Abstract
Many of the most virulent strains of Salmonella enterica produce two distinct Cu,Zn-superoxide dismutases (SodCI and SodCII). The bacteriophage-encoded SodCI enzyme makes the greater contribution to Salmonella virulence. We have performed a detailed comparison of the functional, structural, and regulatory properties of the Salmonella SodC enzymes. Here we demonstrate that SodCI and SodCII differ with regard to specific activity, protease resistance, metal affinity, and peroxidative activity, with dimeric SodCI exhibiting superior stability and activity. In particular, monomeric SodCII is unable to retain its catalytic copper ion in the absence of zinc. We have also found that SodCI and SodCII are differentially affected by oxygen, zinc availability, and the transcriptional regulator FNR. SodCII is strongly down-regulated under anaerobic conditions and dependent on the high affinity ZnuABC zinc transport system, whereas SodCI accumulation in vitro and within macrophages is FNR-dependent. We have confirmed earlier findings that SodCII accumulation in intracellular Salmonella is negligible, whereas SodCI is strongly up-regulated in macrophages. Our observations demonstrate that differences in expression, activity, and stability help to account for the unique contribution of the bacteriophage-encoded SodCI enzyme to Salmonella virulence.
Copper, zinc-superoxide dismutases (Cu,Zn-SODs)3 are metalloenzymes that play a key role in protecting cells from the toxic effects of reactive oxygen species (1). These enzymes are present in nearly all eukaryotic cells and in several bacteria, where they are invariably located in extracytoplasmic compartments (2). This localization has led to the suggestion that Cu,Zn-SOD protects bacteria from extracellular superoxide, for example, as generated by phagocytic cells (2, 3). In agreement with this view, several studies have established that Cu,Zn-SOD confers protection against reactive oxygen species generated in vitro (4–9), facilitates bacterial survival within eukaryotic cells (4, 7, 10–16), and enhances bacterial virulence (4, 5, 7–9, 15, 17, 20–25). It is likely that periplasmic Cu,Zn-SOD plays additional roles that are not directly related to bacterial interaction with animal cells, as the enzyme is also present in non-pathogenic microorganisms. An interesting possibility is that Cu,Zn-SOD scavenges superoxide released in the periplasmic space during aerobic growth (26).
All Salmonella enterica strains possess a conserved chromosomal gene (sodCII) that is the orthologue of Escherichia coli sodC (18). A second gene, sodCI, is carried by the Gifsy-2 bacteriophage found in a large number of highly virulent strains belonging to different serovars including typhimurium, enteritidis, dublin, choleraesuis, and heidelberg (18, 19). Even though a number of studies have evaluated the contribution of sodCI and sodCII to Salmonella physiology and virulence, several controversies concerning the Cu,Zn-superoxide dismutases of Salmonella remain.
One issue concerns the relative contribution of SodCI and SodCII to Salmonella virulence. Whereas all investigations have clearly demonstrated a significant contribution of SodCI to the ability of Salmonella to replicate and cause disease in an infected host, the involvement of SodCII in the host-pathogen interaction has remained a matter of disagreement. Some studies have suggested that inactivation of sodCII significantly reduces pathogenicity of Salmonella (8, 16, 18), implying an additive contribution of the two Salmonella SodC enzymes. In contrast, others have suggested that the role of SodCII in virulence is negligible (21, 23, 24). It has been proposed, but not demonstrated, that the apparent contribution of SodCII to Salmonella virulence observed in some studies might have been due to toxic effects of a truncated sodCII allele (23, 24).
A second issue concerns whether SodCI and SodCII are functionally equivalent. Although these proteins catalyze the same reaction, two groups have reported that the sodCI and sodCII genes are not interchangeable, suggesting that the two proteins have distinctive structural or functional properties (23, 24). SodCI exhibits the highest catalytic rate ever observed in a natural Cu,Zn-SOD (27), but Cu,Zn-SODs in general are characterized by very high catalytic rates (28, 29). More significantly, SodCI is a dimeric protein, whereas SodCII is a monomer (16). Extensive characterization of different bacterial Cu,Zn-SODs has established that monomeric and dimeric Cu,Zn-SODs have very different activity, thermal stability, metal affinity, and resistance to proteases (29, 30). Slauch and co-workers have recently reported that SodCII is more susceptible than SodCI to protease digestion (25), but found other properties of the two enzymes to be identical (23). In addition, these authors reported that the activity of SodCII, but not SodCI, can be released by osmotic shock (23, 25), and hypothesized that SodCI “tethering” to the periplasm might somehow explain different physiological roles of SodCI and SodCII.
Finally, it has been uncertain whether SodCI and SodCII might make different contributions to bacterial virulence as a consequence of differences in gene regulation. Uzzau et al. (21) have shown that only negligible amounts of SodCII can be identified in S. enterica serovar typhimurium (Salmonella typhimurium) cells recovered from cultivated macrophages or from the spleens of infected mice, whereas SodCI accumulates to high levels in the same cells. This observation was recently confirmed for Salmonella enterica serovar enteritidis (Salmonella enteritidis), and the differential accumulation of SodCI and SodCII in intracellular bacteria was ascribed to differences in gene expression levels (24). However, these observations contrast with reports showing that sodCII is actively transcribed within macrophages (31, 32) and within the infected host (23).
In an attempt to clarify these conflicting issues, we have investigated the contribution of sodCI and sodCII to bacterial virulence, the regulation of the two genes under different environmental conditions, and the biochemical properties of the SodC enzymes. We conclude that SodCI makes a greater contribution to Salmonella virulence than SodCII in the murine model of infection as a result of differences in the properties of the two enzymes as well as in intracellular gene expression.
EXPERIMENTAL PROCEDURES
Salmonella Strains and Growth Conditions—Bacterial strains used in this work are listed in Tables 1 and 2. To assess the role of sodCII in S. typhimurium virulence, the wild type and mutant strains from the Fang and Bossi laboratories were used in parallel. Mutant alleles were transduced into wild type backgrounds (BWT or FWT) using phage P22 HT105/1 int-201 (34). The resulting strains are listed in Table 2. All strains were grown in Luria Bertani (LB) medium supplemented with antibiotics when required (100 mg/liter ampicillin, 50 mg/liter kanamycin, 30 mg/liter chloramphenicol). Anaerobic growth was achieved in GasPack anaerobic jars (BBL). Minimal medium was Vogel Bonner E supplemented with 0.2% glucose. Oligonucleotides and plasmids used for mutant construction via the λ-red recombinase method are listed in Table 3.
TABLE 1.
Bacterial strains
| Bacterial strainsa | Relevant genotype | Source or Ref. |
|---|---|---|
| Salmonella enterica serovar typhimurium strains | ||
| Wild type 14028s (BWT) | 19 | |
| MA7039 (B2BWT) | sodCII::phoA::kan | 21 |
| MA7224 | sodCI::3XFLAG-kan ilvI::Tn10dTac-cat::3XFLAG-kan | 21 |
| MA7225 | sodCII::3XFLAG-kan ilvI::Tn10dTac-cat::3XFLAG-kan | 21 |
| MA7537 | 5′CI::sodCII::3XFLAG-kan ΔsodCI ilvI::Tn10dTac-cat::3XFLAG-kan | 24 |
| MA7538 | 5′CII::sodCI::3XFLAG-kan ΔsodCII ilvI::Tn10dTac-cat::3XFLAG-kan | 24 |
| SA120 | sodC1::3XFLAG::scar ilvI::Tn10dTac-cat::3XFlag::scar | This study |
| SA121 | sodC2::3XFLAG::scar ilvI::Tn10dTac-cat::3XFlag::scar | This study |
| SA122 | fnr::kan | This study |
| SA123 | znuA::kan | 54 |
| SA126 | fnr::kan sodCI::3XFLAG ilvI::Tn10dTac-cat::3XFLAG | This study |
| SA127 | fnr::kan sodCII::3XFLAG ilvI::Tn10dTac-cat::3XFLAG | This study |
| SA171 | znuA::kan sodCI::3XFLAG ilvI::Tn10dTac-cat::3XFLAG | This study |
| SA153 | znuA::kan sodCII::3XFLAG ilvI::Tn10dTac-cat::3XFLAG | This study |
| SA130 | pCI-lacZ[amp] | This study |
| SA131 | pCII-lacZ[amp] | This study |
| SA158 | znuA::kan pCII-lacZ[amp] | This study |
| SA175 | znuA::kan pCI-lacZ[amp] | This study |
| Wild type 14028s | pSEsodCI | This study |
| Wild type 14028s | pSEsodCII | This study |
| Wild type 14028s | pPLsodC | This study |
| Wild type 14028s (FWT) | F. F. laboratory collection | |
| F2FWT | sodCI::kan | F. F. laboratory collection |
| F3FWT | sodCII::kan | This study |
| MF1007 (F6OWT)b | sodCII::pRR10(ΔtrfA) | 18 |
| SA167 | sodCI::3XFLAG-kan ilvI::Tn10dTac-cat::3XFLAG-kan | This study |
| SA168 | sodCII::3XFLAG-kan ilvI::Tn10dTac-cat::3XFLAG-kan | This study |
| Escherichia coli strains | ||
| 71/18 | F′ lac Iq Δ(lacZ)M15 proA+B+/Δ(lac-proAB) thi supE | 33 |
| 71/18 | pSEsodCI | This study |
| 71/18 | pSEsodCII | This study |
| 71/18 | pPLsodC | This study |
Nomenclature adopted in this work is indicated in parentheses: each strain is identified by the mutated allele (in italic) and the background strain.
Mutant is in an ATCC14028-derived background in which rpoS is down-regulated (F. F., personal comunication).
TABLE 2.
S. typhimurium strains used in mouse infection studies
| Name | Allele description | P22 donor strain | Recipient strain |
|---|---|---|---|
| B2FWT | sodCII::phoA::kan | B2BWT | FWT |
| F6BWT | sodCII::pRR10(ΔtrfA) | F6OWT | BWT |
| F3BWT | sodCII::kan | F3FWT | BWT |
| F6FWT | sodCII::pRR10(ΔtrfA) | F6OWT | FWT |
TABLE 3.
Oligonucleotides and plasmids
| Name | Sequence or reference |
|---|---|
| Oligonucleotides | |
| Oli-117 | CTGGCGGTTGTGCTATCCATTGCCAGGATTGCAGTATCAGTGTAGGCTGGAGCTGCTTCG |
| Oli-118 | TTTTCTGGAAACGCCCCAGCAGACGGCTAATGGTTTCGACCATATGAATATCCTCCTTAG |
| Oli-123 | ATTGTTGTTCCCTGCCATCG |
| Oli-125 | GATTGTGCGTATCGCGGAGCTGACGGCTAAGCGCGGGCAGGACTACAAAGACCATGACGG |
| Oli-126 | CATGCTGCAGATTGTTGCTGGCGCTGGGACCAGGAAGGCGCATATGAATATCCTCCTTAG |
| MC101 | CTTCATCAAGAGACTTCAGACGCGGCGCAGTAACCGGTTCGCTGGCGATAATCCGGGGTCCGTCGACC |
| MC102 | GCGATTAAGTTTAGCGATGGTGACGCTGCTGGCCTGTGCGGGTGCGCAGGGTGTAGGCTGAGCTGCTTC |
| MC103 | CCGTAATCGCCAACTCGTTAC |
| MC104 | CGCCCACATGGATCATGAGCGC |
| K1 | Datsenko and Wanner (35) |
| Promc1lacZFor | Ammendola et al. (16) |
| Promc1lacZRev | Ammendola et al. (16) |
| Promc2lacZFor | Ammendola et al. (16) |
| Promc2lacZRev | Ammendola et al. (16) |
| Plasmids | |
| Name | Features or reference |
| pSEsodCI | Plasmid overexpressing S. typhimurium SodCI (A. B. lab collection) |
| pSEsodCII | Plasmid overexpressing S. typhimurium SodCII (A. B. lab collection) |
| pPLSOD | Plasmid overexpressing P. leiognathi SodC (D'Orazio et al. (43)) |
| pMC1403 | Cloning plasmid (Ammendola et al. (16)) |
| pKD46 | λ-Red recombinase functions (Datsenko and Wanner (35)) |
| pKD4 | Kanamycin cassette template (Datsenko and Wanner (35)) |
| pCP20 | FLP recombinase expressing plasmid (Datsenko and Wanner (35)) |
All gene replacement experiments were carried out by the λ-red-mediated recombination procedure as described by Datsenko and Wanner (35). After replacement, constructions were moved into the appropriate genetic background by P22 transduction (34). When necessary, antibiotic resistance cassettes were excised using FLP recombinase encoded in plasmid pCP20 (36). The first 368 nucleotides of the sodCII gene (368 of 521 bases, strain F3FWT) were replaced by the kanamycin resistance cassette of plasmid pKD4 (using primers MC101 and MC102). PCR verification of replacement was performed with primers MC103 and MC104. A similar procedure was used to construct strain SA122 carrying a deletion of the fnr coding sequence spanning from nucleotide 119 to 703 (primers oli-117 and oli-118 were used in combination with plasmid pKD4; PCR verification of the construct was performed with primers oli-123 and K1). The fnr::kan allele was transferred into strains SA120 and SA121 to yield strains SA126 and SA127, respectively (Table 1).
For studies concerning the role of zinc availability on sodCI and sodCII expression, a znuA::kan mutant was employed (SA123, Table 1). The znuA::kan allele from strain SA123 was transduced into strains SA120 and SA121, to yield SA171 and SA153.
Cloning of sodC Promoters and β-Galactosidase Activity Assay—The sodC promoter fragments were amplified by PCR from S. typhimurium ATCC14028 genomic DNA by using oligonucleotides Promc1lacZFor and Promc1lacZRev for sodCI and Promc2lacZFor and Promc2lacZRev for sodCII (Table 3). Amplified fragments were digested with EcoRI and BamHI and inserted in the promoter probe plasmid pMC1403 (Table 3), yielding plasmids pPCI-lacZ and pPCII-lacZ. The vectors were introduced by electroporation into S. typhimurium wild type and mutant strain SA123 (znuA::kan). β-Galactosidase activity was measured as described (37).
Extraction of Periplasmic Proteins and Detection of SOD Activity—Overnight cultures of mutant and wild type strains grown in LB medium supplemented with 250 μm CuSO4 and 10 μm ZnSO4 were collected, harvested by centrifugation, and suspended in 20% sucrose, 30 mm Tris-HCl, 1 mm EDTA (1/20 of the culture volume), pH 8.0. Cells were incubated on ice for 10 min, harvested by centrifugation, and resuspended in an equal volume of 0.5 mm MgCl2. After another 10 min incubation on ice, cells were centrifuged 5 min at 17,000 × g, and the supernatant, containing periplasmic proteins released by osmotic shock, was removed. The release of periplasmic proteins by enzymatic lysis was achieved by harvesting bacterial cultures and resuspending in 1/20 volume 30 mm Tris-HCl, 20% sucrose, 1 mm EDTA, pH 8.0, and 1 mg/ml lysozyme (Sigma). After 10 min incubation on ice, cells were centrifuged for 5 min at 17,000 × g before the supernatant containing the periplasmic fraction was collected. Protein content was analyzed according to the method of Lowry (38) and superoxide dismutase activity was measured by the pyrogallol method (39).
Mouse Infection and Competition Assays—Mice were intraperitoneally inoculated with doses of bacteria (as indicated) and monitored. Moribund animals were euthanized. Competition assays were performed as previously described (20).
Experiments were also carried out to compare the lethality of wild type and mutant strains in C3H/HeN and DBA2 mice (obtained from Harlan-Sprague). Animals were inoculated intraperitoneally with 0.2 ml of sterile saline containing 2000 colony forming units of each S. typhimurium strain and mortality was evaluated daily. Kaplan-Meier analysis, carried out by GraphPad Prism 4, was used to determine the statistical significance of differences in survival of mice. p values <0.05 were considered significant.
Cell Cultures—The macrophage-like cell line J774.1 was used to detect SodCI and SodCII accumulation in intracellular bacteria. Cell cultures were grown in Dulbecco's modified medium-high glucose supplemented with 2 mm l-glutamine (Sigma) and 10% fetal calf serum (Euroclone). Infection with Salmonella strains was carried out as previously described (40). The human mononuclear cell line THP-1 was maintained in RPMI 1640 medium containing non-essential amino acids (Sigma), 1 mm sodium pyruvate, 2 mm l-glutamine, and 10% fetal calf serum and differentiated for 48 h by adding 20 ng/ml phorbol myristate acetate (Sigma). The human colonic epithelial cell line Caco-2 was grown in Dulbecco's modified Eagle's medium containing 1 g/liter glucose, non-essential amino acids, 4 mm l-glutamine, and 10% fetal calf serum.
Western Blot and Immunodetection of FLAG Epitope—Preparation of bacterial lysates for Western blot and immunodetection with anti-FLAG monoclonal antibodies (Sigma) was carried out as described (21).
Protein Purification—Recombinant SodCI was purified as described previously (27). SodCII was purified from E. coli 71/18 cells bearing plasmid pSEsodCII, obtained by inserting the sodCII gene in plasmid pSE420 under control of the trc promoter. Expression and purification of SodCII was carried out essentially as described for the monomeric Cu,Zn-SOD from E. coli (41). The purity of the enzyme was checked by SDS-PAGE. The protein appeared to be >98% homogeneous. Protein concentration was evaluated by the method of Lowry (38), whereas paramagnetic copper content was determined by double integration of the EPR spectra using a Cu(II)-EDTA solution as a standard (42).
SodCI has a very high affinity for copper ions and cannot be completely demetallated by the procedures previously described to obtain apoSODs lacking both copper and zinc ions (30, 43). Therefore, to remove the copper, the two StSOD enzymes were treated with potassium ferrocyanide to reduce the metal, then dialyzed for 12 h against 100 mm potassium phosphate buffer, 50 mm KCN, pH 6.0 (44). Subsequently the copper-free enzymes were extensively dialyzed against phosphate buffer to remove KCN. The zinc ion was removed by dialysis against 100 mm acetate buffer, pH 3.8, 2 mm EDTA, following established procedures (44). The metal content of the apoSODs was analyzed by atomic absorption spectroscopy using the PerkinElmer spectrometer AAnalyst 300 equipped with the graphite furnace HGA-800. Copper and zinc saturation of the available active sites was less than 2%.
Assays—To analyze catalase activity, aliquots of bacteria corresponding to 5 × 108 cells were collected from overnight inocula, sonicated, and resuspended in loading buffer without SDS. Samples were run on a 10% polyacrylamide gel under non-denaturing conditions. The gel was rinsed with deionized water, incubated with 0.003% H2O2 for 10 min, and stained with a solution of 2% potassium ferricyanide and 2% ferric chloride (45). Areas of catalase activity appeared as clear bands on a dark background.
Superoxide dismutase activity assays were carried out at pH 8.2 by the pyrogallol method (40). One unit is defined as the amount of SOD necessary to achieve 50% inhibition of pyrogallol autoxidation. Proteinase K susceptibility of the two Salmonella SodC enzymes was assayed by incubating proteins at a concentration of 0.1 mg/ml at 37 °C in 20 mm Tris-HCl, pH 8.0, in the presence of 0.1 mg/ml proteinase K. Aliquots were withdrawn at the indicated times and immediately assayed by the pyrogallol method to measure residual activity.
The effect of EDTA on SodCI and SodCII activity was analyzed as described (30). Cu,Zn-SOD samples at a concentration of 0.04 mg/ml were incubated at 37 °C in 100 mm phosphate buffer, 0.1 mm EDTA, pH 6.2 or 7.8. Aliquots were withdrawn at different times and immediately assayed for residual activity by the pyrogallol method (40).
To evaluate relative copper binding by apo- and copper-free SodCI and SodCII, equal quantities of the two proteins were mixed and incubated for 5 min with a quantity of copper sulfate sufficient to metallate 50% of the active sites of the enzyme. The mixture was then injected onto a HiLoad™ 16/60 Superdex™ 75 FPLC gel filtration column (GE Health-care) equilibrated with 20 mm phosphate buffer, pH 7.0, 150 mm NaCl and eluted with the same buffer to obtain separation of the two proteins. SOD activity of the peak fractions was measured. A contribution from trace metal contamination in the elution buffer was measured by the separation of control mixtures not reconstituted with copper.
The peroxidative activity of Cu,Zn-SODs was evaluated by monitoring the oxidation of 2,2′-azino-bis-[3-ethylbenzothiazoline sulfonate] (ABTS–), according to a described procedure (46). SodCI and SodCII, either in holo or zinc-free forms, were dissolved in 100 mm sodium phosphate, 10 mm H2O2, pH 7.2, at a copper concentration of 6.25 μm. The oxidation of ABTS– to 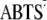 was monitored over time at 415 nm in the presence or absence of 10 mm NaHCO3. Zinc-free enzymes were obtained by incubating the apo-proteins with stoichiometric amounts of copper. Copper incorporation in the active site was monitored by analysis of the EPR spectrum of the enzyme. 1 mm EDTA was added as required.
was monitored over time at 415 nm in the presence or absence of 10 mm NaHCO3. Zinc-free enzymes were obtained by incubating the apo-proteins with stoichiometric amounts of copper. Copper incorporation in the active site was monitored by analysis of the EPR spectrum of the enzyme. 1 mm EDTA was added as required.
RESULTS
Contributions of SodCI and SodCII to Salmonella Virulence in Mice—To ensure that discrepant findings concerning the role of sodCII in Salmonella virulence were not attributable to differences in bacterial strain background, wild type strains and sodCII mutant alleles used in earlier studies as well as newly constructed sodCII deletion mutants were compared. Each sodCII mutant allele was transduced into the wild type strain (denoted FWT and BWT, from the Fang and Bossi laboratories, respectively), to generate the strains listed in Table 2. All wild type and mutant strains exhibited identical growth rates in LB medium and comparable survival in stationary phase (data not shown). Comparable lethal doses were obtained in infection studies with DBA2 mice. No differences were observed in the pattern of expression of catalases, as evaluated by catalase staining of native gels, or in Cu,Zn-superoxide dismutase activity (data not shown). As expression of sodCII, katG, and katE is controlled by σS, these results suggest that the two wild type S. typhimurium ATCC14028 strains BWT and FWT do not functionally differ at the rpoS locus, as is sometimes observed in strains subjected to laboratory passage (47).
To compare the contribution of each SodC to virulence, competition assays were performed in BALB/c mice, using mixed inocula containing wild type and mutant strains. All the strains listed in Table 4 were in the FWT background, but each competition experiment with sodCII alleles was also performed in the BWT background, with comparable results. Three different mutant sodCII alleles were assessed: two carrying a complete replacement of sodCII with a kanamycin or phoA-kanamycin cassette (21), and the third with a pRR10(ΔtrfA) insertion (18) abrogating SodCII activity but leaving a residual truncated sodCII gene of 344 base pairs. As shown in Table 4, only the insertion mutant (sodCII::pRR10(ΔtrfA) exhibited a significant competitive disadvantage in BALB/c mice.
TABLE 4.
Competition assays in BALB/c mice
| Strain A | Strain B | Mean CIa | Number of mice | pb |
|---|---|---|---|---|
| Wild type | sodCII::kan | 1.259 | 5 | 0.4467 |
| Wild type | sodCII::pRR10(ΔtrfA) | 4.384 | 5 | 0.0301 |
| Wild type | sodCII::phoA | 0.747 | 5 | 0.2610 |
| sodCII::kan | sodCII::pRR10(ΔtrfA) | 11.64 | 5 | 0.0027 |
| sodCII::phoA | sodCII::pRR10(ΔtrfA) | 8.312 | 5 | 0.0097 |
Mean CI is calculated by the formula: output (Strain A/Strain B)/inoculum (Strain A/Strain B).
Statistical differences between outputs and inocula with p values ≥0.05 were considered not significant.
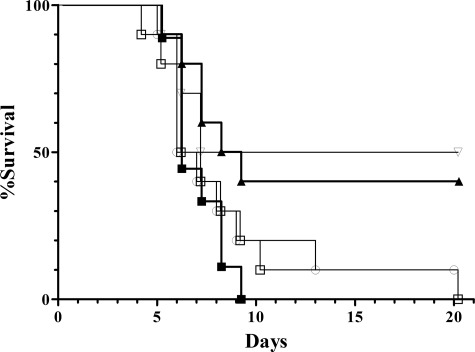
BALB/c mice are highly susceptible to Salmonella infection and are therefore insensitive models for the detection of small differences in bacterial virulence (19). Therefore, resistant mouse strains (C3H/HeN and DBA2) were intraperitoneally infected with wild type and sodCII mutant strains (Fig. 1 and supplemental Fig. 1S, respectively). C3H/HeN mice were administered 2000 colony forming units of wild type (BWT), sodCII mutant, or sodCI mutant strains (sodCI::kan). This infectious dose proved to be lethal for mice infected with the wild type strain (filled squares), whereas deletion of sodCI (open triangles) significantly decreased mortality (p = 0.0405, according to a Kaplan-Meier survival analysis). A comparable loss of virulence (p = 0.032) was observed for the sodCII insertion mutant (closed triangles), whereas sodCII deletion mutants (open squares and circles) displayed only a modest decrease in pathogenicity when compared with wild type. Although the observed difference between wild type and sodCII deletion mutants did not reach statistical significance, it was repeatedly observed in independent experiments. Comparable results were observed in DBA2 mice (supplemental Fig. 1S). In showing that the sodCII insertion causes greater attenuation than the deletion mutations, these results suggest the existence of allele-specific effects. Such effects might help reconcile the previous discrepancies (see “Discussion”).
FIGURE 1.
Virulence of wild type and sodC mutant S. typhimurium in C3H/HeN mice. Survival is shown for groups of 10 mice infected with wild type (▪), sodCII::kan (○), sodCII::phoA (□), sodCII::pRR10(ΔtrfA)(▴), and sodCI::kan (▿) strains.
Release of SodCI and SodCII by Osmotic Shock—Some recent investigations have suggested that SodCI cannot be released from the periplasmic space upon osmotic shock, either when produced in Salmonella or E. coli (23, 25), suggesting that SodCI stably interacts with components of the periplasmic environment. It was proposed that this phenomenon, described as SodCI tethering, might account for the greater contribution of this enzyme to Salmonella virulence. However, others have suggested that SodCI can be easily released from the periplasm when expressed in E. coli (27, 29) or in Salmonella serovars other than typhimurium (16).
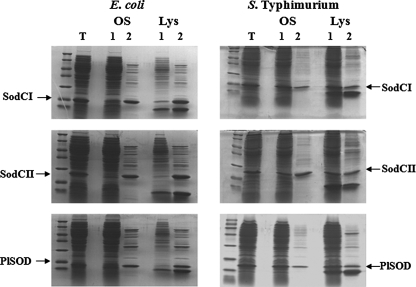
To investigate this issue, SodCI, SodCII, and Photobacterium leiognathi SodC were overproduced in E. coli and in S. typhimurium and their release measured following osmotic shock or lysozyme treatment. Fig. 2 shows that all of these proteins were quantitatively released by osmotic shock from the periplasm of E. coli, whereas a large fraction of the proteins remained in the pellet when produced in S. typhimurium. SodCII was released more efficiently than either SodCI or P. leiognathi SodC, independent of the method used to obtain periplasmic fractions. Attempts to enhance the proportion of SodC proteins released from the periplasm (e.g. by increasing time of lysozyme incubation or concentration of EDTA) were unsuccessful.
FIGURE 2.
SodCI and SodCII release from the periplasm of E. coli and S. typhimurium. SDS-PAGE of total cellular lysates (T), pellet (lane 1) and periplasmic fractions (lane 2) released by osmotic shock (OS) or lysozyme treatment (Lys) from E. coli 71/18 or S. typhimurium ATTC14028 overexpressing SodCI, SodCII, or P. leiognathi SodC.
These results indicate that tethering or retention in the periplasm occurs only when SodCs are expressed in Salmonella and that the phenomenon is not specific for SodCI. Although monomeric SodCII is released more efficiently than dimeric SodCI and P. leiognathi SodC, it is unlikely that protein size is the main factor responsible for SodC tethering. ZnuA, a monomeric periplasmic protein of 31 kDa (a molecular mass similar to that of SodCI) is efficiently released by osmotic shock (data not shown).
Biochemical Properties of SodCI and SodCII—Previous studies have shown that the monomeric Cu,Zn-SOD from E. coli has structural/functional properties distinct from those of dimeric Cu,Zn-SOD enzymes, but a recent investigation of the Brucella abortus SodC has suggested that this may not be true of all monomeric Cu,Zn-SODs (25). To address this problem, we purified SodCI and SodCII to homogeneity and compared their properties in vitro. Although proteins were purified from bacteria grown in media containing high concentrations of copper and zinc, all preparations of SodCI and SodCII contained a large fraction of copper-free inactive enzyme (10–20% for SodCI and 20–30% for SodCII). Therefore, activity was standardized relative to the copper content of the sample. Purified SodCI displayed an activity of 18,600 units/mg, whereas SodCII showed an activity of 6,800 units/mg, confirming that SodCI possesses a very high catalytic rate (27).
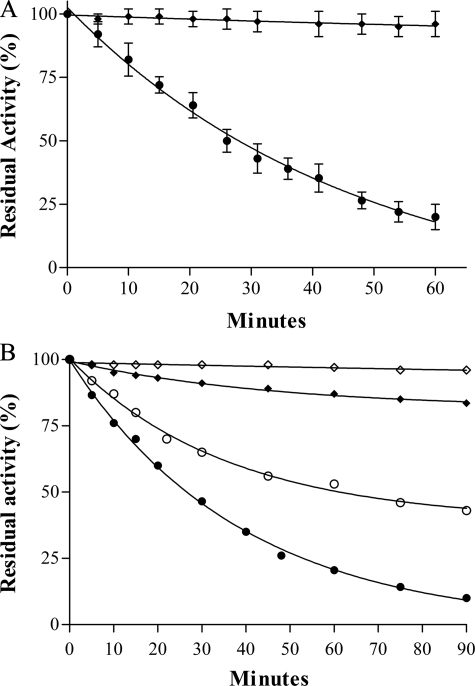
Unlike the eukaryotic Cu,Zn-SODs, which are extremely resistant to protease inactivation, all bacterial Cu,Zn-SODs are prone to proteolysis, due to high flexibility of the loops surrounding the active site channel (48). The monomeric Cu,Zn-SOD of E. coli has been found more susceptible to proteinase K digestion than the dimeric enzymes from P. leiognathi, S. typhimurium, and Vibrio cholerae (29). In agreement with these observations, we found (Fig. 3, panel A) that SodCII (filled circles) is much more susceptible than SodCI (filled diamonds) to inactivation by proteinase K.
FIGURE 3.
A, proteinase K susceptibility of SodCI and SodCII. Proteins were incubated at 37 °C in 20 mm Tris-HCl at pH 8.0, in the presence of 0.1 mg/ml proteinase K. Aliquots were withdrawn at the indicated times and assayed for residual SOD activity by the pyrogallol method. SodCI (♦), SodCII (•). B, EDTA inactivation of SodCI and SodCII. Cu,Zn-SOD samples at a concentration of 0.04 mg/ml were incubated at 37 °C in 100 mm phosphate buffer, 0.1 mm EDTA, pH 6.2 and 7.8. Aliquots were withdrawn at the indicated times and assayed for residual SOD activity as above. Each data point represents the mean of at least three independent measures. SodCI, pH 7.8 (♦), SodCII, pH 7.8 (•), SodCI, pH 6.2 (⋄), SodCII, pH 6.2 (○).
Bacterial Cu,Zn-SODs easily lose their metal cofactors and are therefore irreversibly inactivated in the presence of EDTA (30, 49). The observed rates of EDTA-mediated inactivation vary for different enzymes (30), are affected by the stability of the quaternary structure (43, 49), and are strongly influenced by pH, being faster at alkaline pH (29, 30). Fig. 3, panel B, shows the inactivation rates of SodCI and SodCII in 0.1 mm EDTA, 100 mm potassium phosphate, pH 6.2 (open symbols) and 7.8 (filled symbols). Monomeric SodCII (circles) was found to lose activity rapidly under both conditions tested, with more rapid inactivation at pH 7.8 than at pH 6.2. In contrast, the activity of SodCI (diamonds) was not affected by incubation with EDTA at pH 6.2, and was only slightly decreased at alkaline pH. In disagreement with the results of Krishnakumar et al. (23), our findings suggest that SodCI binds the copper ion with higher strength than SodCII, and the stability of the active sites of these two enzymes is differentially affected by changes in pH. The ability of the two enzymes to bind copper ion was subsequently compared. Equal amounts of copper-free SodCI and SodCII or of apo-SodCI and -SodCII (devoid of both copper and zinc) were mixed together before an amount of copper sufficient to saturate 50% of the enzyme subunits was added. The mixture was applied to a gel filtration chromatography column to separate dimeric SodCI from monomeric SodCII, and each fraction was assayed for SOD activity by the pyrogallol method. The percentage of reconstituted protein was evaluated with reference to samples of the individual proteins reconstituted with the same amount of copper. When copper was added to the mixture of copper-free enzymes, SodCI and SodCII bound the metal with comparable ability (45% was found in SodCI and 55% in SodCII). In contrast, when copper was added to the mixture of proteins lacking both copper and zinc, nearly all copper (84%) was bound by SodCI.
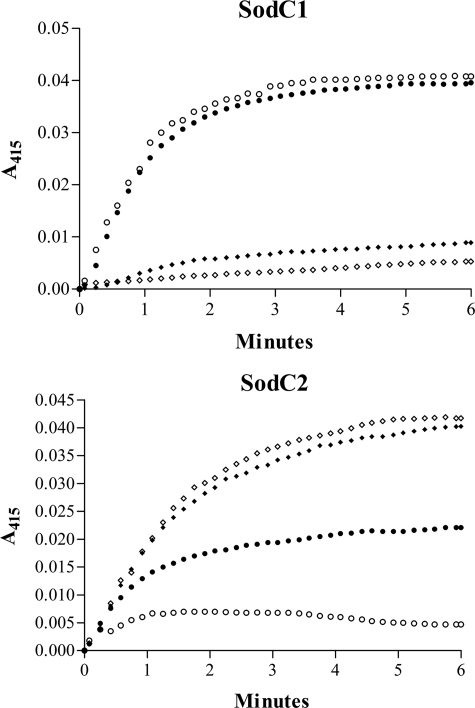
Cu,Zn-SODs can catalyze reactions in addition to superoxide dismutation, including the peroxidation of various substrates (50). Fig. 4 shows a comparison of the peroxidative activity of SodCI and SodCII. Both holoenzymes (diamonds) were able to catalyze the H2O2-dependent peroxidation of ABTS– in the presence of carbonate. No peroxidative activity was observed in the absence of copper (not shown). Holo-SodCII possesses higher peroxidative activity than SodCI. The peroxidative activity of the two enzymes was not affected by the addition of EDTA (open diamonds). Interestingly zinc-free SodCI (filled circles, upper panel) exhibited a noticeable increase in peroxidative activity, suggesting that the loss of zinc causes alterations in copper geometry and/or in its redox properties that affect this secondary activity of the enzyme. The peroxidative activity of zinc-free SodCI (open circles, upper panel) was not affected by the addition of EDTA, indicating that binding of copper to the enzyme is stable in the absence of zinc. In contrast, the peroxidative activity of zinc-free SodCII (filled circles, lower panel) was lower than that of the holoenzyme, possibly due to poor copper binding in the absence of zinc. This was confirmed by nearly complete abrogation of the ability to catalyze ABTS peroxidation upon the addition of EDTA (open circles, lower panel). Our results confirm and extend previous observations showing that copper binding to monomeric Cu,Zn-SOD is unstable (51) and suggest that SodCII can function as an active SOD only when it contains zinc.
FIGURE 4.
Peroxidase activity of SodCI and SodCII. The time-dependent increase in  formation was monitored at 415 nm as a measure of the peroxidase activity of SodCI and SodCII at 25 °C in the presence of 10 mm NaHCO3. Holo-enzyme (♦), holo-enzymes plus EDTA (⋄), zinc-free enzyme (•), and zinc-free enzyme plus EDTA (○) are shown. All bacterial enzymes showed no detectable peroxidative under the conditions tested in the absence of NaHCO3.
formation was monitored at 415 nm as a measure of the peroxidase activity of SodCI and SodCII at 25 °C in the presence of 10 mm NaHCO3. Holo-enzyme (♦), holo-enzymes plus EDTA (⋄), zinc-free enzyme (•), and zinc-free enzyme plus EDTA (○) are shown. All bacterial enzymes showed no detectable peroxidative under the conditions tested in the absence of NaHCO3.
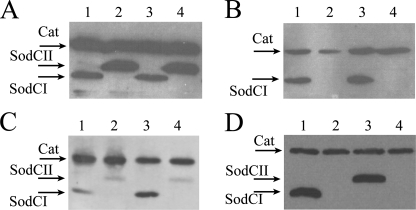
Abundance of SodCI and SodCII in Vitro and within Intracellular Bacteria—Previous work by Uzzau and collaborators (21) showed that whereas both SodCI and SodCII are made at significant rates in bacteria cultured in vitro, SodCI, but not SodCII, accumulates in bacteria proliferating inside macrophages or in infected mouse spleens. These experiments were repeated in the present study upon transferring the epitope-tagged sodCI and sodCII genes from the strains used by Uzzau et al. (21) (MA7224 and MA7225) into the FWT background. Results in Fig. 5 confirm that SodCI and SodCII are both highly expressed in stationary phase Salmonella, with SodCII accumulating at higher levels (panel A). In contrast, SodCII cannot be detected in bacteria harvested from J774.1 macrophages, whereas SodCI abundance was enhanced in the intracellular environment (Fig. 5, panel B). It should be noted that no differences in SodCI and SodCII abundance are observed between FWT and BWT strain backgrounds (Fig. 5, panels A and B), thus confirming the identity of the two strains. Accumulation of SodCI and SodCII was also analyzed in Salmonella internalized by human monocytes (THP-1) or colonic cells (Caco-2) (Fig. 5, panel C). Accumulation of the SodCs in THP-1 cells was similar to the results observed in J774.1 macrophages, but SodCI accumulation was not enhanced in bacteria recovered from Caco-2 cells in comparison to synthetic media, whereas bacterial SodCII levels were decreased in all cell lines examined.
FIGURE 5.
In vitro and in vivo SodCI and SodCII accumulation. Bacterial lysates were loaded on a 12% polyacrylamide gel, which was processed for Western blot analysis. Membranes were probed with anti-FLAG monoclonal antibodies. Strains MA7224 (lane 1), MA7225 (lane 2), SA167 (lanes 3), and SA168 (lane 4) were grown overnight in LB medium (panel A) or collected from J774.1 macrophages 23 h post-infection (panel B). Panel C, strains MA7224 (lanes 1 and 3) and MA7225 (lanes 2 and 4) were harvested from Caco-2 or THP-1 cells 23 h post-infection. Panel D, intracellular accumulation of SodCI and SodCII expressed from MA7224 (lane 1), MA7225 (lane 2) MA7537 (lane 3), and MA7538 (lane 4) in bacteria harvested from J774.1 macrophages 23 h post-infection. Cat, chlorampheniol acetyl transferase.
To exclude the possibility of the observed differences being due to enhanced SodCII protein turnover with respect to SodCI, levels of SodCI and SodCII were measured in J774.1 macrophages infected with strains MA7537 and MA7538, which express the SodCII protein under control of the sodCI promoter and SodCI under control of the sodCII promoter, respectively (24). As shown in Fig. 5D, the SodCII protein was detected at high levels when expressed from the sodCI promoter, whereas no SodCI protein could be detected in cells infected with the strain MA7538. In keeping with earlier observations (24), this confirms that differences in transcriptional regulation can account for the different abundance of SodCI and SodCII in the intracellular environment.
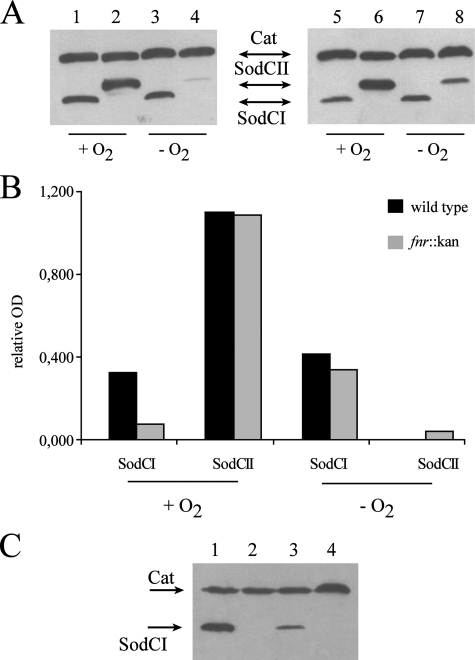
Regulation of sodCI and sodCII by Oxygen—Little is known about the regulation of sodCI and sodCII, other than that sodCII is under the control of σS (18) and sodCI is part of the PhoPQ regulon (32). Although the production of superoxide anion requires the presence of oxygen, some evidence suggests that the macrophage phagosome is a hypoxic environment (52). Therefore, regulation of the sodC genes was investigated under oxygen-limiting conditions. Epitope-tagged SodCI- and SodCII-expressing strains (MA7224 and MA7225) and sodC promoter-exchanged strains (MA7537 and MA7538) were grown overnight in LB with aeration or under anaerobic conditions, harvested, subjected to SDS-PAGE, and immunoblotted. As shown in Fig. 6A (left) a modest increase of SodCI levels can be observed under anaerobic conditions, whereas SodCII synthesis is clearly repressed. The dramatic effect of anaerobiosis on sodCII expression was confirmed by measurement of SodC protein levels in strains in which the promoters of sodCI and sodCII are switched, indicating that regulation of sodC expression in response to oxygen occurs at the level of transcription (data not shown). Anaerobic repression has been reported for the E. coli sodC gene (6, 53), in which the iron-sulfur cluster-containing protein FNR, an oxygen-sensitive transcriptional regulator, has been suggested to be the primary regulator of anaerobic sodC repression (6). Salmonella SodCI and SodCII accumulation were therefore measured in an fnr mutant background. Epitope-tagged PepT, a protease expressed during anaerobic conditions under strict control by FNR, provided a positive control (data not shown). Strain SA126 (fnr::kan sodCI::3XFLAG ilvI::Tn10dTac-cat::3XFLAG) and strain SA127 (fnr::kan sodCII::3XFLAG ilvI::Tn10dTac-cat::3XFLAG-), were constructed as described under “Experimental Procedures.” Analysis of epitope-tagged SodCI and SodCII accumulation by Western blot (Fig. 6, A, right, and B) showed the fnr deletion to cause a slight decrease in SodCI levels under aerobic conditions (compare Fig. 6A, lanes 1 and 5, and Fig. 6B, black and gray bars), whereas no significant difference was seen in anaerobiosis.
FIGURE 6.
Aerobic and anaerobic SodCI and SodCII accumulation in wild type and Δfnr mutant strains. Panel A, approximately 5 × 108 bacteria were grown overnight in LB medium with vigorous aeration (+O2) or in anaerobic conditions (–O2). Bacterial lysates were processed for Western blot analysis as described in the legend to Fig. 5. Strains shown are MA7224 (lanes 1 and 3), MA7225 (lanes 2 and 4), SA126 (lanes 5 and 7), and SA127 (lanes 6 and 8). Panel B, densitometric analysis of Western blots presented in panel A; relative OD are calculated as SodCI/Cat or SodCII/Cat ratios. Panel C, wild type or Δfnr mutant bacteria were collected from J774.1 macrophages 23 h post-infection. Strains shown are MA7224 (lane 1), MA 7225 (lane 2), SA 126 (lane 3), and SA 127 (lane 4). Cat, chlorampheniol acetyl transferase.
In contrast, the fnr deletion did not affect SodCII levels under aerobic conditions while partially relieving the anaerobic repression (compare Fig. 6A, lanes 4 and 8, and Fig. 6B, black and gray bars). Finding this relief to be incomplete suggests that additional factors besides FNR are involved in the repression of sodCII when oxygen is limiting. Finally, the fnr deletion did not restore SodCII expression and caused a marked decrease in SodCI accumulation in bacteria recovered from infected macrophages (Fig. 6C).
Regulation of sodCII by Zinc Availability—The stringent zinc requirement for stable copper binding by SodCII led us to investigate the effects of zinc availability on sodCI and sodCII expression. Recent studies have established in E. coli that, when zinc availability is limited, periplasmic zinc is bound by the periplasmic transporter ZnuA, which prevents zinc binding by Cu,Zn-SOD (51). Moreover, the ability of Salmonella to multiply in mice is dependent on the high affinity zinc transporter ZnuABC, suggesting that zinc is limiting for bacterial growth in host tissues (54, 55).
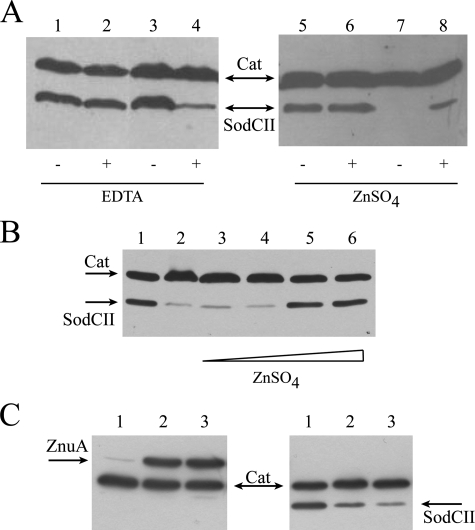
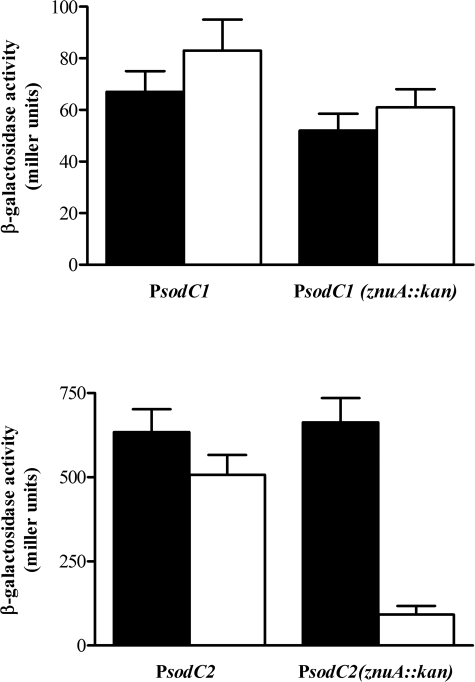
The accumulation of epitope-tagged SodCI and SodCII was measured in bacteria grown in LB with or without supplementation with the divalent metal chelator EDTA, in wild type and znuA mutant strains. SodCI levels were only slightly affected by a znuA mutation (data not shown). In contrast, SodCII accumulation was significantly decreased in a znuA mutant upon the addition of EDTA (Fig. 7A, lanes 1–4). SodC levels were also determined in zinc-poor minimal medium, in which znuA is actively expressed (55), with or without zinc supplementation. Fig. 7A (lanes 5–8) shows that SodCII is expressed in wild type but not znuA mutant bacteria, unless exogenous zinc is provided. EDTA is not a zinc-specific chelator and the lower levels of SodCII could be affected by the unavailability of zinc or other divalent metals. To corroborate these observations, we analyzed the accumulation of SodCII in a strain lacking znuA in LB medium supplemented with 0.5 mm EDTA and variable amounts of zinc. The addition of 50 μm zinc restored normal SodCII levels (Fig. 7B). Moreover, when bacteria were cultivated in the presence of TPEN, a chelating agent with high zinc specificity (55), we observed strong induction of ZnuA and a concomitant decrease of SodCII accumulation (Fig. 7C). Taken together, these results suggest that sodCII is regulated by zinc availability, but do not exclude a post-transcriptional effect of zinc deprivation on SodCII accumulation (e.g. from rapid degradation of the apo-protein). Transcriptional fusions of the sodCI and sodCII promoters to lacZ encoding β-galactosidase were used to show that sodCII transcription is significantly reduced in znuA mutant bacteria grown in the presence of EDTA, whereas sodCI transcription was only modestly affected (Fig. 8).
FIGURE 7.
Effect of zinc availability on SodCII accumulation. Bacterial lysates were processed for Western blot analysis as described in the legend to Fig. 5. A, strains MA 7225 (lanes 1, 2, 5, and 6) and SA 153 (lanes 3, 4, 7, and 8) were grown in LB or LB containing 0.5 mm EDTA (left panel) or minimal medium supplemented or not with 4 μm ZnSO4 (right panel). B, SodCII accumulation in SA153 cells cultivated in LB (lane 1) or LB supplemented with 0.5 mm EDTA (lane 2) and 10, 20, 50, and 100 μm zinc (lanes 3–6, respectively). C, ZnuA (left panel) and SodCII (right panel) accumulation in strains SA 140 and SA 153 cells cultivated in LB (lane 1) or LB plus 50 or 100 μm N,N,N′,N′-Tetrakis(2-pyridylmethyl)ethylenediamine (lanes 2 and 3). Cat, chlorampheniol acetyl transferase.
FIGURE 8.
Effects of metal chelation on sodCI and sodCII transcription. Wild type and ΔznuA strains carrying plasmids pPCI-lacZ or pPCII-lacZ were grown in LB medium or LB containing 0.5 mm EDTA andβ-galactosidase activities were determined as described under “Experimental Procedures.”
DISCUSSION
Previous studies in cultured macrophages (11) and mice (8, 9, 16, 18, 21, 23, 24) have shown a consistent contribution of SodCI to Salmonella virulence. However, some of these studies have found a substantial role of SodCII in virulence as well (8, 12, 16, 18), whereas others have not (21, 23, 24). These discrepancies have been difficult to reconcile.
In the present study, we find that the sodCII::pRR10(ΔtrfA) has a larger effect on Salmonella virulence than the complete deletion of the sodCII gene (Fig. 1), suggesting that the truncated SodCII protein expressed by the sodCII::pRR10(ΔtrfA) mutant has detrimental adventitious effects on Salmonella. The interrupted sodCII gene is predicted to encode a protein lacking the final 34 amino acids of the mature protein. This deletion involves one copper ligand and leaves the four zinc ligands unchanged. Although the mutant protein might still bind metals with altered geometry, it is unlikely that the protein folds in a Cu,Zn-SOD-like conformation due to the loss of some elements of the β-barrel core (supplementary materials Fig. S2). Attempts to purify the overexpressed truncated protein in E. coli were unsuccessful due to protein instability (data not shown). However, it is interesting to note that similar deletions have been identified in Cu,Zn-SODs from human patients affected by familial forms of amyotrophic lateral sclerosis (56). The familial forms of this disease are frequently associated with mutations in Cu,Zn-SOD, which lead to the acquisition of novel toxic functions (mutations are dominant and disease manifestations are not attributable to the loss of enzyme activity) (57). Several mutant Cu,Zn-SODs lacking a C-terminal fragment have been identified in familial forms of amyotrophic lateral sclerosis. These mutations are caused by frameshifts, deletions, or insertions localized between the fourth and fifth exon, leading to production of proteins with length very similar to that produced by the sodCII::pRR10(ΔtrfA) insertion (see supplementary Fig. S3). The most studied allele causes a frameshift after codon 126, producing a protein lacking the last 27 amino acids. This mutant protein is unstable and undetectable in the tissues of patients (58), but nonetheless causes disease. According to recent investigations, toxicity likely derives from aggregation phenomena stimulated by mutant-Cu,Zn-SOD, which damage cellular or mithochondrial membranes (59, 60). The S. typhimurium sodCII::pRR10(ΔtrfA) mutant serendipitously shows that mutations in Cu,Zn-SOD can exert toxic effects in cellular systems unrelated to the nervous system. This may provide an additional model in which to explore the role of Cu,Zn-SOD in amyotrophic lateral sclerosis pathogenesis.
Overall, the different contribution of the sodC genes to Salmonella virulence correlates with levels of the two proteins in Salmonella recovered from infected macrophages and mice. Our results confirm previous reports that sodCII is down-regulated within the host cells (21). We have found that intracellular accumulation of SodCI and SodCII varies in different cell lines (J774.1, THP-1, and Caco-2), but in all cases SodCII is down-regulated, whereas SodCI accumulates at high levels in bacteria internalized by phagocytic cells.
Both sodCI and sodCII are maximally expressed in stationary phase during in vitro growth in rich medium (16, 18, 21), but only sodCII is regulated by the alternative sigma factor σs (RpoS) (16, 18). The Salmonella plasmid virulence genes are regulated by σs (61), and σs-regulated genes are induced soon after bacterial uptake by eukaryotic cells (62). Therefore, the apparent repression of sodCII in infected cells suggests that other regulatory networks might contribute to its regulation in vivo. Here we show that, analogously to the E. coli sodC gene, sodCII exhibits repression under anaerobic conditions. However, sodCII repression is only partially dependent on fnr. Unexpectedly, we have found that sodCI is also regulated by FNR, but in the opposite fashion. Maximal accumulation of SodCI in vitro during aerobic growth or in infected macrophages requires FNR, a protein product inactive under aerobic conditions (63, 64). The sodCI promoter region does not contain a consensus FNR-binding site (data not shown), suggesting that regulation by FNR is indirect. FNR regulates the transition between aerobic and anaerobic metabolism by inducing transcription of a large set of genes required for anaerobic growth (65). In the presence of oxygen, the [4Fe-4S]2+ FNR cluster is reversibly converted into a transcriptionally inactive [2Fe-2S]2+ form, which can undergo further decomposition to an apo-form (66). Recent studies have shown that the iron-sulfur cluster of FNR is disrupted by superoxide (67) or nitric oxide (68). It is therefore tempting to speculate that phagocyte-derived reactive oxygen or nitrogen species might promote SodCI accumulation in intracellular bacteria. It must be acknowledged that a very recent genome-wide transcriptional analysis of the Salmonella FNR regulon genome did not demonstrate significant variation in sodCI and sodCII transcription in an fnr mutant strain (65), suggesting that FNR might regulate SodCI and SodCII at a post-transcriptional level. This could explain the greater intracellular accumulation of SodCI relative to SodCII despite apparent transcription of sodCII in intracellular bacteria (31, 32). The differential regulation of SodCI and SodCII production is consistent with distinct physiological roles of these proteins.
In the present study we have demonstrated important functional differences between SodCI and SodCII that help to account for their different roles. In contrast to earlier investigators who suggested that SodCI might be “tethered” to the periplasm (23), we have observed that both SodCI and SodCII are poorly released following osmotic shock of S. typhimurium. The release of SodCII is slightly more efficient than that of SodCI, possibly due to the smaller size of SodCII. More interestingly we have observed that SodCI and SodCII display markedly different molecular properties, including catalytic efficiency, protease resistance, pH sensitivity, peroxidative activity, and metal affinity. These results are consistent with studies of other bacterial Cu,Zn-SODs, which have established a complex relationship between quaternary structure, interface stability, active site metal occupancy, and pH. Monomeric Cu,Zn-SODs are characterized by lower conformational stability (30), decreased active site affinity for metal ions (29, 30, 51), and susceptibility to protease digestion (29, 48). These features can be rationalized as a consequence of the higher flexibility of the protein loops surrounding the active site of monomeric enzymes, the primary site of proteolytic attack (48). Conversely, single amino acid substitutions at the protein-protein interface of dimeric bacterial Cu,Zn-SODs can decrease metal affinity and modulate catalytic activity (43, 49).
One of our most intriguing findings is the difference between copper binding by SodCI and SodCII in the absence of zinc. Copper in the catalytic center of the enzyme is cyclically oxidized and reduced during successive encounters with superoxide anion. In the physiological pH range of 6.0–8.0, the activity of dimeric Cu,Zn-SODs is not affected by the presence of zinc (69), which appears primarily to confer structural stability. SodCI maintains an ability to stably bind copper in the absence of zinc and retains catalytic activity comparable with that of the holoenzyme (data not shown), although the removal of zinc enhances peroxidative activity (see Fig. 4). In contrast, monomeric SodCII is unable to stably bind copper in the absence of zinc, as previously reported for the E. coli enzyme (51), implying that the enzyme is likely to be inactive under zinc-limited conditions. We have recently observed that mutational inactivation of the ZnuABC high affinity zinc transporter, which is expressed only under severe zinc limitation, strongly attenuates Salmonella virulence, suggesting that zinc availability is restricted in eukaryotic tissues (55). Moreover, znuA is highly expressed in Salmonella recovered from cultured cells or the spleens of infected mice. The down-regulation of sodCII we have observed under zinc-limited conditions (Figs. 7 and 8) is consistent with the large reduction in SodCII accumulation within infected macrophages (21, 24) (Fig. 5). Thus Salmonella appears to selectively express the zinc-independent SodCI enzyme when limited availability of zinc could render SodCII inactive.
Our work definitively establishes that SodCI and SodCII play distinctive roles in Salmonella physiology and pathogenesis, and this is reflected by different regulation of expression and enzymatic properties. The phage-associated sodCI gene is regulated by a network of regulators including PhoPQ and FNR, which promote expression in the intracellular environment to protect Salmonella from reactive oxygen and nitrogen species produced by the host phagocytes (4). The physicochemical properties of SodCI, including a high catalytic rate, resistance to protease attack, high affinity for copper and low zinc requirements, confer resistance to the harsh environmental conditions encountered by bacteria during infection. Interestingly, in some Salmonella serovars other than S. typhimurium (i.e. S. enteritidis and S. choleraesuis), sodCI is embedded in genomic regions containing sequences derived from different phages or phage remnants (16, 24), suggesting a selective pressure to maintain sodCI in Salmonella. In contrast, SodCII is expressed in aerated cultures and carried by non-pathogenic as well as pathogenic bacteria, supporting the hypothesis that this enzyme protects bacteria from endogenous superoxide produced during aerobic metabolism, possibly due to electron leakage from the respiratory chain (26). Selective expression of SodCII in stationary phase corresponds to conditions in which bacterial cells are most susceptible to oxidative damage (70). Although a modest effect of a ΔsodCII mutation in immunocompetent mice was observed, the contribution of SodCII to Salmonella virulence in mice is clearly less important than that of sodCI. This is likely because SodCII is down-regulated in the host environment due to restricted availability of the zinc cofactor required for its activity, and perhaps due to oxygen limitation as well. Earlier studies suggesting a greater role of SodCII in pathogenesis can now be rationalized by the adventitious toxic effects of a nonfunctional truncated SodCII protein in sodCII::pRR10(ΔtrfA) mutant strains.
The redundancy of the Salmonella Cu,Zn-SODs is therefore more apparent than real. SodCI is essential for the intracellular survival of virulent Salmonella within host cells, whereas SodCII is most likely to promote resistance to reactive oxygen species produced during bacterial growth in aerobic extracellular environments.
Supplementary Material
Acknowledgments
We thank Mirko Bongianni and Eleonora Franzè for participation in preliminary experiments.
This work was supported, in whole or in part, by National Institutes of Health Grants AI039557 and AI50660 (to F. F.). This work was also supported by a Murst Prin Grant (to A. B.), Istituto Superiore di Sanità Grants 530/F-A23 and 530-F7/1 (to A. B. and P. P.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviation used is: SOD, superoxide dismutase.
References
- 1.Bannister, J. V., Bannister, W. H., and Rotilio, G. (1987) CRC Crit. Rev. Biochem. 22 111–180 [DOI] [PubMed] [Google Scholar]
- 2.Battistoni, A. (2003) Biochem. Soc. Trans. 6 1326–1329 [DOI] [PubMed] [Google Scholar]
- 3.Lynch, M., and Kuramitsu, H. (2000) Microbes Infect. 2 1245–1255 [DOI] [PubMed] [Google Scholar]
- 4.De Groote, M. A., Ochsner, U. A., Shiloh, M. U., Nathan, C., McCord, J. M., Dinauer, M. C., Libby, S. J., Vazquez-Torres, A., Xu, Y., and Fang, F. C. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 13997–14001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilks, K. E., Dunn, K. L., Farrant, J. L., Reddin, K. M., Gorringe, A. R., Langford, P. R., and Kroll, J. S. (1998) Infect. Immun. 66 213–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gort, A. M., Ferber, D. M., and Imlay, J. A. (1999) Mol. Microbiol. 32 179–191 [DOI] [PubMed] [Google Scholar]
- 7.Piddington, D. L., Fang, F. C., Laessig, T., Cooper, A. M., Orme, I. M., and Buchmeier, N. A. (2001) Infect. Immun. 69 4980–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sansone, A., Watson, P. R., Wallis, T. S., Langford, P. R., and Kroll, J. S. (2002) Microbiology 148 719–726 [DOI] [PubMed] [Google Scholar]
- 9.Pacello, F., Ceci, P., Ammendola, S., Pasquali, P., Chiancone, E., and Battistoni, A. (2008) Biochim. Biophys. Acta 1780 226–232 [DOI] [PubMed] [Google Scholar]
- 10.Battistoni, A., Donnarumma, G., Greco, R., Valenti, P., and Rotilio, G. (1998) Biochem. Biophys. Res. Commun. 243 804–807 [DOI] [PubMed] [Google Scholar]
- 11.Battistoni, A., Pacello, F., Folcarelli, S., Ajello, M., Donnarumma, G., Greco, R., Ammendolia, M. G., Touati, D., Rotilio, G., and Valenti, P. (2000) Infect. Immun. 68 30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sly, L. M., Guiney, D. G., and Reiner, N. E. (2002) Infect. Immun. 70 5312–5315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn, K. L., Farrant, J. L., Langford, P. R., and Kroll, J. S. (2003) Infect. Immun. 71 1604–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Battistoni, A., Ajello, M., Ammendola, S., Rotilio, G., and Valenti, P. (2004) Int. J. Immun. Pharm. 17 71–76 [DOI] [PubMed] [Google Scholar]
- 15.Gee, J. M., Valderas, M. W., Kovach, M. E., Grippe, V. K., Robertson, G. T., Ng, W. L., Richardson, J. M., Winkler, M. E., and Roop, R. M., 2nd (2005) Infect. Immun. 73 2873–2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ammendola, S., Ajello, M., Pasquali, P., Kroll, J. S., Langford, P. R., Rotilio, G., Valenti, P., and Battistoni, A. (2005) Microbes Infect. 7 698–707 [DOI] [PubMed] [Google Scholar]
- 17.Farrant, J. L., Sansone, A., Canvin, J. R., Pallen, M. J., Langford, P. R., Wallis, T. S., Dougan, G., and Kroll, J. S. (1997) Mol. Microbiol. 25 785–796 [DOI] [PubMed] [Google Scholar]
- 18.Fang, F. C., DeGroote, M. A., Foster, J. W., Bäumler, A. J., Ochsner, U., Testerman, T., Bearson, S., Giard, J. C., Xu, Y., Campbell, G., and Laessig, T. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 7502–7507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Figueroa-Bossi, N., and Bossi, L. (1999) Mol. Microbiol. 33 167–176 [DOI] [PubMed] [Google Scholar]
- 20.San Mateo, L. R., Toffer, K. L., Orndorff, P. E., and Kawula, T. H. (1999) Infect. Immun. 67 5345–5351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uzzau, S., Bossi, L., and Figueroa-Bossi, N. (2002) Mol. Microbiol. 46 147–156 [DOI] [PubMed] [Google Scholar]
- 22.Ho, T. D., and Slauch, J. M. (2001) J. Bacteriol. 183 611–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishnakumar, R., Craig, M., Imlay, J. A., and Slauch, J. M. (2004) J. Bacteriol. 186 5230–5238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Figueroa-Bossi, N., Ammendola, S., and Bossi, L. (2006) Microbes Infect. 8 1569–1578 [DOI] [PubMed] [Google Scholar]
- 25.Krishnakumar, R., Kim, B., Mollo, E. A., Imlay, J. A., and Slauch, R. (2007) J. Bacteriol. 189 4343–4352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korshunov, S. S., and Imlay, J. A. (2006) J. Bacteriol. 188 6326–6334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pesce, A., Battistoni, A., Stroppolo, M. E., Polizio, F., Nardini, M., Kroll, J. S., Langford, P. R., O'Neill, P., Sette, M., Desideri, A., and Bolognesi, M. (2000) J. Mol. Biol. 302 465–478 [DOI] [PubMed] [Google Scholar]
- 28.Bordo, D., Pesce, A., Bolognesi, M., Stroppolo, M. E., Falconi, M., and Desideri, A. (2001) in Handbook of Metalloproteins (Messerschmidt, A., Huber, R., Poulos, T., and Wieghardt, K., eds) pp. 1284–1300, John Wiley and Sons, Ltd., Chichester, United Kingdom
- 29.Gabbianelli, R., D'Orazio, M., Pacello, F., O'Neill, P., Nicolini, L., Rotilio, G., and Battistoni, A. (2004) Chem. Biol. 385 749–754 [DOI] [PubMed] [Google Scholar]
- 30.Battistoni, A., Folcarelli, S., Cervone, L., Polizio, F., Desideri, A., Giartosio, A., and Rotilio, G. (1998) J. Biol. Chem. 273 5655–5661 [DOI] [PubMed] [Google Scholar]
- 31.Eriksson, S., Lucchini, S., Thompson, A., Rhen, M., and Hinton, J. C. (2003) Mol. Microbiol. 47 103–118 [DOI] [PubMed] [Google Scholar]
- 32.Golubeva, Y. A., and Slauch, J. M. (2006) J. Bacteriol. 188 7853–7861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Messing, J., Gronenborg, B., Mueller-Hill, B., and Hofschneider, P. H. (1977) Proc. Natl. Acad. Sci. U. S. A., 74 3642–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maloy, S. R., Stewart, V. J., and Taylor, R. K. (1996) Genetic Analysis of Pathogenic Bacteria: A Laboratory Manual, pp. 473–492, Cold Spring Harbor Press, Cold Spring Harbor, NY
- 35.Datsenko, K. A., and Wanner, B. L. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cherepanov, P. P., and Wackernagel, W. (1995) Gene (Amst.) 158 9–14 [DOI] [PubMed] [Google Scholar]
- 37.Miller, J. H. (1972) Experiments in Molecular Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 38.Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951) J. Biol. Chem. 193 265–275 [PubMed] [Google Scholar]
- 39.Marklund, S., and Marklund, G. (1974) Eur. J. Biochem. 47 469–474 [DOI] [PubMed] [Google Scholar]
- 40.Uzzau, S., Figueroa-Bossi, N., Rubino, S., and Bossi, L. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 15264–15269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Battistoni, A., Folcarelli, S., Gabbianelli, R., Capo, C., and Rotilio, G. (1996) Biochem. J. 320 713–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polticelli, F., Bottaro, G., Battistoni, A., Carrì, M. T., Djinovic-Carugo, K., Bolognesi, M., O'Neill, P., Rotilio, G., and Desideri, A. (1995) Biochemistry 34 6043–6049 [DOI] [PubMed] [Google Scholar]
- 43.D'Orazio, M., Battistoni, A., Stroppolo, M. E., and Desideri, A. (2000) Biochem. Biophys. Res. Commun. 272 81–83 [DOI] [PubMed] [Google Scholar]
- 44.Bonaccorsi di Patti, M. C., Giartosio, A., Rotilio, G., and Battistoni, A. (2002) Methods Enzymol. 349 49–61 [DOI] [PubMed] [Google Scholar]
- 45.Sun, Y., Elwell, J. H., and Oberley, L. W. (1988) Free Radic. Res. Commun. 5 67–75 [DOI] [PubMed] [Google Scholar]
- 46.Liochev, S. I., and Fridovich, I. (1999) Free Radic. Biol. Med. 27 1444–1447 [DOI] [PubMed] [Google Scholar]
- 47.Visick, J. E., and Clark, S. (1997) J. Bacteriol. 179 4158–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Falconi, M., Parrilli, L., Battistoni, A., and Desideri, A. (2002) Proteins 47 513–520 [DOI] [PubMed] [Google Scholar]
- 49.Stroppolo, M. E., Pesce, A., D'Orazio, M., O'Neill, P., Bordo, D., Rosano, C., Milani, M., Battistoni, A., Bolognesi, M., and Desideri, A. (2001) J. Mol. Biol. 308 555–563 [DOI] [PubMed] [Google Scholar]
- 50.Hodgson, E. K., and Fridovich, I. (1975) Biochemistry 14 5294–5299 [DOI] [PubMed] [Google Scholar]
- 51.Berducci, G., Mazzetti, A. P., Rotilio, G., and Battistoni, A. (2004) FEBS Lett. 569 289–292 [DOI] [PubMed] [Google Scholar]
- 52.Schnappinger, D., Ehrt, S., Voskuil, M. I., Liu, Y., Mangan, J. A., Monahan, I. M., Dolganov, G., Efton, B., Butcher, P. D., Nathan, C., and Schoolnik, G. K. (2003) J. Exp. Med. 198 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benov, L. T., and Fridovich, I. (1994) J. Biol. Chem. 269 25310–25314 [PubMed] [Google Scholar]
- 54.Campoy, S., Jara, M., Busquets, N., Perez De Rozas, A. M., Badiola, I., and Barbe, J. (2002) Infect. Immun. 70 4721–4725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ammendola, S., Pasquali, P., Pistoia, C., Petrucci, P., Petrarca, P., Rotilio, G., and Battistoni, A. (2007) Infect. Immun. 75 5867–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watanabe, Y., Adachi, Y., and Nakashima, K. (2001) Neuropathology 21 61–66 [DOI] [PubMed] [Google Scholar]
- 57.Valentine, J. S., Doucette, P. A., and Zittin Potter, S. (2005) Annu. Rev. Biochem. 74 563–593 [DOI] [PubMed] [Google Scholar]
- 58.Nakashima, K., Watanabe, Y., Kuno, N., Nanba, E., and Takahashi, K. (1995) Neurology 45 1019–1020 [DOI] [PubMed] [Google Scholar]
- 59.Ferri, A., Cozzolino, M., Crosio, C., Nencini, M., Casciati, A., Gralla, E. B., Rotilio, G., Valentine, J. S., and Carri, M. T. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 13860–13865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shaw, B. F., and Valentine, J. S. (2007) Trends Biochem. Sci. 32 78–85 [DOI] [PubMed] [Google Scholar]
- 61.Fang, F. C., Libby, S. J., Buchmeier, N. A., Loewen, P. C., Switala, J., Harwood, J., and Guiney, D. G. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 11978–11982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen, C. Y., Eckmann, L., Libby, S. J., Fang, F. C., Okamoto, S., Kagnoff, M. F., Fierer, J., and Guiney, D. G. (1996) Infect. Immun. 64 4739–4743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strauch, K. L., Lenk, J. B., Gamble, B. L., and Miller, G. C. (1985) J. Bacteriol. 161 673–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crack, J. C., Green, J., Cheesman, M. R., Le Brun, N. E., and Thomson, A. J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 2092–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fink, R. C., Evans, M. R., Porwollik, S., Vazquez-Torres, A., Jones-Carson, J., Troxell, B., Libby, S. J., McClelland, M., and Hassan, H. M. (2007) J. Bacteriol. 189 2262–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sutton, A., Buencamino, R., and Eisenstark, A. (2000) J. Bacteriol. 182 4375–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sutton, V. R., Stubna, A., Patschkowski, T., Munck, E., Beinert, H., and Kiley, P. J. (2004) Biochemistry 43 791–798 [DOI] [PubMed] [Google Scholar]
- 68.Cruz-Ramos, H., Crack, J., Wu, G., Hughes, M. N., Scott, C., Thomson, A. J., Green, J., and Poole, R. K. (2002) EMBO J. 21 3235–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pantoliano, M. W., Valentine, J. S., Burger, A. R., and Lippard, S. J. (1982) J. Inorg. Biochem. 17 325–341 [DOI] [PubMed] [Google Scholar]
- 70.Nyström, T. (2004) Annu. Rev. Microbiol. 58 161–181 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.