Abstract
APOBEC3G (A3G) restricts HIV-1 infection by catalyzing processive C → U deaminations on single-stranded DNA (ssDNA) with marked 3′ → 5′ deamination polarity. Here we show that A3G exists in oligomeric states whose composition is dictated primarily by interactions with DNA, with salt playing an important, yet secondary, role. Directional deaminations correlate with the presence of dimers, tetramers, and larger oligomers observed by atomic force microscopy, and random deaminations appear to correlate mainly with monomers. The presence of a 30-nt weakly deaminated “dead” zone located at the 3′-ssDNA end implies the presence of a preferred asymmetric direction for A3G catalysis. Single turnover reaction rates reveal a salt-dependent inhibition of C deamination toward the 3′-ssDNA region, offering a molecular basis underlying A3G deamination polarity. Presteady state analysis demonstrates rapid diffusion-limited A3G-ssDNA binding, a slower salt-dependent conformational change, possibly indicative of DNA wrapping, and long (5–15 min) protein-DNA complex lifetimes. We suggest that diverse A3G oligomerization modes contribute to the human immunodeficiency virus, type 1, proviral DNA mutational bias.
In 2002, Sheehy et al. (1) determined that APOBEC3G (A3G), originally called CEM15, a proposed cytidine deaminase based on sequence analysis, is the nonpermissive host factor that blocks virion infectivity factor-defective (Δvif) HIV-1 infection of T cells. The experimental determination that CEM15 was a cytidine deaminase belonging to the APOBEC family followed soon after with experiments demonstrating G → A-induced hypermutation of proviral DNA in a Δvif HIV-1 virion (2, 3). A3G has a duplicated deaminase domain structure, but only the C-terminal domain is responsible for the single-stranded DNA (ssDNA)2 deamination activity (4, 5).
Apart from A3G-catalyzed deamination, A3G may also have the capacity for blocking reverse transcription, (+)-DNA synthesis, and provirus formation either by interacting with RNA or DNA of HIV-1 (6–8) or by possibly interacting with HIV-1 proteins (9, 10). However, these noncatalytic effects on HIV inhibition may be attributable to the overexpression of A3G and may not be occurring during normal infection (11–13). It is important to bear in mind that any actions of A3G on DNA, catalytic and possibly noncatalytic, are balanced in vivo against cellular RNA binding, which forms a high molecular mass A3G-RNA complex, which may prevent A3G incorporation into virions (7, 14–16), and against HIV RNA binding, which forms an intravirion A3G complex in which A3G must be activated by HIV RNase H for DNA deamination to ensue (16).
We have shown that A3G-catalyzed deamination occurs processively while exhibiting a 3′ → 5′ polarity favoring deamination toward the 5′-region of ssDNA (15). Directional deamination is an intrinsic property of A3G, occurring in the absence of an obvious source of energy (e.g. ATP or GTP) (15) and can in principle contribute to the HIV-1 G → A mutational bias, increasing in a 3′-direction in the RNA genome (17, 18). A3G processivity appears consistent with a three-dimensional facilitated diffusion process involving sliding and jumping motions along the DNA (15), where jumping involves microscopic dissociation and reassociation events occurring with the same DNA molecule, rather than diffusion into the surrounding bulk solution (19–21). Three-dimensional processive movement has been documented for a variety of site-specific DNA-targeting proteins (e.g. restriction endonucleases) (21–24), but in contrast to A3G, catalysis along the DNA is random. Random processive deaminations also occur with activation-induced cytidine deaminase (AID), an APOBEC family member with a single deaminase domain (25).
We have previously suggested that A3G deamination is strongly favored toward the 5′-direction, perhaps because of a catalytic orientation specificity and structural asymmetries in the enzyme (15). The presence of multiple catalytic and binding domains in A3G contained in an elongated rod-shaped oligomeric structure found in this deaminase family (26, 27) has the potential to impose catalytic directionality. A3G has been structurally characterized in solution as a tail-to-tail dimer (26), but monomeric forms are also active (28–30). When bound to DNA, A3G may exist in different oligomeric states, as implied by multiple A3G-DNA bands in gel shift analyses (15, 31).
In the present work, using human A3G produced from baculovirus-infected Sf9 insect cells, we have applied presteady state kinetic analyses in combination with atomic force microscopy (AFM) to investigate the dynamic and structural determinants of A3G processivity and catalytic polarity. We provide evidence showing that the 3′ → 5′ bias in deamination of C → U appears to be determined by the oligomeric state of the enzyme, with populations of dimers, tetramers, and perhaps larger oligomers favoring directionally biased deamination and most likely monomers catalyzing random deamination. Both the directional and random deamination modes occur with high processivity, readily observable on a time scale of a few seconds.
EXPERIMENTAL PROCEDURES
Substrates—DNA oligonucleotides were synthesized on an Applied Biosystems 3400 DNA/RNA synthesizer and are listed in supplemental Table S1. Fluorescein-dT and pyrrolo-dC were purchased from Glen Research. The partially dsDNA substrates were constructed by annealing with a 1.4-fold molar excess of complementary ssDNA. A circular DNA substrate was constructed by 5′-end phosphorylation of a 118-nt ssDNA substrate with T4 polynucleotide kinase (New England Biolabs) and subsequent ligation using a linker DNA oligonucleotide. The circular product was then purified after resolution on a 10% denaturing polyacrylamide gel.
A3G and AID Expression and Purification—A baculovirus-expressed GST-A3G protein was constructed as described previously (15). A3G recombinant virus was obtained as described previously (32). Sf9 cells were infected with recombinant A3G virus at a multiplicity of infection of 1 and harvested after 72 h. Cells were lysed as described previously (15) in the presence of 50 μg/ml RNase A. Cleared lysates were then incubated with glutathione-Sepharose resin (GE Healthcare) and subjected to a series of salt washes (0.25–1 m NaCl) before elution in buffer as previously described (15). A3G was treated with 0.02 units/μl thrombin (GE Healthcare) for 16 h at 21 °C to release the glutathione S-transferase tag, and subsequently the sample was diluted to 50 mm NaCl and loaded onto a DEAE FF column (GE Healthcare) equilibrated with 50 mm Tris-HCl, pH 8.9, 50 mm NaCl, 1 mm dithiothreitol, and 10% glycerol. A3G was eluted at ∼200 mm NaCl by using a linear gradient from 75 to 1000 mm NaCl. Fractions were collected and stored at -70 °C. A3G is ∼95% pure. AID was purified as previously described (32).
Deamination Assays—A3G activity on ssDNA substrates was measured in reactions containing 50 nm 32P-labeled DNA, 5 or 500 nm A3G in buffer (50 mm HEPES, pH 7.3, 1 mm dithiothreitol, and varied amounts of MgCl2 or other salts where indicated) and incubated for 2.5–60 min at 37 °C. ssDNA substrates (0.3–0.5 μm) with an internal fluorescein label were incubated with A3G or AID (20–300 nm), and deaminations were carried out as described above. All reactions with partial dsDNA substrates also contained 30 mm NaCl to stabilize the duplex region. Reactions were terminated by an extraction with phenol/chloroform/isoamyl alcohol (25:24:1), and deaminations were detected by the uracil DNA glycosylase assay, as described previously (15). Circular substrates were linearized before PAGE resolution by annealing to a 30-nt complementary oligonucleotide containing an MseI site and incubating with 5 units of MseI for 60 min at 37 °C. Gel band intensities were visualized by phosphorimaging or with a FX fluorescence scanner (Bio-Rad) and then measured with ImageQuant software (Amersham Biosciences).
Presteady state reactions were mechanically sampled and quenched using a KinTek quench-flow instrument. A solution of DNA and A3G at twice the final intended concentration was rapidly mixed in equal volumes and allowed to react for a given amount of time (0.1–180 s) before quenching with phenol/chloroform/isoamyl alcohol (25:24:1). Samples were then treated as described above.
Analysis of Processive Deamination by a Single A3G or AID— The analysis of processive deamination described previously (15, 25) was used. This entails the quantification of integrated band intensity of fractions of substrates with a single deamination at the 5′ Cor3′ C or a double deamination at both the 5′ C and 3′ C. The predicted fraction of independent double deaminations is then calculated by multiplying the fraction of all deaminations occurring at the 5′ C (sum of the integrated gel band intensities from deaminations occurring at 5′ C and double deaminations (5′ C and 3′ C)) to the fraction of all deaminations occurring at the 3′ C (sum of the integrated gel band intensities from deaminations occurring at 3′ C and double deaminations (5′ C and 3′ C)). The ratio of the observed fraction of double deaminations (occurring at both 5′ C and 3′ Con the same molecule) to the predicted fraction of independent double deaminations is the “processivity factor” and was used to determine processive (i.e. correlated double) deamination of two target motifs. A ratio larger than 1 indicates that a majority of double deaminations are caused by the same A3G or AID molecule acting processively on both target motifs.
Presteady State Anisotropy and Fluorescence Assays—Reactions were at 37 °C in buffer containing 50 mm HEPES, pH 7.3, 1 mm dithiothreitol, and 0 or 5 mm MgCl2.
Stopped flow anisotropy measurements were performed using an Applied Photophysics Π*180 spectrofluorometer equipped for anisotropy by exciting with vertically polarized light at 494 nm (2.5-nm slit width) from a xenon lamp and monitoring emissions using a 515-nm cut-off filter. To determine on-rates, 0.8–2 μm A3G was incubated in one syringe, and 200 nm fluorescein-labeled 69-nt ssDNA substrate was incubated in a second syringe, before rapidly mixing 50 μl from each syringe and monitoring the change in anisotropy for 0.2 s. The anisotropy change as a function of time was fit to a double exponential equation to obtain on-rates. To determine off-rates, in one syringe, 100 nm fluorescein-labeled 69-nt ssDNA substrate was preincubated with 400 nm A3G. The second syringe contained 10 or 20 μm unlabeled ssDNA substrate. The reaction was initiated by mixing equal volumes of the syringes (50 μl) and monitoring the change in anisotropy for 500–1000 s. The anisotropy change as a function of time was fit to a double exponential decay to obtain off-rates.
Stopped flow fluorescence measurements were performed using an Applied Photophysics Π*180 spectrofluorometer equipped for fluorescence. The ssDNA containing pyrrolo-C was excited at 335 nm (2-nm slit width) with a xenon-mercury lamp, and emissions were monitored using a 435-nm cut-off filter. To determine fluorescence changes, measured in volts, 2 μm A3G was incubated in one syringe, and 200 nm pyrrolo-C-labeled 69-nt ssDNA substrate was incubated in a second syringe, before rapidly mixing 50 μl from each syringe and monitoring the change in fluorescence emission for 0.6 s. The fluorescence change as a function of time was fit to a double exponential equation to obtain rates.
Atomic Force Microscopy—A3G imaging buffer (50 mm HEPES, pH 7.3) either with or without 5 mm MgCl2 was heated to 65 °C and allowed to cool slowly to dissolve salts. A3G was then diluted to 100 nm in A3G imaging buffer, and 0.1 mm dithiothreitol (final concentration) was added to the sample. Ten microliters of the protein sample were then deposited onto freshly cleaved mica (Spruce Pine Mica Company, Spruce Pine, NC). The sample was rinsed immediately with nanopure water, excess water was wicked from the surface using filter paper, and the surface was then dried using a stream of nitrogen before being placed in the AFM for imaging. For experiments done in the presence of DNA substrate, 200 nm (final concentration) of ssDNA substrate was added to the A3G sample before depositing as for the protein alone.
All AFM images were captured in air using either a Nanoscope III or IIIa (Digital Instruments, Santa Barbara, CA) microscope in tapping mode. Pointprobe Plus tapping mode silicon probes (Agilent, Tempe, AZ) with resonance frequencies of ∼170 kHz were used for imaging. Images were collected at a speed of 2–3 Hz with an image size of 1 × 1 μm at 512 × 512 pixel resolution. Each experiment was repeated at least twice. Volume analysis was done as previously described (33, 34). The program Kaleidagraph (Synergy Software, Reading, PA) was used to generate statistical plots.
RESULTS
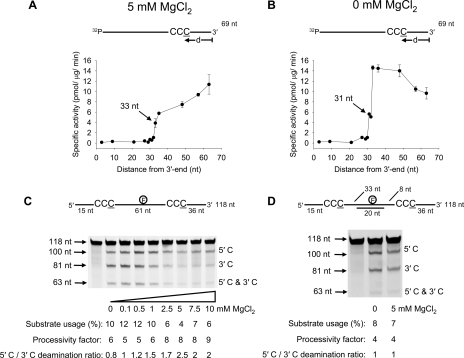
Electrostatic Influence on A3G Deamination Polarity and Processivity— We previously observed that A3G deamination on linear ssDNA exhibits a marked deamination bias, where the rate of deaminating the third C in the 5′-CCC target to U is enhanced significantly as the motif is located further away from the ssDNA 3′-end (15). The earlier experiments were performed in the presence of salt (MgCl2) (15). To study A3G deamination bias at higher resolution, with and without salt, we used 11 ssDNA substrates, each with a 5′-A(G/T)A CCC AAA hot spot motif in a different location along a 69-nt ssDNA molecule (Fig. 1, A and B).
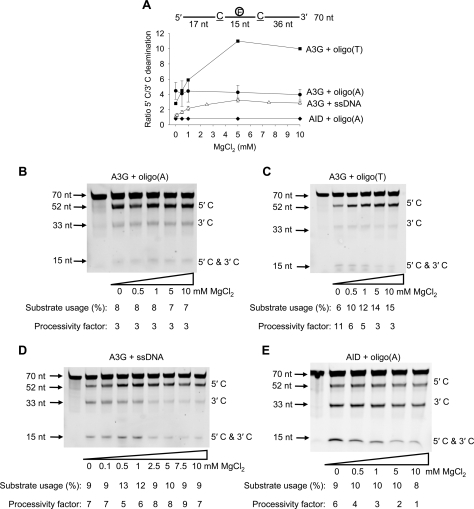
FIGURE 1.
A3G deamination activity, polarity, and processivity on ssDNA is dependent on MgCl2 and the presence of a dsDNA region. A and B, specific activity of A3G (5 nm) is plotted as a function of the distance (d) of the CCC motif from the 3′-end of a 69-nt ssDNA substrate (50 nm) deaminated in the presence of 5 mm MgCl2 (A) or absence of MgCl2 (B). Error bars, ±1 S.E. from three independent experiments. C and D, A3G deamination of a 118-nt internally fluorescein-labeled ssDNA substrate at an enzyme/DNA ratio of 1:20 in the absence (C) or presence (D) of a 20-nt complementary oligonucleotide annealed between the two C targets. Single deaminations of the 5′ C and 3′ C are detected as the appearance of labeled 100- and 81-nt fragments, respectively; double deamination of both Cs on the same molecule results in a 63-nt labeled fragment (5′ C and 3′ C). Substrate usages (%), processivity factors, and ratios of 5′ C/3′ C deaminations are shown below the gels. For deamination reactions with partially dsDNA, 30 mm NaCl was present to stabilize the duplex region. The reaction on ssDNA shown in C is unaffected by the presence of 30 mm NaCl.
A “dead” zone containing 30 nt was present at the 3′-end of the ssDNA substrate, where the specific activity was <1 pmol/μg/min irrespective of target position or salt concentration (Fig. 1, A and B). We speculate that the 30-nt dead zone may be the optimal space A3G needs at a 3′-ssDNA end to encounter a CCC motif in an orientation allowing catalysis. There was a monotonic increase in A3G specific activity, in the presence of 5 mm MgCl2, as the target motif was moved out of the dead zone toward the 5′-end, achieving a maximum specific activity of 11 pmol/μg/min (Fig. 1A). In the absence of MgCl2 the activity of A3G reached a peak near the center of the ssDNA (∼15 pmol/μg/min) and then remained roughly constant before decreasing from about 50 to 69 nt from the 3′-end (0 mm MgCl2) (Fig. 1B).
A3G processivity and deamination polarity can be measured simultaneously by placing two CCC motifs on the same ssDNA substrate and having a fluorescein label in between (15, 25) (Fig. 1C). The processivity factor (see “Experimental Procedures”) measures the fraction of correlated double deaminations occurring at two CCC motifs located on the same ssDNA substrate by a single enzyme molecule compared with uncorrelated double deaminations catalyzed independently by two different A3G molecules (15, 25). The analysis requires that less than ∼15% of the substrate be used to maintain single-hit kinetics (35). A3G processivity did not change significantly as a function of MgCl2 concentration, where the processivity factors are in the range of 5–9 (Fig. 1C). A measurement of A3G deamination polarity on the two C targets, which are out of the deamination dead zone, as a function of salt concentration, shows that deamination at the 5′ C target (100 nt from the 3′-end) is favored over the 3′ C target (36 nt from the 3′-end) by roughly 2-fold at MgCl2 concentrations of >5 mm (5′ C/3′ C deamination ratio; Fig. 1C). The deamination polarity decreased with less salt until, in the near absence of MgCl2, A3G deaminated the C targets with no preference (5′ C/3′ C deamination ratio ∼1; Fig. 1C).
The directional bias of A3G deamination was clearly electrostatic in origin as opposed to a specific interaction with Mg2+, because favored deamination toward the 5′-end also occurred with CaCl2, MnCl2, and NaCl (supplemental Fig. S1). In the case of monovalent NaCl, a 20-fold higher concentration was required to obtain a similar 5′-end deamination bias observed for divalent salts.
We reexamined the polar and processive properties of A3G on the 118-nt ssDNA (Fig. 1C) but with a 20-nt oligonucleotide annealed between the deamination targets (Fig. 1D). Since A3G bound dsDNA with a low GC content at ∼5-fold lower affinity than ssDNA (supplemental Fig. S2) and does not bind dsDNA with a high GC content (15), a loss in the sliding component of the processive motion is expected (15, 31). However, the jumping component of processivity appeared to be retained, since the processivity factor was reduced from 8 to 4 (Fig. 1D). The lack of deamination bias in the absence of MgCl2 (5′ C/3′ C deamination ratio is 1; Fig. 1D) was expected (see Fig. 1C, 0 mm MgCl2). However, the annealed dsDNA region between the two deamination targets also resulted in a loss of deamination bias in the presence of 5 mm MgCl2 (5′ C/3′ C deamination ratio is ∼1; Fig. 1, compare C and D), indicating that the dsDNA between the deamination sites decoupled the two sides of the substrate. The deamination in each single-stranded region depended on the target size for each region, the 36-nt target length from the 3′-end of the 118 nt substrate and the 33-nt target length region between the 5′ C and the dsDNA (Fig. 1D). There is, however, an important distinction to be made regarding the occurrence of the dead zone (Fig. 1, A and B). The ∼30-nt dead zone occurs only at the 3′-end of the ssDNA; there is no internal dead zone created by the dsDNA region, but instead deamination is dependent of the nucleotide target size (15).
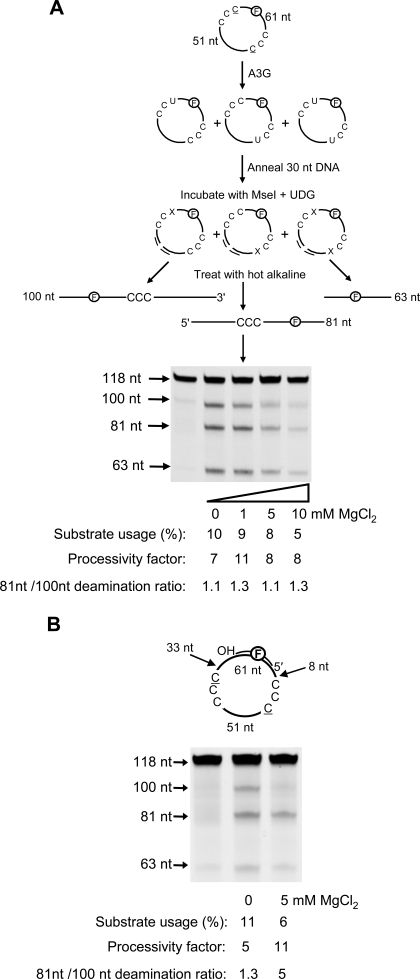
Deamination Polarity on Circular ssDNA Requires an Intervening dsDNA Region—To examine what effect the ssDNA ends have on A3G deamination bias, ends were eliminated by circularizing the molecule depicted in Fig. 1C (Fig. 2A, sketch). A3G shows essentially no deamination bias either with or without MgCl2 on the circular molecule (81 nt/100 nt deamination ratio ∼1.1–1.3; Fig. 2A). Although deamination was slightly inhibited in the presence of MgCl2, the apparent binding constants of the circular DNA compared with its linear counterpart were the same (Kd ∼ 160 nm). A3G was processive on the circular substrate with processivity factors similar to those on the linear substrate (compare Fig. 2A with Fig. 1C).
FIGURE 2.
A3G directional bias is absent on circular ssDNA and present on partially double-stranded circular DNA. A3G deamination bias and processivity were characterized on circular fluorescein-labeled DNA substrates containing two CCC motifs using an enzyme/DNA ratio of 1:20. A (sketch), deamination events on a 118-nt circular ssDNA are visualized after a 30-nt complementary oligonucleotide is annealed to the circular DNA to enable cutting by MseI (sketch). This procedure linearizes the circular DNA so that all possible single and double deamination events can be detected using the uracil DNA glycosylase (UDG) assay (see “Experimental Procedures”). An X represents an abasic site. A, A3G exhibits processive but not directionally biased deamination on circular DNA as a function of MgCl2. B, A3G deamination of circular substrate in the presence of a 20-nt complementary oligonucleotide annealed between the two C targets is processive, and the directional bias is restored with 5 mm MgCl2. Substrate usages (%), processivity factors, and ratios of 81-nt fragment to 100-nt fragment are shown below the gels. For deamination reactions with partially dsDNA, 30 mm NaCl was present to stabilize the duplex region. The reaction on circular ssDNA shown in A is unaffected by the presence of 30 mm NaCl.
We examined the effect on deamination of an oligonucleotide annealed to the circular DNA substrate to form a partially dsDNA circle (Fig. 2B). To ensure that A3G had enough room to deaminate both C targets equally (see Fig. 1B), we placed the dsDNA at least 33 nt from the 3′-side of each CCC motif. A3G was able to act processively on partially dsDNA circles (processivity factor between 5 and 11; Fig. 2B). Without MgCl2, A3G deaminated each C target with about equal preference (81 nt/100 nt deamination ratio of 1.3; Fig. 2B). However, with 5 mm MgCl2, A3G regained the ability to deaminate with a directional bias (81 nt/100 nt deamination ratio of 5; Fig. 2B).
The dsDNA segment, in the presence of MgCl2, may reinstate the deamination bias by limiting the available nucleotide target size. That A3G now favors the C target with the largest target size for binding (i.e. that furthest from the 3′-end of the dsDNA segment) (87 nt; Fig. 2B) implies that there is an intrinsic 3′ → 5′ directional catalytic constraint for A3G deamination so that deamination occurs predominantly when A3G approaches a C target with a 3′ → 5′ orientation on ssDNA, consistent with our previous data (15). A3G showed no deamination bias on a completely single-stranded circle (Fig. 2A), presumably because the enzyme can encounter either of the two C targets in a 3′ → 5′ orientation without a nucleotide target size limitation imposed by linear ssDNA ends. A3G did not use a terminal 3′-OH for directional orientation (supplemental Fig. S3). Deamination assays with a biotin/streptavidin block at the 3′-or5′-end showed no difference in the deamination activity or directional bias, which suggests that preferential binding at an ssDNA end is probably not occurring (data not shown).
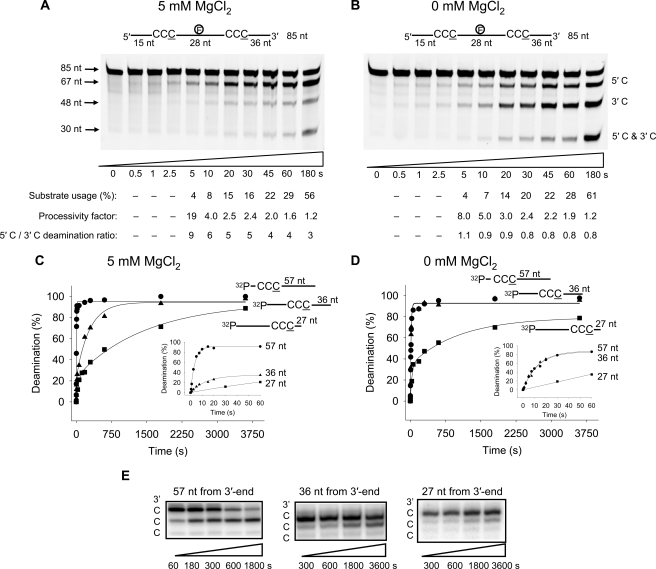
Temporal Analysis of A3G Deamination Polarity and Processivity—To make a finer tuned analysis of the two deamination modes of A3G, directionally biased or random, we used rapid-quench kinetics to investigate deamination polarity and processivity on short time scales (several seconds). We had previously established that during “long” (10-min) incubations, A3G-catalyzed C deaminations occur processively and with 3′ → 5′ polarity in the presence of salt (15) (Fig. 1C). Notably, the same degree of processivity and deamination polarity in the presence of salt was observed during “short” incubations (Fig. 3, A and B). The reactions with 5 mm MgCl2 (Fig. 3A) show that at the earliest time that deaminations are detectable (∼5 s), deamination of the 5′ C is favored over the 3′ C by a factor of 9. In the absence of MgCl2, A3G deaminated each C target with no preference; the “3′ C” was 36 nt from the 3′-end and was fully accessible for deamination (Fig. 3B). The processivity factors decreased substantially, approaching ∼1 (no processivity) as substrate usage went from 4 to >50% (Fig. 3, A and B). The loss of processivity at high substrate usage probably resulted from multiple encounters of A3G with the ssDNA substrates, leading to independent double deaminations (15), whereas, at low substrate usage, each ssDNA substrate is expected to encounter A3G only once, giving rise to processive correlated double deaminations.
FIGURE 3.
Presteady state analysis of polarity, processivity and deamination rates. A and B, processive deamination occurs on a presteady state time scale. A3G was incubated with an 85-nt internally fluorescein-labeled ssDNA substrate with two CCC motifs 28 nt apart and at an enzyme/DNA ratio of 1:1. Single deaminations of the 5′ C and 3′ C are detected as the appearance of labeled 67- and 48-nt fragments, respectively; double deamination of both Cs on the same molecule results in a 30-nt labeled fragment (5′ C and 3′ C). Processive deamination was determined in the presence of 5 mm MgCl2 (A) and absence of MgCl2 (B). Substrate usages (%), processivity factors, and the ratios of 5′ C/3′ C deaminations are shown below the gels. C and D, single turnover reaction rates for A3G were determined on ssDNA substrates with CCC motifs 57 (•), 36 (▴), or 27 nt (▪) from the 3′-end of a 69-nt substrate under enzyme saturating conditions (500 nm A3G, 50 nm DNA). C, with 5 mm MgCl2, the deamination of a CCC motif 57 nt from the 3′-end, measured as a function of time, was fit to a single exponential equation to determine a single turnover rate of 0.25/s. The deamination of a CCC motif 36 nt from the 3′-end measured as a function of time was fit to a double exponential equation to determine a minor “fast” rate component of 0.12/s (23% amplitude) and a major “slow” rate component of 0.004/s (72% amplitude). The deamination of a CCC motif 27 nt from the 3′-end measured as a function of time was fit to a double exponential equation to determine a minor “fast” rate component of 0.02/s (24% amplitude) and a major “slow” rate component of 0.0007/s (70% amplitude). Inset graph, the first 60 s of the reaction. D, reaction rates determined as in C but in the absence of MgCl2. The data for substrates with CCC motif 57 and 36 nt from the 3′-end were fit to a single exponential equation to obtain rates of 0.07/s and 0.08/s, respectively. The data for substrate with CCC motif 27 nt from the 3′-end were fit to a double exponential equation with an initial “fast” rate of 0.03/s (34% amplitude) and a second “slow” rate of 0.001/s (45% amplitude). Inset graph, the first 60 s of the reaction. E, examination of how CCC motifs are deaminated. Deamination of the 3′ and middle C residues in the motif 5′-CCC-3′ is influenced by distance from the 3′-end of the ssDNA. Deamination of the 5′ C residue in the motif 5′-CCC-3′ is barely observed. The gel reaction buffers contained 5 mm MgCl2.
To characterize the mechanism of deamination polarity and to obtain A3G single turnover deamination rates at different target motif locations along the ssDNA substrate, we employed three of the 69-nt ssDNA molecules used previously (see Fig. 1, A and B) with one 5′-A(G/T)A CCC AAA hot spot motif either 57, 36, or 27 nt from the 3′-DNA end in reactions with a large excess of A3G over substrate DNA (A3G/DNA ∼ 10:1) (Fig. 3, C and D). The ssDNA molecules were bound by A3G with the same apparent Kd (100 nm) (supplemental Fig. S4).
The single turnover rate for a reaction containing 5 mm MgCl2 for a C target 57 nt from the 3′-end fits well to a single exponential and yields a deamination rate of 0.25/s (Fig. 3C), with a maximum of ∼100% substrate deamination within 20 s (Fig. 3C, inset). In contrast, the C targets 36 or 27 nt from the 3′-end required 1500 and 3600 s to reach 100% substrate deamination, respectively (Fig. 3C). These data are not fit well by single exponentials and require double exponential fits with a fast and a slow phase. For ssDNA with a C target near the middle (36 nt from the 3′-end), the rate of the fast phase is 0.12/s but only accounts for 23% of the deaminations taking place (Fig. 3C, inset), whereas the slower rate, 0.004/s (one deamination every 250 s) accounts for 77% of the deamination (Fig. 3C). The C target in the ssDNA 3′-end dead zone, 27 nt from the 3′-end, was deaminated the slowest, with a fast rate of 0.02/s occurring for about 24% of the deaminations (Fig. 3C, inset) and a slower rate of 0.0007/s for the remainder (Fig. 3C). Biphasic deamination rates under single turnover conditions suggest that multiple deamination modes of A3G exist that catalyze deaminations at different rates.
In the absence of MgCl2, A3G showed greater ability to deaminate at the center of the ssDNA (Fig. 3D), which is consistent with specific activities determined at various locations along the DNA substrate (Fig. 1B). The deamination rates for substrates with C targets 57 or 36 nt from the 3′-end were essentially the same, with deaminations occurring every 14 s (∼0.08/s; Fig. 3D, inset). On the substrate with a C target in the ssDNA 3′-end dead zone, 27 nt from the 3′-end, the initial rate (0.03/s) limited deaminations to ∼30 s for each and was far slower than the other C targets (Fig. 3D, inset). The second reaction rate representing 45% of the amplitude for a C target located 27 nt from the 3′-end was 0.001/s (Fig. 3D).
When we probed deaminations occurring at individual C sites within each motif, it was evident that the third C in the 5′-CCC was favored for deamination, the middle C (5′-CCC) could also be deaminated on a longer time scale, and the first C (5′-CCC) was barely deaminated (Fig. 3E). The general pattern of deaminations within a motif (Fig. 3E) was consistent with A3G 3′ → 5′ directional polarity in which triplet motifs undergo more rapid deamination nearer the 5′-end of the ssDNA substrate (Fig. 3, A and C).
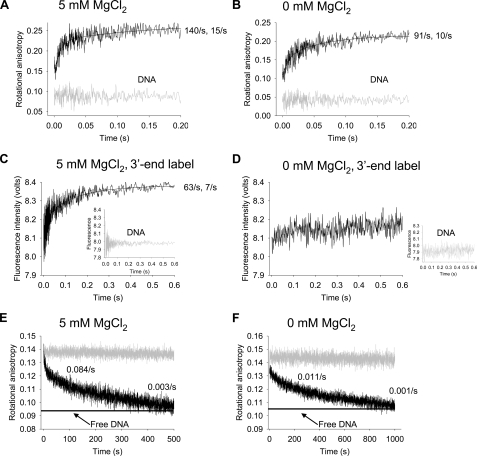
Rates of A3G Binding and Release of ssDNA are Biphasic—It is essential to view A3G deamination time scales (Fig. 3) within the broader milieu of time scales required for A3G-ssDNA complex formation and dissociation. We used presteady state kinetic analyses of rotational anisotropy and fluorescence emission to measure A3G on-rates and off-rates and to investigate possible conformational changes of the ssDNA substrate (Fig. 4).
FIGURE 4.
Stopped-flow fluorescence presteady state interactions of A3G with ssDNA. A and B, the on-rate of A3G (1 μm) to a 69-nt fluorescein-labeled ssDNA substrate (100 nm) was determined by monitoring changes in rotational anisotropy of free ssDNA (gray line) following the addition of A3G (black line) as a function of time. The on-rate was fit to a double exponential equation. A, for A3G in buffer with 5 mm MgCl2 the rapid rate of 140 ± 14/s (62% amplitude) was followed by a slower rate of 15 ± 2/s (38% amplitude). B, for A3G in the absence of MgCl2, the rapid rate of 91 ± 10/s (53% amplitude) was followed by a slower rate of 10 ± 1/s (47% amplitude). C and D, changes in fluorescence of 100 nm pyrrolo-C-labeled ssDNA (inset graph) upon the addition of 1 μm A3G (black line) were monitored as a function of time. The pyrrolo-C label was placed 10 nt from the 3′-end of the 69 nt ssDNA. The fluorescence change was fit to a double exponential equation. C, under conditions with 5 mm MgCl2 the fluorescence change began with a rapid rate of 63 ± 8/s (40% amplitude) and was followed by a slower rate of 7 ± 0.3/s (60% amplitude). D, in the absence of MgCl2, no fluorescence change was detected. E and F, the off-rate of A3G (0.2 μm) from a 69-nt fluorescein-labeled ssDNA substrate (50 nm) was determined by preincubating A3G with ssDNA (gray line) and monitoring changes in rotational anisotropy following the addition of 100–200-fold excess unlabeled 69-nt ssDNA as trap (black line). The anisotropy of free fluorescein-labeled ssDNA mixed with unlabeled ssDNA trap is denoted with a horizontal line. The off-rate was fit to a double exponential decay. E, in the presence of 5 mm MgCl2, the reduction in the rotational anisotropy signal, representing A3G dissociating from fluorescein-labeled DNA and binding the unlabeled trap DNA, had an initial rate of decay fitting to 0.084 ± 0.005/s (20% amplitude) and a second decay rate of 0.00279 ± 0.00006/s (80% amplitude). F, in the absence of MgCl2, the reduction in the rotational anisotropy signal had an initial rate of decay fitting to 0.011 ± 0.001/s (25% amplitude) and a second decay rate of 0.0010 ± 0.0002/s (75% amplitude).
The on-rate of A3G, determined with a 10-fold excess of A3G to DNA in the presence and absence of MgCl2, was extremely rapid, exhibiting an initial fast phase and slower second phase (Fig. 4, A and B). About half of the ssDNA was bound within the instrument dead time (1.1 ms). The estimated on-rates under these conditions are 140/s (Fig. 4A; 5 mm MgCl2) and 91/s (Fig. 4B; without MgCl2). These measurements were also made as a function of A3G concentration (0.4–0.8 μm) to calculate the bimolecular rate constants, which approach the diffusion limit, with or without MgCl2 (5 mm MgCl2, 1 × 108/m/s; 0 mm MgCl2, 9 × 107/m/s). Each on-rate experiment, in the presence or absence of MgCl2, confirmed that the A3G on-rate is biphasic with an initial rapid phase (∼60% amplitude) and secondary slow phase (∼40% amplitude). It may be that the two distinct on-rates reflect different interactions of a single A3G form with DNA or that multiple oligomeric forms of A3G exist and interact differently with DNA.
It is likely that when an A3G molecule, which has two DNA binding domains per monomer, binds to ssDNA, there will be multiple binding interactions along the substrate, which may cause a conformational change in the structure of the ssDNA. To detect conformational changes in the ssDNA, such as base unstacking, we measured the presteady state change in fluorescence intensity of a 2′-deoxycytidine analogue, pyrrolo-dC, which was located 10 nt from the 3′-end of a 69-nt ssDNA (Fig. 4, C and D). Notably, biphasic changes in fluorescence were observed with 5 mm MgCl2 (Fig. 4C), having a fast rate of 63/s (40% amplitude) and slower rate of 7/s (60% amplitude). The data at 5 mm MgCl2 suggest that there was a conformational change in the ssDNA occurring at a considerably slower rate (63/s; Fig. 4C) than A3G-ssDNA binding (140/s; Fig. 4A). In contrast, there was almost no detectable fluorescence change in the absence of MgCl2 of a pyrrolo-C residue located 10 nt from the 3′-end (Fig. 4D).
An estimate of the spatial properties of the conformational change can be made by measuring the change in steady state fluorescence intensity for pyrrolo-C residues placed at different locations along the ssDNA (supplemental Fig. S5A). There was a significant increase in fluorescence at all locations along the DNA in the presence of MgCl2, with maxima occurring at 10 nt from the 5′- and 3′-ends, corresponding to the locations of the fluorophores in the presteady state analysis (Fig. 4C and supplemental Fig. S5, A and B). In the absence of MgCl2, there was essentially no change in fluorescence intensity along the DNA except when the pyrrolo-C was located 10 nt in from the 5′-end (supplemental Fig. S5, A and C). The presteady state and steady state fluorescence intensity measurements are mutually supportive and suggest that a conformational change in the DNA, perhaps indicative of DNA wrapping, occurs most strongly with 5 mm MgCl2 (supplemental Fig. S5A) at ∼15 ms after A3G binds to DNA (Fig. 4, A and C).
A3G must remain bound to the same ssDNA substrate long enough for it to act processively (see Fig. 3, A and B). To measure the residence time of A3G bound to a single DNA substrate molecule (Fig. 4, E and F), A3G was prebound to fluorescein-labeled ssDNA, and a large excess (100–200-fold) of the identical unlabeled ssDNA was added as a trap to bind A3G once it dissociated, observed as a decrease in anisotropy from the fully bound starting point. We verified that the off-rates were independent of the trap concentration, suggesting that the unlabeled DNA acts as a passive trap, not as a competitor for A3G binding.
The off-rate of A3G was biphasic with a rapid initial off-rate occurring during the first few seconds (∼20% amplitude), followed by a much slower off-rate occurring on a time scale of minutes (∼80% amplitude) (Fig. 4, E and F). The residence time (reciprocal of the off-rate) for the A3G rapidly dissociating small amplitude phase was ∼12 s (0.084/s) with salt (Fig. 4E) and about 8-fold longer (∼90 s) without salt (0.011/s; Fig. 4F). The second much more stable binding mode was characterized by residence times of about 300 s and 1000 s, in the presence and absence of MgCl2, respectively (Fig. 4, E and F). It is likely that the two distinct off-rates reflect the stability of different A3G-DNA complexes. The key point, however, is that A3G can remain bound to the DNA on the order of minutes, which provides ample time to catalyze the correlated deaminations that take place within5sofa single A3G-DNA encounter (Fig. 3, A and B).
A3G Deamination Polarity Dependence on DNA Topology— Target CCC motifs were embedded in homopolymer A and T regions to investigate the effect of DNA topology on A3G deamination bias (Fig. 5). Since A3G can cause a conformational change in DNA in the presence of MgCl2 (Fig. 4C) we expected that A3G would be sensitive to DNA topology. Oligo(A) ssDNA substrates are known to have more rigid conformations than oligo(T) or ssDNA (36). When acting on an oligo(A) substrate, (A15C-CCA6T1A6CCCA36, where T contains fluorescein label), A3G deaminated the 5′ C with the same 5′ C/3′ C deamination ratio (∼4) regardless of MgCl2 concentration (Fig. 5, A and B). This result is in contrast to ssDNA, where A3G showed no deamination bias in the absence of MgCl2 (Fig. 5, A and D; see Fig. 1C). When acting on the more flexible oligo(T) substrate, (T15CCCT13CCCT36), A3G exhibited a deamination bias in the presence or absence of MgCl2, but in contrast to oligo(A) and ssDNA, there was a marked increase in the deamination bias with increasing MgCl2, going from a 5′ C/3′ C deamination ratio of 2.8 (no salt) to 10 (10 mm MgCl2) (Fig. 5, A and C). In contrast, the APOBEC family member, AID, deaminated ssDNA in an unbiased manner in the presence and absence of MgCl2 on ssDNA (25) or on the oligo(A) substrate (A16GCA7T1A6GCA36; Fig. 5, A and E).
FIGURE 5.
A3G and AID deamination of oligo(A) and oligo(T) substrates containing two target motifs. A3G or AID was incubated with a 70-nt internally fluorescein-labeled ssDNA substrate with two CCC or AGC motifs 13 nt apart and at an enzyme/DNA ratio of 1:20. Single deaminations of the 5′ C and 3′ C are detected as the appearance of labeled 52- and 33-nt fragments, respectively; double deamination of both Cs on the same molecule results in a 15-nt labeled fragment (5′ C and 3′ C). A, A3G deamination bias is affected by the sequence composition. The ratio of 5′ C/3′ C deamination is shown as a function of MgCl2 concentration for each variation of the 70-nt internally fluorescein-labeled ssDNA substrate tested. Error bars show ±1 S.E. from three independent experiments. Shown is A3G deamination on an oligo(A) sequence (A15CCCA6T1A6CCCA36) (B), an oligo(T) sequence (T15CCCT13CCCT36) (C), and an ssDNA (D). E, AID deamination on an oligo(A) sequence (A16GCA7T1A6GCA36). Substrate usages (%) and processivity factors are shown below the gels.
The Oligomerization State of A3G Depends on MgCl2 and DNA—The presteady state kinetic data revealed biphasic interactions of A3G with DNA (Fig. 4) and for some conditions biphasic deamination rates (Fig. 3, C and D). This could mean that a homogenous population of A3G dimers (26) interacts with the DNA in two modes or that different oligomeric states of A3G are present and exhibit different kinetic rates.
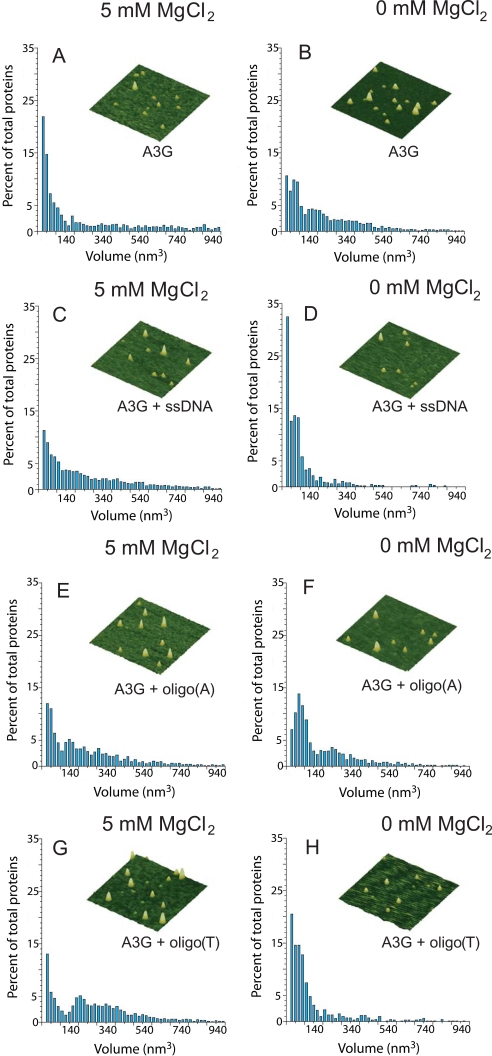
AFM, which yields topographic images of molecules deposited on a surface, can be used effectively for examining the oligomerization states of proteins (33, 34, 37–42). Specifically, it has been demonstrated that the volume of proteins (V) measured by AFM exhibits a linear dependence on molecular weight (MW) with V = 1.2 × MW - 14.7 (33, 34). Accordingly, we used AFM to characterize the oligomeric state of A3G (45.6 kDa) on ssDNA, oligo(A), and oligo(T) substrates, in the presence and absence of MgCl2 (Fig. 6).
FIGURE 6.
MgCl2 and DNA binding modulate the oligomerization state of A3G. AFM was used to determine the oligomeric state of A3G alone in the presence of 5 mm MgCl2 (A), alone in the absence of MgCl2 (B), in the presence of ssDNA and 5 mm MgCl2 (C), and in the presence of ssDNA and absence of MgCl2 (D). The oligomeric state of A3G was also determined in the presence of oligo(A) and 5 mm MgCl2 (E), in the presence of oligo(A) and absence of MgCl2 (F), in the presence of oligo(T) and 5 mm MgCl2 (G), and in the presence of oligo(T) and absence of MgCl2 (H). The 70-nt ssDNA was added to 200 nm final concentration. A3G was added to 100 nm final concentration. Volume distributions of A3G are plotted against the percentage of total proteins. Total proteins counted were as follows: 1354 (A), 3168 (B), 4231 (C), 663 (D), 1563 (E), 1224 (F), 6738 (G), and 706 (H). Representative AFM images of A3G are shown for each condition. Images are 300 × 300 nm with a height scale of 5 nm.
A3G alone (i.e. in the absence of ssDNA) in the presence of salt exists as a mix of monomers (∼40%) and a considerably smaller population of dimers (∼14%), along with higher order oligomers (∼46%) (Fig. 6A and Table 1). A3G alone in the absence of salt behaved differently, showing about a 2-fold reduction in monomers and an increase in the combined fractions of dimers and higher order oligomers (Fig. 6B and Table 1). These AFM data, which visualize A3G in the absence of DNA, are consistent with gel filtration data showing that A3G exists in multiple forms (7, 15). A3G dimers were observed with small angle x-ray scattering in the presence of NaCl and MgCl2, and we speculate that the absence of higher order oligomers may reflect differences in buffer conditions (26).
TABLE 1.
Distribution of A3G oligomerization states in response to MgCl2 and DNA Data are summarized from AFM experiments shown in Fig. 6.
|
AFM conditions
|
Distribution of oligomerization states
|
||
|---|---|---|---|
| Monomers (0–70 nm3) | Dimers (70–120 nm3) | Higher order oligomers (> 120 nm3) | |
| % | % | % | |
| A3G alone, 5 mm MgCl2 | 40 | 14 | 46 |
| A3G alone, 0 mm Mg Cl2 | 23 | 19 | 58 |
| A3G + ssDNA, 5 mm Mg Cl2 | 23 | 15 | 62 |
| A3G + ssDNA, 0 mm Mg Cl2 | 51 | 26 | 23 |
| A3G + oligo(A), 5 mm Mg Cl2 | 26 | 11 | 63 |
| A3G + oligo(A), 0 mm Mg Cl2 | 24 | 27 | 49 |
| A3G + oligo(T), 5 mm Mg Cl2 | 21 | 7 | 71 |
| A3G + oligo(T), 0 mm Mg Cl2 | 43 | 28 | 29 |
A3G in the presence of salt and ssDNA resulted in about a 2-fold reduction in monomers (∼23%) and an increase in higher order oligomers (62%), relative to the absence of DNA (Fig. 6, compare A with C, and Table 1). These data (Fig. 6C) are consistent with equilibrium binding data for A3G association with ssDNA, which show a slight cooperativity in A3G binding to DNA (15) (supplemental Fig. S4). The polydispersed nature of A3G on DNA (Fig. 6C) is also in agreement with previous data showing multiple bands in gel shift analyses (15, 31). In contrast, in the absence of MgCl2, the addition of ssDNA caused a shift in the A3G distribution, resulting in 2-fold more monomers (51%) than dimers (26%), whereas in the absence of salt and DNA, the monomer/dimer ratio was ∼1 (Fig. 6, B and D, and Table 1). The 2-fold increase in monomer/dimer ratio was accompanied by a concomitant 2-fold reduction in higher order oligomers. These data indicate that in the presence of salt, binding to ssDNA influences A3G to associate into higher order oligomers, whereas, in the absence of salt, binding of ssDNA causes the A3G to dissociate from higher order oligomers to predominantly monomers and dimers.
It appears that different oligomeric states may be responsible for the biphasic ssDNA binding and deamination kinetics. Taken together with Fig. 5D, the AFM data (Fig. 6, C and D) suggest that the different oligomeric states present when A3G binds ssDNA under different salt conditions influence whether A3G acts in either a directional or random manner, outside the 30-nt dead zone (Fig. 1, A and B). We confirmed that this correlation is also found when the AFM conditions, A3G/DNA ratio of 1:2, are used for the deamination reaction (supplemental Fig. S6). We further speculate that oligomers promote directional deamination and monomers promote random deamination (compare Fig. 6, C and D, with Fig. 5D; see Table 1).
The observation that the oligo(A) and oligo(T) ssDNA substrates promote the deamination polarity of A3G without the addition of MgCl2 (Fig. 5A) suggests that binding to these DNA substrates may influence the oligomeric state of A3G. To test this idea, we investigated the oligomeric states of A3G upon binding to both oligo(A) and oligo(T) ssDNA substrates in the presence and absence of MgCl2. The distributions of A3G with oligo(A) were similar in the presence and absence of MgCl2 (Fig. 6, E and F, respectively). The monomeric species were similar, as were the sum of dimeric and higher order species (Table 1). The data are consistent with A3G exhibiting comparable deamination polarities on oligo(A) in both the presence and absence of salt (Fig. 5B and supplemental Fig. S6). A comparison of the data in the absence of MgCl2 for no DNA (Fig. 6B), oligo(A) (Fig. 6F), and ssDNA (Fig. 6D) reveals that ssDNA, but not oligo(A), promotes the dissociation of A3G to a predominantly monomer population (Table 1) (2-fold fewer monomers with oligo(A) than ssDNA) in accord with the lack of polarity for A3G deamination of ssDNA in the absence of MgCl2 (Fig. 5D). In contrast to the oligo(A) data, the distributions of A3G with oligo(T) in the presence and absence of MgCl2 are different (Figs. 6, G and H, respectively), with a far higher proportion of molecules being present in higher order oligomeric states in the presence of salt (71%) compared with the absence of salt (29%) (Table 1). These relative distributions agree with the large increase in deamination polarity for A3G on oligo(T) with increasing MgCl2 concentration (Fig. 5A, supplemental Fig. S6). In the absence of MgCl2, the distribution for A3G binding to oligo(T) DNA is similar to that for A3G binding to ssDNA, except that the monomer peak is reduced for oligo(T) relative to ssDNA (43% versus 51%, Table 1; Fig. 6, compare H and D). This shift to higher order oligomeric states is consistent with A3G showing polarity on oligo(T) in the absence of MgCl2; the ratio of 5′ C/3′ C deamination is ∼2.8 (Fig. 5A).
DISCUSSION
We have previously shown that A3G binds randomly to ssDNA and is able to deaminate C residues processively with a 3′ → 5′ bias (15). In this paper, we have investigated the mechanisms of processivity and favored deamination in the 5′-ssDNA direction using a combination of kinetics and AFM analysis. Both attributes of A3G, its polarity in the presence of salt and its processivity in the presence and absence of salt, occur at the shortest times (∼5 s) for which deamination can be detected (Fig. 3, A and B).
Aside from the importance of understanding the mechanisms underlying A3G processivity and especially polarity, which occur in the absence of an external energy source such as ATP or GTP, the intrinsic biochemical properties of A3G are important biologically. The time scales of A3G-DNA complexes, which rapidly form and remain for 5–15 min (Fig. 4, A, B, E, and F) suggest that in vivo A3G has the potential to catalyze sizable numbers of processive deaminations on (-)-cDNA within the limited time that it is single-stranded. Notably, in vivo data reveal twin 5′ → 3′ G to A mutational gradients of the HIV-1 genome, which means that deaminations are increasing toward the 5′-end of the (-)-cDNA in two regions marked by reverse transcription primer sites on the (-)-cDNA (17, 18). It has been suggested that the mutational gradients are attributable to the temporal effects of reverse transcription acting in combination with RNase H, so that regions that remain single-stranded over the longest time frame are most susceptible to A3G-catalyzed deaminations (17, 18). A recent analysis of edited HIV-1 genomes, however, suggests that the intrinsic 3′ → 5′ deamination bias of A3G (15) makes a substantial contribution to local regions of HIV-1 mutational polarity in vivo (17).
Our AFM results demonstrate that interactions of A3G with ssDNA and salt can determine A3G oligomeric forms. Binding of A3G to ssDNA at high salt promotes the formation of higher order oligomers, whereas binding at low salt favors the presence of monomers (Table 1). Monomeric forms of A3G observed by AFM in the presence of ssDNA and absence of salt (Fig. 6D and Table 1) correlate with random deamination (Fig. 5D), whereas oligomeric forms that occur in the presence of ssDNA and salt (Fig. 6C and Table 1) correlate with a 3′ → 5′ bias (Fig. 5D). However, the polydispersed nature of the larger oligomers of A3G, at 5 mm MgCl2 (Fig. 6C and Table 1), precludes the identification of a single oligomeric state responsible for the deamination polarity. Nevertheless, the role of dimers, tetramers, and higher oligomeric forms of A3G in governing the deamination bias is strongly supported by deaminations of CCC motifs embedded within oligo(A) or oligo(T) sequences. A3G deaminates with a 3′ → 5′ deamination bias even in the absence of salt on the oligo(A) and oligo(T) (Fig. 5, B and C) but not on ssDNA substrates (Fig. 5D), and the higher oligomeric states of A3G are more stable on oligo(A) and oligo(T) than on the ssDNA in the absence of MgCl2 (Fig. 6, compare F and H with D; see Table 1). It is conceivable that larger oligomeric states form concatamers of the dimeric form (26) and probably involve C-terminal as well as N-terminal interactions, in accord with the APOBEC2 crystal structure (27), but we cannot resolve such oligomeric interactions with AFM data.
The single turnover deamination rates, determined with A3G in a 10-fold molar excess over ssDNA, show that the MgCl2 concentration dictates the intrinsic catalytic rates of C targets at different locations along an ssDNA molecule. Virtually identical intrinsic deamination rates exist in the absence of salt for target motifs located outside of the deamination dead zone at 57 and 36 nt from the 3′-end (Fig. 3D). In the presence of salt, however, the single turnover deamination rates differ substantially for target motifs located 57 and 36 nt from the 3′-end (Fig. 3C, inset). Comparing the deamination rates with the slowest off-rate measured in the presence of salt (0.003/s; Fig. 4E), a C target 57 nt from the 3′-end of ssDNA, having a deamination rate of 0.25/s (Fig. 3C) can easily be deaminated before A3G falls off the DNA. However, this is not generally true for the C target 36 nt from the 3′-end of an ssDNA, which is characterized by biphasic deamination kinetics (Fig. 3C). Roughly 25% of the substrates are deaminated at 0.12/s, whereas 75% are deaminated at 0.004/s (Fig. 3C), the latter rate being about the same as the slowest off-rate (Fig. 4E). Thus, in the presence of salt, there is a reduction in intrinsic rates of deamination occurring in targets nearer the 3′-end that seems to account for the 3′ → 5′ polarity.
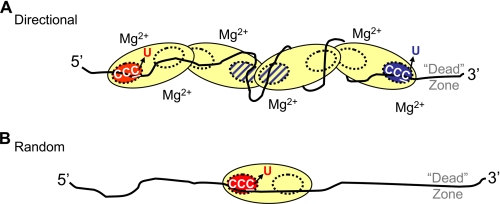
In the presence of salt, the slower catalytic rate for C-targets closer to the 3′-ssDNA end (Fig. 3C) may stem, in part, from reduced access of the CCC motifs to enzyme active sites. The reduced catalytic rate toward the 3′-DNA region occurs in conjunction with the formation of larger oligomers in the presence of salt (Fig. 6C and Table 1). We speculate that active sites situated within an oligomer, such as a tetramer, are unable to access and deaminate C targets efficiently. However, we hypothesize that A3G oligomers, including dimers, are directionally biased. If multiple binding interactions were to wrap the DNA around A3G (Fig. 4C) and restrict free diffusion of the oligomer, then a second level of bias is added. The intact oligomer would still be able to diffuse bidirectionally, encompassing sliding and jumping motions, but three-dimensional diffusion would become increasingly constricted as the oligomer size increased, limiting sampling of the ssDNA to find all possible C targets. We have incorporated these ideas in a schematic model (Fig. 7A). The elongated shape of the A3G protein contained in the sketch is based on small angle x-ray scattering of A3G in aqueous solution (26).
FIGURE 7.
Model for oligomeric regulation of A3G directional and random deamination. A, directional 3′ → 5′ deamination in the presence of “large” A3G oligomers. Larger oligomers, shown here as a tetramer, although dimers may also be present, are stable when binding ssDNA in the presence of Mg2+ (Fig. 6C). In the presence of Mg2+, there is a reduction in the intrinsic rates of C-deamination toward the 3′-DNA end, which accounts for the 3′ → 5′ polarity (Fig. 3, C and D). The catalytically inactive N-terminal domains are depicted as yellow dashed circles. The hot active site (red circle) is aligned in the catalytically favored 3′ → 5′ orientation and closest to the preferential C for deamination in a 5′-CCC target motif, whereas the cold active site (blue circle) is aligned in the opposite orientation nearest the barely deaminated C in a 5′-CCC target motif. Active sites located internally (cross-hatched circles) cannot effectively access CCC motifs and are essentially noncatalytic. The combination of inaccessible CCC motifs and the cold active site (blue circle) located toward the 3′-DNA end results in an inhibition of deamination relative to the easily accessible CCC motifs toward the 5′-DNA end. CCC motifs located within the ∼32-nt dead zone at the 3′-DNA end are deaminated with at least a 10-fold lower activity compared with motifs outside the dead zone located near the 5′-DNA end (Fig. 1A). We envision that although the A3G oligomer can slide and jump bidirectionally along the ssDNA, multiple A3G-DNA binding interactions may wrap the DNA around A3G (Fig. 4C) and restrict free diffusion of the oligomer, thus reducing the possibility for resampling nondeaminated C targets. B, random deamination performed by a monomer. Larger oligomers are disrupted upon binding ssDNA in the absence of Mg2+ (Fig. 6D). The A3G monomers can diffuse freely along the ssDNA and thereby deaminate each CCC motif with roughly similar efficiencies outside the dead zone at the 3′-DNA end. The monomer retains partial catalytic asymmetry by deaminating with at least a 15-fold lower activity on CCC motifs within the ∼30-nt dead zone at the 3′-DNA end compared with motifs outside the dead zone (Fig. 1B). The dead zone at the 3′-DNA end appears to provide an optimal binding region, allowing A3G to align in a catalytically favored 3′ → 5′ orientation to access the preferred C of a 5′-CCC motif.
We further suggest that there is a polarity of the C-terminal active sites at each end of the A3G oligomer so that they do not deaminate with equal rates (Fig. 7A, red circle (high rate) and blue circle (low rate)). There are three observations, in addition to previous data (15), that support an asymmetric catalytic orientation of A3G so that active sites facing the 5′-DNA end (Fig. 7A, red circle) catalyze C → U deaminations much more efficiently than those facing the 3′-DNA end (Fig. 7A, blue circle). First, there is a 30-nt region that is deaminated with extremely low efficiency (deamination dead zone) located at the 3′-end of linear ssDNA (Fig. 1, A and B). The 30-nt dead zone may provide an optimal space for A3G to encounter a CCC motif in an orientation allowing catalysis. Second, directionally biased deamination occurs on a partially dsDNA circle in the presence but not absence of salt, whereas directional deamination does not occur on an ssDNA circle either with or without salt (Fig. 2), implying that A3G needs to approach a CCC motif with a specific orientation. Third, there is a biased orientation in each 5′-CCC motif, where A3G prefers to deaminate the 3′-C residue (5′-CCC) much more than the 5′-C residue (5′-CCC) even under single turnover conditions (Fig. 3E). Therefore, we have depicted the “hot” active site facing the 5′-DNA end and aligned closest to the favored C for deamination in a 5′-CCC motif (Fig. 7A). Conversely, the “cold” active site is facing the 3′-DNA end and aligned closest to the barely deaminated C in a 5′-CCC motif (Fig. 7A). Although A3G has not been previously modeled with C-terminal domains external to an oligomer (26), there is evidence that a “monomeric” C-terminal deaminase domain of A3G is catalytically active (29, 30), which suggests that trans subunit complementation common to the cytidine deaminase family (43) is not required. We favor this model, because it seems to be the most probable asymmetric arrangement that can account for all aspects encompassing the biochemical characteristics of A3G 3′ → 5′ deamination polarity (Fig. 7A).
Random deaminations take place in the absence of salt. Concomitantly, A3G oligomers that form in the absence of salt (Fig. 6B) favor the formation of monomers in the presence of ssDNA (Fig. 6D and Table 1). A3G is illustrated as a monomer in the absence of salt (Fig. 7B), although A3G dimers may also form (Fig. 6D and Table 1). It is likely that the A3G monomers are free to diffuse randomly on the ssDNA substrate and deaminate CCC motifs essentially irrespective of target location, since there is not a strong conformational change indicative of DNA wrapping, in the absence of MgCl2 (Fig. 4D), which would otherwise limit resampling of undeaminated C targets (Fig. 7A).
For our study, the high salt state has provided mechanistic insights into the determinants for 3′ → 5′ deamination polarity. In vivo, there are likely to be multiple forms of A3G in higher and lower order oligomeric states. There is evidence that A3G exists in high molecular mass or intravirion A3G complex and low molecular mass states when associated with and when dissociated from RNA, respectively (7, 16, 26). Although A3G is incorporated into virions as an intravirion A3G complex, unable to deaminate (-)-cDNA, RNase H releases the inhibitory HIV RNA during reverse transcription to form the active low molecular mass form, characterized as monomers and/or dimers by gel filtration (16). However, the form that the low molecular mass takes when binding DNA has not been functionally characterized apart from our study, and it is likely that in vivo, the dimeric and larger oligomeric states formed with A3G, DNA, and salt (Fig. 6C) are relevant.
In vivo, the intrinsic polarity of A3G deamination makes a substantial contribution to local regions of HIV-1 mutational polarity (17), which suggests that higher order oligomers of A3G are acting on (-)-cDNA. Apart from deamination activity, A3G may also have the capacity for blocking reverse transcription. A recent in vitro study has shown that A3G can block reverse transcriptase DNA elongation by competitively binding RNA or ssDNA segments of the HIV-1 genome or (-)-cDNA with a higher affinity than reverse transcriptase (8), a finding supported by A3G inhibition of HIV-1 in resting CD4+ T cells (7). Our data suggest that a potentially key aspect of the mechanism inhibiting reverse transcriptase would involve the formation of higher oligomeric states of A3G. Higher oligomeric forms of A3G can presumably interact with many regions on the DNA, which is likely to inhibit access by other enzymes, including reverse transcriptase. Here the assembly of variably sized oligomeric forms offers two distinct ways that A3G can inhibit viral propagation when binding to (-)-cDNA, a catalytic mode creating biased “hot spots” for deamination (17) to ensure mutational inactivation of HIV and a noncatalytic mode involving higher ordered structures, which can block the action of reverse transcriptase (8).
Supplementary Material
Acknowledgments
We thank Drs. Phuong Pham, Jeffrey Bertram, and Peter Calabrese for insightful criticisms and discussions concerning this study.
This work was supported, in whole or in part, by National Institutes of Health Grants ESO13192 and R37GM21422 (to M. F. G.) and Grant GM79480. This work was also supported by American Cancer Society Grant RSG-03-047 (to D. A. E.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S6.
Footnotes
The abbreviations used are: ssDNA, single-stranded DNA; AFM, atomic force microscopy; dsDNA, double-stranded DNA; HIV, human immunodeficiency virus; nt, nucleotide(s).
References
- 1.Sheehy, A. M., Gaddis, N. C., Choi, J. D., and Malim, M. H. (2002) Nature 418 646-650 [DOI] [PubMed] [Google Scholar]
- 2.Zhang, H., Yang, B., Pomerantz, R. J., Zhang, C., Arunachalam, S. C., and Gao, L. (2003) Nature 424 94-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mangeat, B., Turelli, P., Caron, G., Friedli, M., Perrin, L., and Trono, D. (2003) Nature 424 99-103 [DOI] [PubMed] [Google Scholar]
- 4.Navarro, F., Bollman, B., Chen, H., Konig, R., Yu, Q., Chiles, K., and Landau, N. R. (2005) Virology 333 374-386 [DOI] [PubMed] [Google Scholar]
- 5.Hache, G., Liddament, M. T., and Harris, R. S. (2005) J. Biol. Chem. 280 10920-10924 [DOI] [PubMed] [Google Scholar]
- 6.Li, X. Y., Guo, F., Zhang, L., Kleiman, L., and Cen, S. (2007) J. Biol. Chem. 282 32065-32074 [DOI] [PubMed] [Google Scholar]
- 7.Chiu, Y. L., Soros, V. B., Kreisberg, J. F., Stopak, K., Yonemoto, W., and Greene, W. C. (2005) Nature 435 108-114 [DOI] [PubMed] [Google Scholar]
- 8.Iwatani, Y., Chan, D. S., Wang, F., Maynard, K. S., Sugiura, W., Gronenborn, A. M., Rouzina, I., Williams, M. C., Musier-Forsyth, K., and Levin, J. G. (2007) Nucleic Acids Res. 35 7096-7108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo, K., Wang, T., Liu, B., Tian, C., Xiao, Z., Kappes, J., and Yu, X. F. (2007) J. Virol. 81 7238-7248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo, F., Cen, S., Niu, M., Yang, Y., Gorelick, R. J., and Kleiman, L. (2007) J. Virol. 81 11322-11331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyagi, E., Opi, S., Takeuchi, H., Khan, M., Goila-Gaur, R., Kao, S., and Strebel, K. (2007) J. Virol. 81 13346-13353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schumacher, A. J., Hache, G., Macduff, D. A., Brown, W. L., and Harris, R. S. (2008) J. Virol. 82 2652-2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mbisa, J. L., Barr, R., Thomas, J. A., Vandegraaff, N., Dorweiler, I. J., Svarovskaia, E. S., Brown, W. L., Mansky, L. M., Gorelick, R. J., Harris, R. S., Engelman, A., and Pathak, V. K. (2007) J. Virol. 81 7099-7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiu, Y. L., Witkowska, H. E., Hall, S. C., Santiago, M., Soros, V. B., Esnault, C., Heidmann, T., and Greene, W. C. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 15588-15593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chelico, L., Pham, P., Calabrese, P., and Goodman, M. F. (2006) Nat. Struct. Mol. Biol. 13 392-399 [DOI] [PubMed] [Google Scholar]
- 16.Soros, V. B., Yonemoto, W., and Greene, W. C. (2007) PLoS Pathog. 3 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suspene, R., Rusniok, C., Vartanian, J. P., and Wain-Hobson, S. (2006) Nucleic Acids Res. 34 4677-4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu, Q., Konig, R., Pillai, S., Chiles, K., Kearney, M., Palmer, S., Richman, D., Coffin, J. M., and Landau, N. R. (2004) Nat. Struct. Mol. Biol. 11 435-442 [DOI] [PubMed] [Google Scholar]
- 19.Berg, O. G., Winter, R. B., and von Hippel, P. H. (1981) Biochemistry 20 6929-6948 [DOI] [PubMed] [Google Scholar]
- 20.von Hippel, P. H., and Berg, O. G. (1989) J. Biol. Chem. 264 675-678 [PubMed] [Google Scholar]
- 21.Halford, S. E., and Marko, J. F. (2004) Nucleic Acids Res. 32 3040-3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanford, N. P., Szczelkun, M. D., Marko, J. F., and Halford, S. E. (2000) EMBO J. 19 6546-6557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jack, W. E., Terry, B. J., and Modrich, P. (1982) Proc. Natl. Acad. Sci. U. S. A. 79 4010-4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terry, B. J., Jack, W. E., and Modrich, P. (1985) J. Biol. Chem. 260 13130-13137 [PubMed] [Google Scholar]
- 25.Pham, P., Chelico, L., and Goodman, M. F. (2007) DNA Repair (Amst.) 6 689-692 [DOI] [PubMed] [Google Scholar]
- 26.Wedekind, J. E., Gillilan, R., Janda, A., Krucinska, J., Salter, J. D., Bennett, R. P., Raina, J., and Smith, H. C. (2006) J. Biol. Chem. 281 38122-38126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prochnow, C., Bransteitter, R., Klein, M. G., Goodman, M. F., and Chen, X. S. (2007) Nature 445 447-451 [DOI] [PubMed] [Google Scholar]
- 28.Opi, S., Takeuchi, H., Kao, S., Khan, M. A., Miyagi, E., Goila-Gaur, R., Iwatani, Y., Levin, J. G., and Strebel, K. (2006) J. Virol. 80 4673-4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen, K. M., Martemyanova, N., Lu, Y., Shindo, K., Matsuo, H., and Harris, R. S. (2007) FEBS Lett. 581 4761-4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen, K. M., Harjes, E., Gross, P. J., Fahmy, A., Lu, Y., Shindo, K., Harris, R. S., and Matsuo, H. (2008) Nature 452 116-119 [DOI] [PubMed] [Google Scholar]
- 31.Iwatani, Y., Takeuchi, H., Strebel, K., and Levin, J. G. (2006) J. Virol. 80 5992-6002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bransteitter, R., Pham, P., Calabrese, P., and Goodman, M. F. (2004) J. Biol. Chem. 279 51612-51621 [DOI] [PubMed] [Google Scholar]
- 33.Ratcliff, G. C., and Erie, D. A. (2001) J. Am. Chem. Soc. 123 5632-5635 [DOI] [PubMed] [Google Scholar]
- 34.Yang, Y., Wang, H., and Erie, D. A. (2003) Methods 29 175-187 [DOI] [PubMed] [Google Scholar]
- 35.Creighton, S., Bloom, L. B., and Goodman, M. F. (1995) Methods Enzymol. 262 232-256 [DOI] [PubMed] [Google Scholar]
- 36.Goddard, N. L., Bonnet, G., Krichevsky, O., and Libchaber, A. (2000) Phys. Rev. Lett. 85 2400-2403 [DOI] [PubMed] [Google Scholar]
- 37.Bao, K. K., Wang, H., Miller, J. K., Erie, D. A., Skalka, A. M., and Wong, I. (2003) J. Biol. Chem. 278 1323-1327 [DOI] [PubMed] [Google Scholar]
- 38.Tessmer, I., Moore, T., Lloyd, R. G., Wilson, A., Erie, D. A., Allen, S., and Tendler, S. J. (2005) J. Mol. Biol. 350 254-262 [DOI] [PubMed] [Google Scholar]
- 39.Brar, S. S., Sacho, E. J., Tessmer, I., Croteau, D. L., Erie, D. A., and Diaz, M. (2008) DNA Repair (Amst.) 7 77-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xue, Y., Ratcliff, G. C., Wang, H., Davis-Searles, P. R., Gray, M. D., Erie, D. A., and Redinbo, M. R. (2002) Biochemistry 41 2901-2912 [DOI] [PubMed] [Google Scholar]
- 41.Schlacher, K., Leslie, K., Wyman, C., Woodgate, R., Cox, M. M., and Goodman, M. F. (2005) Mol. Cell 17 561-572 [DOI] [PubMed] [Google Scholar]
- 42.Verhoeven, E. E., Wyman, C., Moolenaar, G. F., and Goosen, N. (2002) EMBO J. 21 4196-4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie, K., Sowden, M. P., Dance, G. S., Torelli, A. T., Smith, H. C., and Wedekind, J. E. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 8114-8119 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.