octyl-glucoside) (12). Results presented in this manuscript now demonstrate that the dissociation of the BMP-7 complex is a reversible process. Because bacterially expressed BMP-7 pd was used, the results also show that the absence of carbohydrate chains had no influence on assembly of the complex. In addition, velocity sedimentation of the bacterially expressed BMP-7 pd and the pds of the other growth factors examined in this study clearly demonstrated a single peak in the gradient, indicating that the bacterially expressed pds are soluble and not aggregated into a variety of species that would spread throughout the gradient.
Our results with GDF-8 are consistent with previously published reports showing that GDF-8 forms a noncovalently associated complex with its pd in vivo (25) and that the GDF-8 complex can be reconstituted in vitro from separately expressed components, as suggested by size exclusion chromatography (26). Interestingly, we found that the GDF-8 gfd appears to aggregate in the absence of its pd, suggesting that the GDF-8 gfd requires its pd for solubility and may be poorly soluble or even less active in its uncomplexed form. Complex formation by bacterially expressed BMP-7 and GDF-8 pds and by a nonhomogeneous species of GDF-8 gfd strongly indicates that the most stable and soluble form of these components is a complex of pd and gfd.
Comparison of the other experimental components with the control BMP-7 and GDF-8 complexes in the velocity sedimentation assay demonstrated that BMP-4, BMP-10, and GDF-5 pds and gfds form complexes, suggesting that these complexes might also exist in vivo as has been shown not only for TGF-β, GDF-8, and BMP-7 but also for GDF-11 (27) and BMP-9 (28). Support for the in vivo existence of a BMP-4 complex comes from the recent finding that BMP-4 is present as a 100-kDa complex in fetal bovine serum (29).
In contrast to BMP-4, -7, and -10 and GDF-5 and -8, BMP-2 appeared to form much less stable complexes. This result is consistent with previously published data showing that the BMP-2 pd was more abundant in the medium of transfected cells than the processed BMP-2 gfd, suggesting that the majority of recombinant BMP-2 gfd was not associated with its pd (30).
It is possible that the failure of some components to form complexes is due to the artificial presence of histidine tags on the recombinant pds. However, the presence of 6×histidine tags either on the C- or N-terminal end did not prevent the BMP-7 pd from binding to its cognate gfd in enzyme-linked immunosorbent assay solid phase interaction assays (data not shown). Also the presence of a 10×histidine tag on the C-terminal end of the GDF-8 pd did not seem to hinder interaction with its cognate gfd.
Attempts to form heterologous complexes demonstrated that BMP-4 and -7 complex formation by their individual components was likely due to specific interactions and not to spurious interactions of propeptides with tendencies to aggregate. The formation of heterologous pd-gfd complexes at least among members of the same subgroup of the TGF-β family seemed plausible given the high sequence identity of the gfds (86% for the BMP-2/-4 subgroup and 71–80% for the BMP-5/-6/-7 subgroup) and the pds (54% for the BMP-2/-4 subgroup and 54–69% for the BMP-5/-6/-7 subgroup). However, because all attempts to form heterologous complexes failed, successful complex formation by individual components is likely to be driven by specific interactions.
Because the BMP-7 propeptide binds to fibrillin-1 and targets BMP-7 to fibrillin microfibrils (12), we investigated whether propeptides of BMP-2, -4, and -10 and GDF-5 and -8 interact with fibrillin. All propeptides, with the exception of GDF-8, bound with high affinity to the N-terminal recombinant fibrillin-1 peptide called rF23. A comparable recombinant fibrillin-2 peptide, rF86, interacted equally well with these propeptides, suggesting that fibrillin-2 microfibrils also serve as extracellular targets for BMP-2, -4, -7, and -10 and GDF-5. These studies are the first to show that in addition to BMP-7, propeptides of various other TGF-β family members have the ability to bind fibrillin and that different BMPs bind with high affinity to the same region in fibrillin-1 and fibrillin-2.
The N-terminal region of fibrillin-1 contained in rF23 displayed high binding affinities (7–34 nm) for all tested propeptides. Further investigations revealed that all tested propeptides compete for the same binding site within rF23 and that this binding site is located in the N-terminal “unique” region of fibrillin-1. This conclusion was supported by rotary shadowing electron microscopy showing molecules of BMP-7 complex bound at the very end of peptides composed of the N-terminal half of fibrillin-1. Furthermore, the rotary shadowing electron microscopy data provided the additional evidence that binding of the BMP-7 pd to fibrillin results in the targeting of the BMP-7 complex, including the BMP-7 gfd, to fibrillin.
In addition to the universal binding site located in the N terminus, we also identified two other regions containing interaction sites for propeptides. These other sites displayed different affinities for the tested propeptides (Table 3). The C-terminal region (represented by rF63) showed high binding affinity only to the BMP-10 propeptide (30 nm) and weak or no binding to the propeptides of BMP-2 (2 μm), BMP-4 (3 μm), BMP-7 (no binding), and GDF-5 (no binding). The region represented by rF45 demonstrated strong to moderate binding (50–312 nm) to all tested propeptides with the exception of the BMP-7 propeptide, which showed no binding. A comparable fibrillin-2 peptide, rF47, showed moderate to weak binding (150 nm-20 μm) to the pds of BMP-2 and -4 but no binding to BMP-7 pd, suggesting that a similar binding site might also be present in fibrillin-2. Another fibrillin-2 peptide, rF37, that lacks the N terminus did not bind to any of the tested BMP/GDF pds. This finding suggests that, analogous to fibrillin-1, the high affinity binding site for the BMP/GDF pds is located in the N terminus of fibrillin-2. However, rF37 should also contain the weaker binding site found in rF47. An explanation for this discrepancy might be that the weak binding site within the larger rF37 construct was not fully accessible.
Previously, we showed that guanidine-extracted microfibrils contained the binding site for BMP-7, in contrast to collagenase-digested microfibrils, which failed to bind to BMP-7 (22). Our new data that locate the BMP-7 binding site to the N terminus are consistent with these earlier findings, since crude collagenase cleavage sites that we previously identified were very close to the N terminus (22). In accord with the microfibril model in which monomeric fibrillin molecules are staggered with their N-terminal halves on the outside of the microfibril and C-terminal halves forming the core of the microfibril (22), we propose that the N-terminal high affinity BMP/GDF prodomain binding site is located in the globular bead region of the microfibril (Fig. 11). This site in the N terminus is likely exposed and available (22) to serve as a universal docking site for BMP growth factor complexes. Other potential binding sites in the C terminus and in the region represented by rF45 are mapped in the schematic diagram of fibrillin molecules within a microfibril (shaded regions, Fig. 11A). The C-terminal binding site for BMP-10 is positioned in the globular bead region of the microfibril, but it is unknown whether this site is exposed and available within the structure of the microfibril. The region represented by rF45 is likely to be on the surface of the microfibril and available for binding.
FIGURE 11.
Model of BMP/GDF growth factor complexes bound to fibrillin-containing microfibril networks. A, in this model of microfibrils, fibrillin-1 molecules are staggered with N-terminal halves on the outside of the microfibril and C-terminal halves forming the core of the microfibril (22). Binding sites for BMP/GDF growth factor complexes can be mapped to the shaded fibrillin-1 domains shown in the schematic representation. B, fibrillin microfibril networks with associated LTBPs sequester latent complexes of TGF-β (4). In addition, cells secrete BMPs as growth factor complexes (white butterflies), which are then targeted by prodomain/fibrillin interactions to specific positions on microfibrils. Cells receiving positional information through integrins or other receptors that interact with microfibril components determine whether to send or receive growth factor signals. Through this exchange of positional and signaling information, cells may either utilize growth factors immediately or store them on the microfibrils for later use. LAP, latency-associated peptide.
Because an 8-Cys domain in LTBPs binds to the propeptides of TGF-βs, our initial hypothesis was that specific 8-Cys domains in fibrillins would mediate binding to growth factors in the TGF-β superfamily. However, to our surprise, the universal high affinity binding site was localized to the N terminus of fibrillin-1. This region, which contains four cysteines, is homologous to the N termini of fibrillins and LTBPs, raising the possibility that LTBPs may also mediate binding to BMPs. In addition, although it has been reported that fibrillins do not interact with TGF-βs (3), in view of other recent data strongly implicating fibrillin in the TGF-β signaling pathway (6–8), it may be that 8-Cys domains function to generally mediate binding to TGF-β propeptides. However, binding of BMPs to rF45 could be mediated by either of the two 8-Cys domains present in this region. Further study is required to more precisely define the binding site(s) in this region.
Immunofluorescence microscopy of human fetal tissues demonstrated the presence of both the BMP-7 propeptide and BMP-7 growth factor in fibrillar patterns co-localized with fibrillin-1 (12). In this study we used commercially available antibodies to growth factors and immunofluorescence microscopy of early postnatal mouse tissues to determine the localization of any of the selected growth factors. Antibody specific for BMP-7 (R&D Systems) gave immunolocalization patterns similar to the fibrillar patterns generated by our mAb 6. Antibodies to BMP-2, GDF-5, and GDF-8 (R&D Systems) were negative using the conditions, tissues, and time points tested. However, antibody specific for the BMP-4 growth factor (R&D Systems) demonstrated fibrillar staining in blood vessels, dermis, and peripheral nerves. Because BMP-4 is known to be expressed in other tissues, it was surprising that strong immunofluorescence was not found in, for example, the perichondrium.
In contrast to BMP-7, the BMP-4 propeptide contains a second cleavage site which controls the activity and signaling range of BMP-4 (31). Moreover, studies in mice in which the S2 site is mutated to prevent cleavage showed that cleavage of this site is required for normal development and that cleavage at this site is tissue-specific (32). Our in vitro velocity sedimentation assay for complex formation indicates that the BMP-4 pd shortened at the S2 site cannot form a complex with the BMP-4 gfd, in contrast to the full-length BMP-4 pd, which can form a complex. These results are consistent with co-immunoprecipitation experiments showing noncovalent association of S1-cleaved BMP-4 pd with gfd but no association of S2 cleaved BMP-4 pd with gfd (33). All together, we suggest that cleavage at the S2 site can regulate the activity of BMP-4 by releasing the BMP-4 gfd from noncovalent linkage with its pd, that immunolocalization of the BMP-4 gfd in certain tissues may represent stored BMP-4 complexes, and that the absence of abundant stores of BMP-4 in other tissues may indicate tissue-specific activation of BMP-4 in these other tissues (e.g. perichondrium). Stored BMP-4 complexes, presumably cleaved only at the S1 site, must also escape the lysosomal targeting and degradation proposed by Degnin et al. (33).
In summary, we have provided evidence that the extracellular regulation of BMP growth factor signaling includes the stability of the pd·gfd complex and the targeting of these complexes to the connective tissue by interactions between propeptides and fibrillin. Variations in the stability of pd·gfd complexes in pd sequences that may be recognized by specific activators and in binding sites and affinities for fibrillin may be mechanisms by which these very similar growth factors function differently in a tissue-specific manner. Studies of fibrillin-1 mutant mice have demonstrated dysregulated TGF-β signaling (6–8), consistent with a model of the microfibril network in which LTBPs are stabilized by interaction with fibrillin microfibrils (Fig. 11) (4). Ongoing studies of fibrillin mutant mice indicate that BMP signaling is perturbed.3 These studies may provide in vivo evidence for an essential role performed by fibrillin in the extracellular regulation of BMP signaling. Future studies should be directed toward the integration of these complex signaling pathways by the extracellular physical scaffold and toward the roles performed by cells as they interact coordinately with stored growth factors and with the scaffold (Fig. 11).
Acknowledgments
We thank Dr. Kerry Maddox and the Analytical Core Facility of the Portland Shriners Hospital for protein sequencing and amino acid analysis, and we acknowledge Bruce A. Boswell, Sara Tufa, Valerie Carlberg, Steve Chalberg, and Glen Corson for help with the experimental procedures and with engineering the fibrillin recombinant peptides. We appreciate the helpful discussions and comments on the manuscript from Dr. Jan L. Christian (Oregon Health and Science University).
This work was supported, in whole or in part, by National Institutes of Health Grants P01 AR049698 and RO1AR46811 (to L. Y. S.). This work was also supported by the Shriners Hospitals for Children (to L. Y. S., D. R. K., and H. P. B.) and by Deutsche Forschungsgemeinschaft Forschungsstipendium SE1115/1-1 (to G. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TGF-β, transforming growth factor β; LTBP, latent TGF-β-binding protein; BMP, bone morphogenetic protein; 8-Cys, 8 cysteine; GDF, growth and differentiation factor; mAb, monoclonal antibody; pAb, polyclonal antibody; rF, recombinant fibrillin; TBS, Tris-buffered saline; pd, prodomain; gfd, growth factor dimer.
G. Sengle, V. M. Carlberg, N. L. Charbonneau, D. R. Keene, F. Ramirez, and L. Y. Sakai, unpublished observations.
References
- 1.De Crescenzo, G., Grothe, S., Zwaagstra, J., Tsang, M., and O'Connor-Mc-Court, M. D. (2001) J. Biol. Chem. 276 29632-29643 [DOI] [PubMed] [Google Scholar]
- 2.Miyazono, K., Hellman, U., Wernstedt, C., and Heldin, C. H. (1988) J. Biol. Chem. 263 6407-6415 [PubMed] [Google Scholar]
- 3.Saharinen, J., and Keski-Oja, J. (2000) Mol. Biol. Cell 11 2691-2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isogai, Z., Ono, R. N., Ushiro, S., Keene, D. R., Chen, Y., Mazzieri, R., Charbonneau, N. L., Reinhardt, D. P., Rifkin, D. B., and Sakai, L. Y. (2003) J. Biol. Chem. 278 2750-2757 [DOI] [PubMed] [Google Scholar]
- 5.Rifkin, D. B. (2005) J. Biol. Chem. 280 7409-7412 [DOI] [PubMed] [Google Scholar]
- 6.Neptune, E. R., Frischmeyer, P. A., Arking, D. E., Myers, L., Bunton, T. E., Gayraud, B., Ramirez, F., Sakai, L. Y., and Dietz, H. C. (2003) Nat. Genet. 3 407-411 [DOI] [PubMed] [Google Scholar]
- 7.Habashi, J. P., Judge, D. P., Holm, T. M., Cohn, R. D., Loeys, B. L., Cooper, T. K., Myers, L., Klein, E. C., Liu, G., Calvi, C., Podowski, M., Neptune, E. R., Halushka, M. K., Bedja, D., Gabrielson, K., Rifkin, D. B., Carta, L., Ramirez, F., Huso, D. L., and Dietz, H. C. (2006) Science 312 117-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohn, R. D., van Erp, C., Habashi, J. P., Soleimani, A. A., Klein, E. C., Lisi, M. T., Gamradt, M., ap Rhys, C. M., Holm, T. M., Loeys, B. L., Ramirez, F., Judge, D. P., Ward, C. W., and Dietz, H. C. (2007) Nat. Med. 13 204-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corson, G. M., Chalberg, S. C., Dietz, H. C., Charbonneau, N. L., and Sakai, L. Y. (1993) Genomics 17 476-484 [DOI] [PubMed] [Google Scholar]
- 10.Gleizes, P. E., Beavis, R. C., Mazzieri, R., Shen, B., and Rifkin, D. B. (1996) J. Biol. Chem. 271 29891-29896 [DOI] [PubMed] [Google Scholar]
- 11.Yuan, X., Downing, A. K., Knott, K., and Handford, P. A. (1997) EMBO J. 16 6659-6666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregory, K. E., Ono, R. N., Charbonneau, N. L., Kuo, C. L., Keene, D. R., Bachinger, H. P., and Sakai, L. Y. (2005) J. Biol. Chem. 280 27970-27980 [DOI] [PubMed] [Google Scholar]
- 13.Hollister, D. W., Godfrey, M., Sakai, L. Y., and Pyeritz, R. E. (1990) N. Engl. J. Med. 323 152-159 [DOI] [PubMed] [Google Scholar]
- 14.Reinhardt, D. P., Sasaki, T., Dzamba, B. J., Keene, D. R., Chu, M.-L., Gohring, W., Timpl, R., and Sakai, L. Y. (1996) J. Biol. Chem. 271 19489-19496 [DOI] [PubMed] [Google Scholar]
- 15.Keene, D. R., Jordan, C. D., Reinhardt, D. P., Ridgway, C. C., Ono, R. N., Corson, G. M., Fairhurst, M., Sussman, M. D., Memoli, V. A., and Sakai, L. Y. (1997) J. Histochem. Cytochem. 45 1069-1082 [DOI] [PubMed] [Google Scholar]
- 16.Reinhardt, D. P., Gambee, J. E., Ono, R. N., Bachinger, H. P., and Sakai, L. Y. (2000) J. Biol. Chem. 275 2205-2210 [DOI] [PubMed] [Google Scholar]
- 17.Reinhardt, D. P., Ono, R. N., Notbohm, H., Muller, P. K., Bachinger, H. P., and Sakai, L. Y. (2000b) J. Biol. Chem. 275 12339-12345 [DOI] [PubMed] [Google Scholar]
- 18.Charbonneau, N. L., Dzamba, B. J., Ono, R. N., Keene, D. R., Corson, G. M., Reinhardt, D. P., and Sakai, L. Y. (2003) J. Biol. Chem. 278 2740-2749 [DOI] [PubMed] [Google Scholar]
- 19.Pereira, L., Andrikopoulos, K., Tian, J., Lee, S. Y., Keene, D. R., Ono, R. N., Reinhardt, D. P., Sakai, L. Y., Jensen-Biery, N., Bunton, T., Dietz, H. C., and Ramirez, F. (1997) Nat. Genet. 17 218-222 [DOI] [PubMed] [Google Scholar]
- 20.Reinhardt, D. P., Keene, D. R., Corson, G. M., Poschl, E., Bachinger, H. P., Gambee, J. E., and Sakai, L. Y. (1996) J. Mol. Biol. 258 104-116 [DOI] [PubMed] [Google Scholar]
- 21.Kohfeldt, E., Maurer, P., Vannahme, C., and Timpl, R. (1997) FEBS Lett. 414 557-561 [DOI] [PubMed] [Google Scholar]
- 22.Kuo, C. L., Isogai, Z., Keene, D. R., Hazeki, N., Ono, R. N., Sengle, G., Bachinger, H. P., and Sakai, L. Y. (2007) J. Biol. Chem. 282 4007-4020 [DOI] [PubMed] [Google Scholar]
- 23.Mayer, U., Poschl, E., Gerecke, D. R., Wagman, D. W., Burgeson, R. E., and Timpl, R. (1995) FEBS Lett. 365 129-132 [DOI] [PubMed] [Google Scholar]
- 24.Schmid, B., Fürthauer, M., Connors, S. A., Trout, J., Thisse, B., Thisse, C., and Mullins, M. C. (2000) Development 127 957-967 [DOI] [PubMed] [Google Scholar]
- 25.Hill, J. J., Davies, M. V., Pearson, A. A., Wang, J. H., Hewick, R. M., Wolfman, N. M., and Qiu, Y. (2002) J. Biol. Chem. 277 40735-40741 [DOI] [PubMed] [Google Scholar]
- 26.Thies, R. S., Chen, T., Davies, M. V., Tomkinson, K. N., Pearson, A. A., Shakey, Q. A., and Wolfman, N. M. (2001) Growth Factors 18 251-259 [DOI] [PubMed] [Google Scholar]
- 27.Ge, G., Hopkins, D. R., Ho, W. B., and Greenspan, D. S. (2005) Mol. Cell. Biol. 14 5846-5858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown, M. A., Zhao, Q., Baker, K. A., Naik, C., Chen, C., Pukac, L., Singh, M., Tsareva, T., Parice, Y., Mahoney, A., Roschke, V., Sanyal, I., and Choe, S. (2005) J. Biol. Chem. 280 25111-25118 [DOI] [PubMed] [Google Scholar]
- 29.Kodaira, K., Imada, M., Goto, M., Tomoyasu, A., Fukuda, T., Kamijo, R., Suda, T., Higashio, K., and Katagiri, T. (2006) Biochem. Biophys. Res. Commun. 345 1224-1231 [DOI] [PubMed] [Google Scholar]
- 30.Israel, D. I., Nove, J., Kerns, K. M., Moutsatsos, I. K., and Kaufman, R. J. (1992) Growth Factors 7 139-150 [DOI] [PubMed] [Google Scholar]
- 31.Cui, Y., Hackenmiller, R., Berg, L., Jean, F., Nakayama, T., Thomas, G., and Christian, J. L. (2001) Genes Dev. 15 2797-2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldman, D. C., Hackenmiller, R., Nakayama, T., Sopory, S., Wong, C., Kulessa, H., and Christian, J. L. (2006) Development 133 1933-1942 [DOI] [PubMed] [Google Scholar]
- 33.Degnin, C., Jean, F., Thomas, G., and Christian, J. L. (2004) Mol. Biol. Cell 15 5012-5020 [DOI] [PMC free article] [PubMed] [Google Scholar]