Abstract
Exposing cells to conditions that modulate growth can impair endoplasmic reticulum (ER) protein folding, leading to ER stress and activation of the transcription factor, ATF6. ATF6 binds to ER stress response elements in target genes, inducing expression of proteins that enhance the ER protein folding capacity, which helps overcome the stress and foster survival. To examine the mechanism of ATF6-mediated survival in vivo, we developed a transgenic mouse model that expresses a novel conditionally activated form of ATF6. We previously showed that activating ATF6 protected the hearts of ATF6 transgenic mice from ER stresses. In the present study, transcript profiling identified modulatory calcineurin interacting protein-1 (MCIP1), also known as regulator of calcineurin 1 (RCAN1), as a novel ATF6-inducible gene that encodes a known regulator of calcineurin/nuclear factor of activated T cells (NFAT)-mediated growth and development in many tissues. The ability of ATF6 to induce RCAN1 in vivo was replicated in cultured cardiac myocytes, where adenoviral (AdV)-mediated overexpression of activated ATF6 induced the RCAN1 promoter, up-regulated RCAN1 mRNA, inhibited calcineurin phosphatase activity, and exerted a striking growth modulating effect that was inhibited by RCAN1-targeted small interfering RNA. These results demonstrate that RCAN1 is a novel ATF6 target gene that may coordinate growth and ER stress signaling pathways. By modulating growth, RCAN1 may reduce the need for ER protein folding, thus helping to overcome the stress and enhance survival. Moreover, these results suggest that RCAN1 may also be a novel integrator of growth and ER stress signaling in many other tissues that depend on calcineurin/NFAT signaling for optimal growth and development.
About 35% of all proteins, including secreted, cell surface and most organelle-targeted proteins are synthesized in the ER (1). Conditions that perturb ER calcium, protein glycosylation or redox status, impair nascent protein folding, triggering the ER stress response (ERSR),4 also known as the unfolded protein response (2, 3). The ERSR has central roles in all metazoan cells, and has been shown to be important in the differentiation and function of secretory cells, including B lymphocytes, β-cells of the pancreas and the liver. Moreover, ER stress has been associated with neurodegenerative diseases (4), as well as pathologies that are precipitated upon impaired oxygen and nutrient delivery (i.e. ischemia) resulting from cardiovascular disease. Ischemia has been shown to activate the ERSR in brain, kidney, liver, and heart, as well as in tumor tissue in vivo (5-9).
Initially, ER stress leads to remodeling of the ER protein biosynthetic machinery in attempts to overcome the stress. This initial phase of remodeling includes the transcriptional induction of genes that encode ER-targeted chaperones and folding enzymes that help proteins fold in the ER lumen. In addition, ER stress up-regulates genes encoding components of an ER-associated degradation system, which is responsible for the ubiquitin/proteasome-mediated degradation of terminally misfolded ER proteins. Accordingly, the initial phase of the ERSR is designed to resolve and/or overcome the stress, therefore supporting cell survival. However, if the stress is too severe to be overcome, prolonged activation of the ERSR leads to induction of various cell destruction pathways, including apoptosis (10).
The accumulation of misfolded proteins in the ER lumen is sensed by 3 ER transmembrane proteins, protein kinase R-like ER kinase (PERK), inositol-requiring protein-1 (IRE-1), and activating transcription factor 6 (ATF6), each of which facilitates signaling from the ER to the nucleus. In the early stages of ER stress, the PERK and IRE-1 pathways are oriented toward recovery from the stress, which enhance survival. However, upon prolonged ER stress, both of these branches of the ERSR activate cell death pathways.
ATF6 is a 670-amino acid ER trans-membrane protein that translocates to the Golgi during ER stress, where it is cleaved by site-1 and site-2 proteases (11). The N-terminal fragment of ATF6 that is generated on the cytosolic face of the Golgi translocates to the nucleus, where it binds to ERSEs and induces ERSR gene expression (12-14). In contrast to the PERK and IRE-1 branches, the ATF6 branch of the ERSR has rarely been associated with ER stress-induced apoptosis; thus, it is believed that, in general, ATF6-regulated genes foster resolution of the stress and, thus, cellular protection (15).
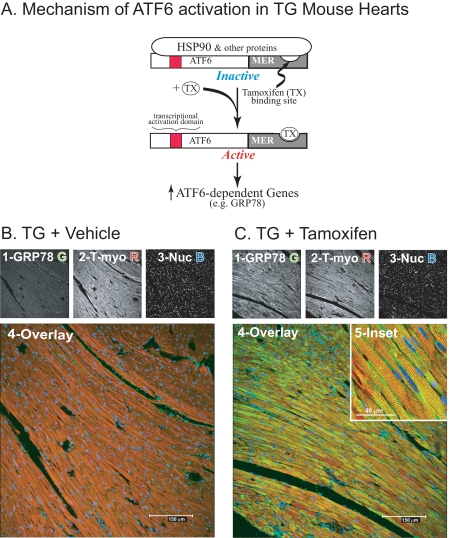
To study the mechanism of ATF6-mediated protection in vivo, we generated an animal model that would allow us to selectively activate ATF6 without activating other branches of the ERSR. This model features transgenic (TG) mice that express the N-terminal active fragment of ATF6 fused to the mutant mouse estrogen receptor (MER), ATF6-MER (16). The transgene is driven by the α-myosin heavy chain promoter; thus, ATF6-MER is expressed constitutively in the hearts of TG mice, beginning at birth. However, due to the binding of Hsp90 and other proteins to the MER portion of the ATF6-MER chimera, the transcriptional activation domain of ATF6 is masked, which disrupts ATF6-mediated gene induction (Fig. 1A, Inactive). Tamoxifen displaces MER-binding proteins, which unmasks the transcriptional activation domain of the N-terminal fragment of ATF6, rendering it able to induce ATF6-dependent genes, such as glucose-regulated protein 78 (GRP78), which encodes a cytoprotective, ER-targeted chaperone (Fig. 1A, Active). Accordingly, ATF6-MER TG mice are well suited for studying the effects of selectively activating the ATF6 branch of the ERSR, in vivo.
FIGURE 1.
Effect of tamoxifen on the ATF6-inducible gene, GRP78, in ATF6-MER transgenic mouse hearts. A, the mechanism of tamoxifen-mediated ATF6 activation and induction of ATF6-inducible ER stress response genes, such as GRP78, in ATF6-MER TG mice is shown. The transcriptional activation domain of ATF6, shown in red, is masked by proteins, such as Hsp90, which bind to the MER fragment in the absence of tamoxifen, rendering the ATF6-MER fusion protein inactive. Tamoxifen displaces these proteins, unmasking the transcriptional activation domain, conferring transcriptional activation to ATF6-MER. B and C, TG mice were treated with vehicle (panel B) or tamoxifen (panel C), as previously described (16), n = 3 mice per treatment, one heart/treatment is shown in this figure. Heart sections were stained for GRP78 protein (green) (1), tropomyosin (red) (2), or treated with TOPRO-3 to identify nuclei (blue) (3). Samples were viewed by laser scanning confocal fluorescence microscopy. Images 1-3 show each stain separately, image 4 shows an enlarged overlay of images 1-3. Image 5 in panel C was acquired from a different section and is shown at higher magnification. All immunofluorescence and confocal microscopy procedures have been previously described (6).
We previously showed that ATF6-MER TG mouse hearts exhibited tamoxifen-dependent protection from ischemia-mediated ER stress, suggesting that ATF6-regulated genes provide a pre-conditioning, protective effect. To begin to delineate the mechanism of ATF6-mediated protection, microarray analysis of ATF6-MER TG mouse hearts was carried out in the present study. The expression levels of 607 genes changed upon ATF6 activation, in vivo, with 381 genes exhibiting increases in expression that ranged from 2- to 120-fold. In addition to the induction of numerous well known ERSR genes, additional genes not previously characterized as ATF6-regulated were induced. One such gene encodes modulatory calcineurin-interacting protein-1 (MCIP1), which has recently been renamed regulator of calcineurin 1 (RCAN1) (17), a known modulator of the growth and development of many different tissues and cell types (18). The results of this study suggest that ATF6 might serve a growth-modulating role, which would reduce the need for protein synthesis in the cytosol and the ER, thus indirectly supporting recovery of the ER protein folding capacity during ER stress.
EXPERIMENTAL PROCEDURES
Animals
The transgenic mice used in this study have been described previously (16). Approximately 100 neonatal rats and 24 adult male mice were used in this study. All procedures involving animals were in accordance with the San Diego State University Institutional Animal Care and Use Committee.
Microarrays
RNA Preparation and Hybridization—Non-transgenic (NTG) and TG mice were treated with vehicle or tamoxifen, n = 3 mice per treatment group, as described previously (16). RNA was extracted from mouse heart ventricles, as described below. Each mouse heart mRNA sample was analyzed on a single microarray chip. Gene expression analysis was performed in accordance with the Affymetrix GeneChip® Expression Analysis Technical Manual (Affymetrix, Inc., Santa Clara, CA). cRNA was fragmented for target preparation, and then hybridized onto Affymetrix mouse 430 2.0 whole genome arrays (Affymetrix, Inc., part 900496), which allow for analysis of roughly 40,000 transcripts. Array chips were scanned on an Affymetrix GeneChip Scanner 3000 using Affymetrix GeneChip Operating Software (GCOS) version 1.1.1.
Statistics and Data Analysis—Statistical analyses were performed using the BioConductor packages in R. The “.CEL” files were input directly into R, and mRNA expression values were obtained using the RMA algorithm (1.10.0) (19). Statistical pairwise comparisons between treatment groups were carried out in R, using the local pooled error method (20). Genes that exhibited significant changes in expression (i.e. p ≤ 0.01) were included for further study. Genes that were differentially expressed in vehicle versus tamoxifen-treated TG mouse hearts by 2-fold, or more, were selected for further study. Because tamoxifen may affect gene expression independently of its ability to activate ATF6-MER, genes that were differentially expressed in vehicle versus tamoxifen-treated NTG mouse hearts were excluded from further study.
Real Time Quantitative-PCR
Real time quantitative-PCR was performed as previously described (16). The following rat primers were used: ANP, 5′-GCGAAGATCAAGCTGCTTCG and 3′-CTCTGGGCTCCAATCCTGTC; BNP, 5′-GTGCTGCCCCAGATGATTCT and 3′-CAGCGGCGACAGATTAAGGA; Dnajb11, 5′-AGGCCCCACTGAACACACAT and 3′-GGGCCTCTTTGAGCTCGTT; ERp72, 5′-TGACATCACCAACGACCGAT and 3′-TCTGTTGCCACCCTCAAACTT; GRP78, 5′-CACGTCCAACCCGGAGAA and 3′-ATTCCAAGTGCGTCCGATG; Hyou1, 5′-TGCGGGATGCTGTCATTTAC and 3′-CCACCTCCCTTGTGAACTCC; RCAN1, 5′-CGGAGGCCAGAGTACACACC and 3′-GGTCAGTGTGCCTGTTCAGCT; and XBP1, 5′-CAGCAAGTGGTGGATTTGGAA and 3′-ATCCCCAAGCGTGTCCTTAAC.
The following mouse primers were used: ANP, 5′-TTGTGGTGTGTCACGCAGCT and 3′-TGTTCACCACGCCACAGTG; BNP, 5′-AAGTCGGAGGAAATGGCCC and 3′-TTGTGAGGCCTTGGTCCTTC; Dnajb11, 5′-CGCAGAACCTGAGCACCTTC and 3′-CAGTCCCGATGAGGTACAGCA; ERp72, 5′-GCCAATGACATCACCAACGA and 3′-GCCAATGACATCACCAACGA and 3′-TCTCTGTTGCCACCCTCAAAC; GRP78, 5′-CCTGTTCCGCTCTACCATGAA and 3′-TGGAATTCGAGTAGATCCGCC; Hyou1, 5′-GTGATAGTGCAGCCGGCAT and 3′-AACGGAGCGTAGCCTTTGG; RCAN1, 5′-CCTCCAGCTTGGGCTTGAC and 3′-GGCAGACGCTTAACGAACGA; and XBP1, 5′-CTCACGGCCTTGTGGTTGA and 3′-TCCATTCCCAAGCGTGTTC.
Activated and Dominant Negative ATF6 Adenovirus
Adenovirus encoding only green fluorescent protein (AdV-Con), green fluorescent protein and constitutively active ATF6 (AdV-ATF6), or green fluorescent protein and dominant negative ATF6 (AdV-DN-ATF6), which encode proteins of the expected physical characteristics, were prepared as previously described (6).
Cultured Cardiac Myocytes
Primary neonatal rat ventricular myocyte cultures (NRVMC) were prepared and maintained in culture, as previously described (6).
RCAN1-luciferase and RCAN1-M-luciferase Constructs
A region of the 5′-flanking sequence preceding exon 4 of the human RCAN1 gene from -984 to +30 was isolated by PCR and cloned into a pGL2 luciferase reporter vector (Clontech). The nucleotides from -329 to -311 in the human RCAN1 gene are CCATT(N)9CAAAG, which exhibits about 73% homology to a consensus ERSE, CCAAT(N)9CCACG (21). The same region of the mouse RCAN1 promoter has the following ERSE-like sequence CCACC(N)9CAGAG, which also exhibits about 73% homology to a consensus ERSE. Using PCR-based mutagenesis, the region from -329 to -311 of human RCAN1-luciferase was changed to AACGG(N)9CCCTT, creating RCAN1-M-luciferase, which has a mutated ERSE.
Reporter Enzyme Assay
Reporter enzyme assays for luciferase and β-galactosidase were carried out as previously described (6).
Analysis of Cultured Cardiac Myocyte Size
Cultured cardiac myocyte area was determined as previously described (22). At least 250 cells were analyzed per condition, and the mean ± S.E. was calculated for the surface area.
Small Interfering RNA Treatment of Cultured Cardiac Myocytes
NRVMC were plated on 12-well plates at 1 m cells per well, then transfected with a combination of 3 different Stealth™ small interfering (si) RNA oligoribonucleotides targeted against rat RCAN1 (Invitrogen, catalog number 130003) or a validated Stealth RNA interference negative control (Invitrogen, catalog number 12935300). Each well was transfected with 10 pmol of each Stealth siRNA using TransMessenger™ Transfection Reagent (Qiagen, Valencia, CA). After ∼20 h, cells were infected with control adenovirus, or an adenovirus encoding ATF6 for 5 h in minimal media. Twenty-four h after infection, cells were then treated ± phenylephrine (50 μm) in minimal media for 48 h. Transcript levels were then assessed using qRT-PCR, and cell size was analyzed using ImageJ; both methods are described above.
Leucine Incorporation
Radiolabeled incorporation of leucine into cultured cardiac myocyte protein was performed as previously described (23).
Statistical Analyses
Data are reported as mean ± S.E. and analyzed via one-way analysis of variance with Newman-Keuls post hoc analysis using SPSS version 11.0. Unless otherwise stated in the figure legends, *, #, or § = p ≤ 0.05 different from all other values.
Calcineurin Activity Assay
A calcineurin phosphatase activity assay was performed essentially as previously described (24). Calcineurin activity was measured using a calcineurin cellular assay kit plus (BIOMOL, Plymouth Meeting, PA, catalog number AK-804) according to the manufacturer's protocol. Briefly, NRVMC were collected in 400 μl of lysis buffer (50 mm Tris, pH 7.5, 0.1 mm EDTA, 0.1 mm EGTA, 1 mm dithiothreitol, 0.2% Nonidet P-40). Free phosphate was removed by passing the lysates through a desalting column before assaying. Calcineurin phosphatase activity was measured spectrophotometrically by detecting free phosphate released from the synthetic RII phosphopeptide.
RESULTS
Induction of the ATF6 Target Gene, GRP78, in ATF6-MER Mice—In initial studies of tamoxifen-dependent ATF6 activation in vivo, the expression of the well known ATF6-target gene, GRP78, was examined in ATF6-MER TG mouse hearts by confocal immunofluorescence microscopy. Sections from vehicle-treated TG mouse hearts showed that GRP78 expression in cardiomyocytes was relatively low under these conditions (Fig. 1B, panels 1 and 4, green). Similar results were found in sections from tamoxifen-treated NTG mouse hearts (not shown). In contrast, sections from tamoxifen-treated TG mouse hearts exhibited robust GRP78 expression (Fig. 1C, panel 1), which was localized to most of the cardiomyocytes, as indicated by co-staining for GRP78 and tropomyosin (Fig. 1C, panel 4). For the most part, GRP78 exhibited a perinuclear staining pattern in cardiomyocytes, which is typical of ER-localized proteins (Fig. 1C, panel 5, green). In addition, GRP78 was expressed in other regions of cardiomyoyctes, exhibiting a sarcomeric staining pattern, which has been previously observed (6). These results demonstrated that tamoxifen effectively up-regulated this ATF6-target gene in the majority of the myocytes in ATF6-MER TG mouse hearts, supporting the utility of this model for analyses of the effects of ATF6 on the induction of other genes, in vivo.
Microarray Analyses of ATF6-inducible Genes—To identify additional ATF6-regulated genes, RNA samples from the hearts of NTG and TG mice treated with vehicle or tamoxifen were subjected to whole genome microarray analysis. To ensure the inclusion of genes that were induced as a result of tamoxifen-mediated activation of ATF6, genes that were differentially expressed in untreated NTG versus tamoxifen-treated NTG mice were excluded. After these exclusions, 607 genes exhibited differential expression of at least 2-fold (p ≤ 0.01), with 381, or about 63% of the genes exhibiting increased expression (see GEO accession number GSE8322 and supplemental Table 1). The differentially expressed genes were analyzed using the Gene Ontology (GO) classification system, which organizes genes on the basis of the molecular and biological function (25). Using this approach, as well as other techniques, 23 known and 14 putative ERSR genes were identified (Table 1), the latter of which were defined as genes with published characteristics similar to known ERSR genes, but not previously shown to be ATF6-inducible. Moreover, 36 of the 37 known and putative ERSR genes were up-regulated by levels ranging from 2- to 46-fold.
TABLE 1.
Known and putative ERSR genes induced by ATF6 activation in the heart
All differentially expressed genes are sorted by -fold change. Also shown is the gene symbol for each gene. More information about each gene, including common names, can be obtained by entering the gene symbol in Mouse Genome Informatics (MGI 3.54). The NCBI Reference Sequence ID, or accession number for each gene is shown. All genes were sorted into Gene Ontology Biological Process categories. Several of the categories were found to be of interest and the numbers in the notes column refer to genes assigned to the following groupings of Gene Ontology Biological Process categories. The 23 known and 14 putative ERSR genes from the microarray analysis that were induced by activated ATF6 in ATF6-MER TG mouse hearts are shown. The columns display the Mouse Genome Informatics (MGI 3.54) gene symbol, the mouse or human alias or protein name, the NCBI Reference Sequence ID or accession number and the -fold increase observed in the microarray. X in the “Valid” column indicates that the microarray results for these genes have been validated by RT-qPCR using the same RNA that was used in the microarray.
| MGI symbol | Alias or protein names | NCBI RefSeq | -Fold up | Valid | |
|---|---|---|---|---|---|
| Known ERSR genes | |||||
| 1 | Derl3 | Degraded in ER protein 3 | NM_024440 | 18.50 | X |
| 2 | Pdia4 | Cai | NM_009787 | 16.37 | |
| 3 | DNAJC3 | P58; P58IPK | NM_008929 | 12.75 | X |
| 4 | DNAJB11 | ERdj3; HEDJ; HSP40 | NM_026400 | 12.39 | X |
| 5 | Socs3 | STAT Induced STAT inhibitor | NM_007707 | 9.68 | |
| 6 | Asns | Asparagine synthetase | NM_012055 | 9.26 | |
| 7 | P4hB | PDI associated 1; ERp59; Thbp | NM_011032 | 9.17 | |
| 8 | Armet | Arginine-rich, mutated early stage tumors | NM_029103 | 8.21 | X |
| 9 | Hsp90b1 | Tral; GRP94 | NM_011631 | 7.07 | X |
| 10 | Trib3 | Nip kinase; Ifld2 | NM_144554 | 6.66 | |
| 11 | Edem1 | EDEM | NM_138677 | 5.36 | X |
| 12 | Calr | Calreticulin | NM_007591 | 5.29 | X |
| 13 | Atf4 | ATF4 | NM_009716 | 4.93 | |
| 14 | Pdia3 | Protein-disulfide isomerase 3, GRP8; ERp61 | NM_007952 | 4.72 | |
| 15 | Hspa5 | GRP78 | NM_022310 | 4.71 | X |
| 16 | Syvn1 | Synoviolin 1 | NM_028769 | 4.60 | |
| 17 | Eif2Ak3 | PERK | NM_010121 | 4.18 | |
| 18 | Pin1 | DOD; UBL5; Rotamase | NM_023371 | 4.12 | |
| 19 | Hyou1 | GRP170; ORP150 | NM_021395 | 3.86 | X |
| 20 | Ugcgl1 | UDP-glucose ceramide glucosyltransferase-like 1 | NM_198899 | 3.70 | |
| 21 | XBP1 | X-box binding protein 1 | NM_13842 | 2.95 | X |
| 22 | Herpud1 | HERP; Mif1; SUP | NM_022331 | 2.43 | X |
| 23 | Herpud2 | HERPUD family member 2 | NM_020586 | 0.23 | |
| Putative ERSR genes | |||||
| 24 | Ptx3 | Pentaxin-related gene | NM_008987 | 46.31 | X |
| 25 | IL6 | Interleukin-6 | NM_031168 | 38.45 | |
| 26 | Sdf2l1 | Stromal cell derived factor 1 | NM_022324 | 17.41 | |
| 27 | GADD45g | Growth arrest DNA damage-inducible protein 45 | NM_011817 | 10.52 | |
| 28 | Rtn4 | Reticulon 4 | NM_024226 | 9.28 | |
| 29 | Azin1 | Ornithine decarboxylase; Oazin | NM_018745 | 5.96 | |
| 30 | Gas5 | Growth arrest specific 5 | AK206770 | 5.90 | |
| 31 | Snord22 | Small nucleolar RNA, C/D box 22 | AK051045 | 4.48 | |
| 32 | Txnrd1 | Thioredoxin reductase | NM_015762 | 4.32 | |
| 33 | RCAN1 | Regulator of calcineurin 1 | NM_019466 | 4.29 | X |
| 34 | Serpinh1 | Serpin peptidase inhibitor, HSP47 | NM_009825 | 3.10 | |
| 35 | Gsk3β | Glycogen synthase kinase-3β | NM_019827 | 3.00 | |
| 36 | Sec11a | Signal peptidase complex | NM_019951 | 2.53 | |
| 37 | H47 | VCP-interacting membrane protein; Vimp | NM_024439 | 2.53 |
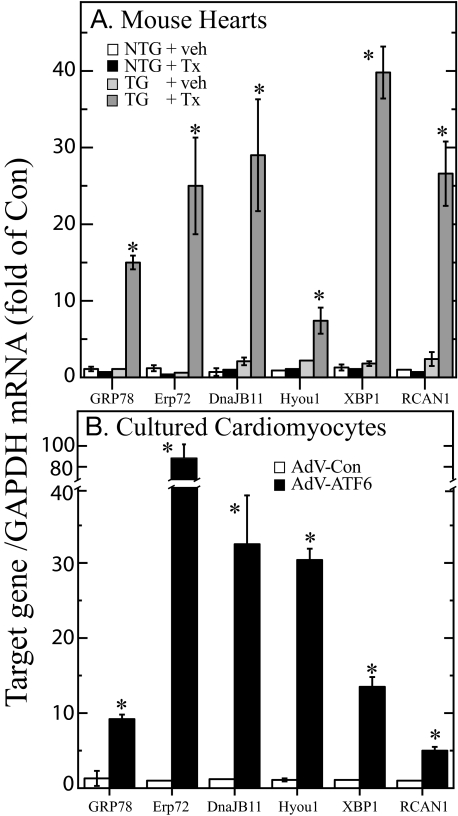
Real time quantitative PCR (RT-qPCR) was used to validate the microarray results for 13 of the known and putative ERSR genes. All 13 genes examined were found to be up-regulated in RNA prepared from tamoxifen-treated TG mouse hearts, but not in RNA from any of the other treatment groups (see “X” in Table 1). An example of this validation for 6 of the genes shows induction of 5 known ERSR genes (GRP78, Erp72, DnajB11, Hyou1, and XBP1) and one putative ERSR gene (RCAN1) (Fig. 2A). We also determined whether these 6 genes were induced upon overexpression of activated ATF6 in primary NRVMC. All 6 of the genes examined were up-regulated in cells infected with an adenovirus that encodes activated ATF6 (AdV-ATF6) (Fig. 2B). These findings provided further validation of the microarray results, and demonstrated that these 6 genes could be induced in vivo and in cultured cells using two different approaches for overexpressing activated ATF6. This also established a cell culture model for examining the functional characteristics of known and putative ATF6-inducible ERSR genes.
FIGURE 2.
Validation of microarray results using real time quantitative PCR. A, the RNA samples that were used for the microarray analysis were subjected to RT-qPCR to examine the levels of the mRNAs encoded by the GRP78, Erp72, DnaJB11, Hyou1, XBP1, and RCAN1 genes. Shown are the mean ± S.E. for each target gene (n = 3 mouse hearts per treatment). Veh, vehicle; Tx, tamoxifen. B, NRVMC were infected with either AdV-Con or AdV-ATF6 (n = 3 cultures per treatment). 48 h after infection, cultures were extracted and the RNA was subjected to RT-qPCR to examine the levels of mRNA for the same target genes described in panel A. Shown are the mean ± S.E. for each target gene (n = 3 cultures per treatment). * = p ≤ 0.05 different from all other values for each target gene.
RCAN1 Is an ATF6-inducible Gene—One of the putative ERSR genes, RCAN1 (entry 33 in Table 1), is of particular interest, because it regulates calcium/calcineurin A-mediated growth and development in numerous tissue types (18). When activated by calcium, the cytosolic phosphatase, calcineurin A, dephosphorylates NFAT, which translocates to the nucleus and activates genes that contribute to growth (26). RCAN1 binds to and inhibits calcineurin A, thus modulating NFAT-mediated growth under certain conditions (27, 28). Moreover, RCAN1 is induced by NFAT, suggesting that it exerts a feedback inhibition of NFAT-mediated gene induction. Because of its wide range of effects in many different cell types, we used cultured cardiac myocytes as a cell model to determine whether RCAN1 is an ATF6-inducible ERSR gene and whether it modulates growth when induced by ATF6.
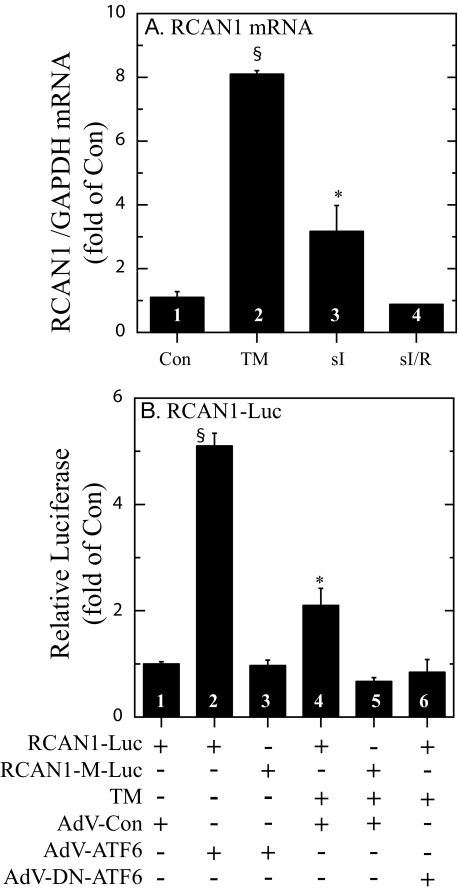
When NRVMC were treated with the prototypical ER stressor, tunicamycin, which inhibits ER protein glycosylation, RCAN1 mRNA increased by about 8-fold (Fig. 3A, bars 1 and 2), similar to previous findings for GRP78 gene induction in NRVMC (6). Simulated ischemia (sI), which activates ER stress and induces GRP78 gene expression in the heart and in NRVMC (6, 16, 29), increased RCAN1 mRNA by about 3-fold (Fig. 3A, bar 3). However, when sI was followed by simulated reperfusion (sI/R), RCAN1 mRNA levels returned to control values (Fig. 3A, bar 4), which was also previously observed for GRP78 (6). These results demonstrated that RCAN1 gene expression is induced by ER stress in NRVMC.
FIGURE 3.
Effect of various treatments on RCAN1 mRNA and RCAN1 promoter activity in cultured cardiac myocytes. A, NRVMC were treated ± tunicamycin (TM, 10 μg/ml, bar 2) for 16 h, or subjected to sI for 24 h (bar 3), or sI for 20 h followed by simulated reperfusion for 20 h (sI/R, bar 4); n = 3 cultures for each treatment. sI and sI/R were carried out as previously described (6). RNA was isolated from culture extracts and subjected to RT-qPCR to determine the levels of mRNAs encoded by the GAPDH and RCAN1 genes. The primers used for RCAN1 were designed to amplify a region of exon 4. Shown are the mean values of RCAN1/GAPDH mRNA, expressed as the -fold of control (bar 1) ± S.E. for each treatment (n = 3 cultures per treatment). B, the human RCAN1 gene promoter (-984 to +30 of the region located to the 5′ of exon 4) and a version harboring a mutation in a putative ERSR element located at -329 to -311 (RCAN1-M), were cloned into a luciferase expression construct. NRVMC were transfected with RCAN1-luciferase or RCAN1-M-luciferase and CMV-β-galactosidase and then infected with AdV-Con, AdV-ATF6, or AdV-DN-ATF6, as previously described (6). 24 h after infection, cultures were treated ± TM, as shown and as described in panel A. 16 h later, cultures were extracted and luciferase and β-galactosidase reporter enzyme activities were determined, as previously described (6). AdV-DN-ATF6 alone had no effect on RCAN1-luciferase, or RCAN1-M-luciferase activation (not shown). Shown are the mean relative luciferase (luciferase/β-galactosidase), expressed as the -fold of control (bar 1) ± S.E. for each treatment (n = 3 cultures per treatment). * and §, p ≤ 0.05 different from all other values.
Because ATF6 is a transcriptional inducer of ERSR genes, the effect of overexpressing the constitutively active, N-terminal fragment of ATF6 on the RCAN1 gene promoter was examined. Infecting NRVMC with recombinant adenovirus encoding activated ATF6 (AdV-ATF6) induced the RCAN1 promoter by about 5-fold, compared with control adenovirus (AdV-Con) (Fig. 3B, bars 1 and 2). However, when a putative ERSE, located between positions -329 to -311 of the RCAN1 promoter, was mutated, promoter activation by ATF6 was lost (Fig. 3B, bar 3). Tunicamycin also induced the native RCAN1 promoter by about 2-fold; this induction was also lost when the putative ERSE was mutated, or when cells were infected with recombinant adenovirus encoding a dominant-negative form of ATF6 (AdV-DN-ATF6) (Fig. 3B, bars 4-6). Thus, the RCAN1 promoter is activated in cultured cardiac myocytes by ATF6 or tunicamycin through an ERSE that is similar to that observed in previously characterized ERSR genes, including GRP78 (21).
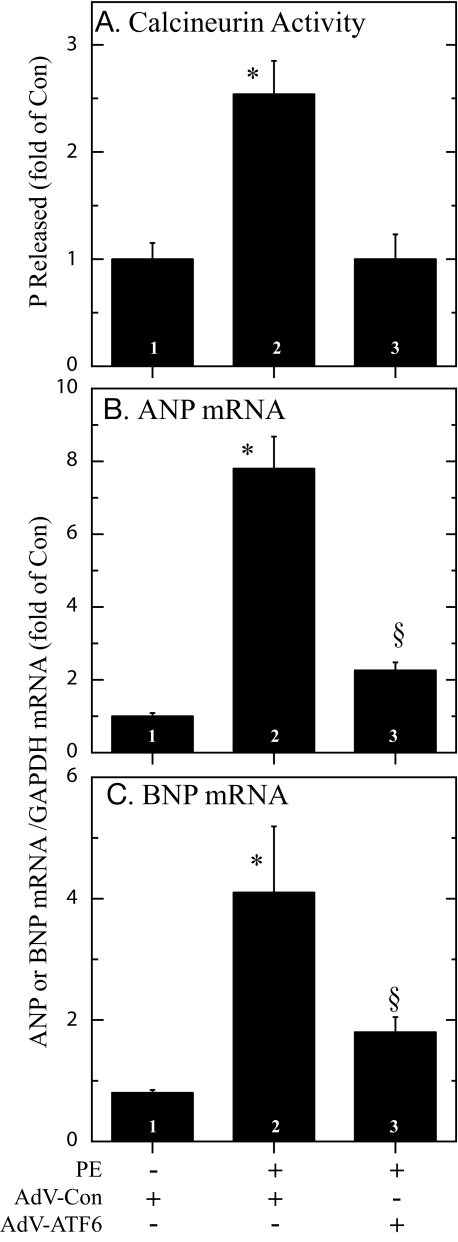
Effect of ATF6 on Calcineurin Activity and Genetic Markers of Cardiac Myocyte Growth—The α1-adrenergic receptor agonist, phenylephrine (PE), activates calcineurin/NFAT signaling in NRVMC (30), but is not known to activate ER stress (31). Accordingly, we examined the effects of ATF6 on the activation of calcineurin by PE. PE conferred an approximate 3-fold increase in calcineurin activation, as expected; however, calcineurin activation was completely blocked by AdV-ATF6 (Fig. 4A). We also examined the effects of activated ATF6 on two well characterized, NFAT-inducible genes, atrial natriuretic peptide (ANP) and B-type natriuretic peptide (BNP). PE treatment of AdV-Con-infected cells increased ANP and BNP and RCAN1 mRNA by 8- and 4-fold, respectively (Fig. 4, B and C, bars 1 and 2). In contrast, AdV-ATF6 decreased the effects of PE on ANP and BNP gene expression (Fig. 4, B and C, bar 3). These results are consistent with the hypothesis that ATF6 induces RCAN1, which in turn, inhibits calcineurin phosphatase activity, as well as calcineurin/NFAT-mediated ANP and BNP gene induction.
FIGURE 4.
Effect of phenylephrine on calcineurin phosphatase activity, and on ANP and BNP induction. NRVMC were infected with AdV-Con or AdV-ATF6, and 24 h later, cultures were treated ± PE (50 μm) for 48 h. Cultures were then examined for calcineurin activity (panel A), ANP mRNA (panel B), or BNP mRNA (panel C). Shown are the mean values of calcineurin phosphatase activity, or for ANP or BNP mRNA/GAPDH mRNA expressed as the -fold of control (bar 1) ± S.E. for each treatment (n = 3 cultures per treatment). * and §, p ≤ 0.05 different from all other values.
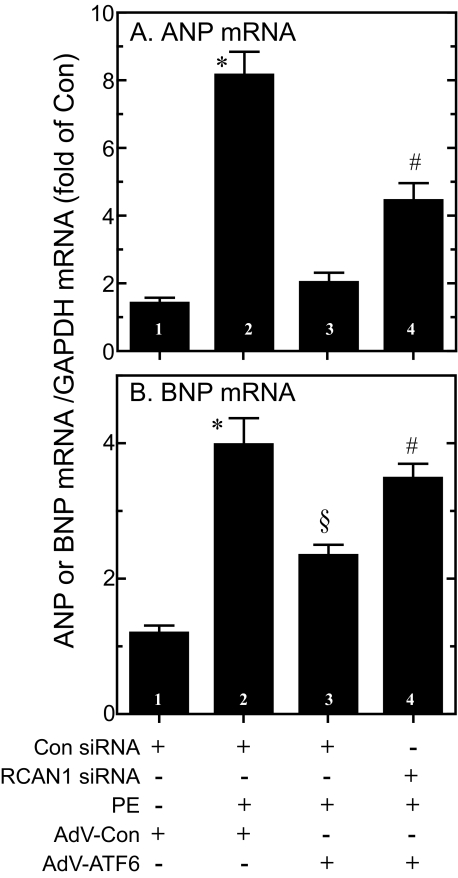
To determine whether RCAN1 was responsible for some of the effects of ATF6, NRVMC were transfected with RCAN1 siRNA using a method that results in nearly 100% transfection efficiency (32). In preliminary experiments, it was shown that RCAN1 mRNA/GAPDH mRNA decreased significantly (p < 0.01) from 100 ± 7 to 39 ± 8% (average ± S.E.) in cells treated with control siRNA, or RCAN1 siRNA, respectively (not shown). As expected, the control siRNA had no effect on PE-mediated ANP and BNP gene induction, or the ability of ATF6 to modulate this induction (Fig. 5, A and B, bars 1-3). However, the modulating effect of ATF6 on ANP and BNP gene induction was attenuated significantly, albeit, not completely, in cells transfected with RCAN1 siRNA (Fig. 5, A and B, bar 4). These results are consistent with the hypothesis that ATF6-mediated modulation of ANP and BNP gene induction is mediated partly by RCAN1.
FIGURE 5.
Effect of RCAN1 siRNA and activated ATF6 on ANP and BNP gene induction by phenylephrine. NRVMC were transfected with either control siRNA (Con siRNA) or RCAN1 siRNA, and then infected with AdV-Con or AdV-ATF6. 24 h later, cultures were treated ± phenylephrine (50 μm) for 24 h. Cultures were then extracted and ANP (panel A), or BNP (panel B) mRNA levels were determined by RT-qPCR. Shown are mean values of each target gene/GAPDH mRNA, expressed as the -fold of control (bar 1) ± S.E. for each treatment (n = 3 cultures per treatment). * and §, p ≤ 0.05 different from all other values.
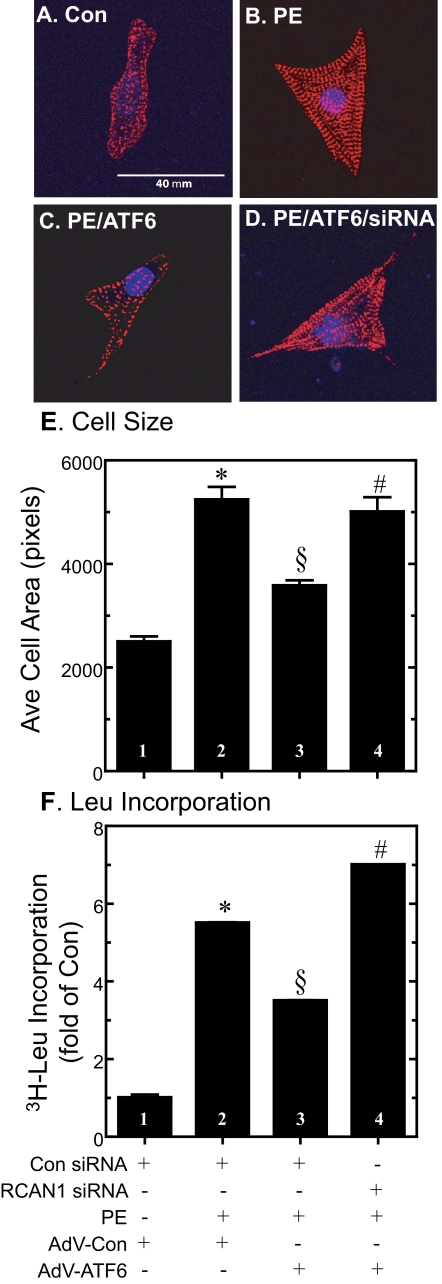
Role of RCAN1 in ATF6-mediated Inhibition of Cell Growth—NRVMC do not divide in culture, but respond to growth factors by increasing in size, exhibiting hypertrophic growth that mimics cardiac myocytes in the adult heart. Growth factors, such as PE, increase cell size and protein synthesis, both of which mimic hypertrophic growth, which is mediated in part by calcineurin/NFAT signaling (33, 34). Accordingly, the effects of ATF6 on myocyte size and protein synthesis were examined in NRVMC. As expected, in AdV-Con-infected cells, PE increased the myocyte area, which serves as a measure of hypertrophic growth (Fig. 6, A and B, and E, bars 1 and 2). PE also increased the incorporation of [3H]leucine into cellular protein, which serves as an indirect estimate of protein synthesis (Fig. 6F, bars 1 and 2). The effects of PE on cell size (Fig. 6, C and E, bar 3) and [3H]leucine incorporation (Fig. 6F, bar 3) were reduced by AdV-ATF6. However, RCAN1 siRNA significantly modulated these effects of ATF6, suggesting that RCAN1 was at least partly responsible for ATF6-mediated growth modulation in cultured cardiac myocytes. These results support the hypothesis that RCAN1 may play a role in coordinating ER stress and growth pathways, and that by modulating cell growth, RCAN1 reduces the protein folding load in the ER.
FIGURE 6.
Effect of activated ATF6, RCAN1 siRNA, and phenylephrine on cardiomyocyte area and [3H]leucine incorporation into protein. A-D, NRVMC were transfected with control siRNA or RCAN1 siRNA, infected with AdV-Con or AdV-ATF6, and treated ± PE, as described in the legend to Fig. 5. Cultures were then fixed and immunostained for α-actinin protein (red) and the nuclei were identified using TOPRO-3 (blue), as previously described (6). Fluorescence confocal micrographs show samples of cells after exposure to each treatment. Bar in panel A = 40 μm. E, cell images obtained from micrographs similar to those shown in panels A-C were quantified densitometrically for area, which provides an estimate of cell size. n = at least 250 cells counted for each treatment; each treatment was carried out on 3 different cultures. Shown is the mean of the relative cell area ± S.E., normalized to the control, which was set at 100% (n = 3 cultures per treatment). *, §, and #, p ≤ 0.05 different from all other values. F, cells were cultured and treated as described in panels A-D, except they were incubated with [3H]leucine and the amount of radiolabel incorporated into trichloroacetic acid-precipitable protein was examined, as described under “Experimental Procedures.” Shown is the mean ± S.E. of labeled protein, normalized to the control. n = 3 cultures/treatment/experiment. *, §, and #, p ≤ 0.05 different from all other values.
DISCUSSION
In this study we examined the effects of selectively activating the ATF6 branch of the ERSR on gene expression, in vivo. Whole genome microarray analysis showed that 381 genes were up-regulated, whereas 236 genes were down-regulated in response to activated ATF6 in the heart (supplemental Table S1). Sorting of the differentially expressed genes using the GO biological process classification system showed that of the 60 genes assigned to the GO category entitled “regulation of transcription, DNA-dependent,” 42 were up-regulated in ATF6-MER TG mouse hearts (notes number 1, supplemental Table S1), consistent with the known roles of ATF6 as a transcriptional inducer. Of the 24 genes assigned to the categories of “protein folding,” “response to unfolded protein,” and “ER overload response,” 19 were up-regulated, consistent with the central role of ATF6 as an inducer of chaperones. Of 19 genes assigned to the category “small GTPase-mediated signal transduction,” many of which are involved in vesicle-mediated transport between the ER and Golgi, 16 were up-regulated, consistent with the expansion of the ER and Golgi networks that takes place during ER stress. Moreover, all 12 genes assigned to the GO categories “negative regulators of apoptosis” and “cell death” were up-regulated, consistent with the decreased apoptosis observed previously in ATF6 TG mouse hearts (16). Somewhat surprising, however, was the finding that 4 of 5 genes in the negative regulation of cell growth category were up-regulated, whereas 35 of 45 genes in the “fatty acid” and “glucose metabolism” categories were down-regulated. These findings suggest that ATF6 activation affects the transcriptome in ways expected to modulate growth. Consistent with this hypothesis was the up-regulation of RCAN1 (supplemental Table S1, entry 148), which is known to inhibit cell growth.
Because of the widespread importance of NFAT as a regulator of growth and development in essentially all tissue types (18), we focused on RCAN1 as one of the genes through which ATF6 might exert its growth-regulating effects. We found that activated ATF6 induced RCAN1 gene expression in cultured cardiac myocytes in a manner that required a putative ERSE located in the regulatory region of the RCAN1 gene. Moreover, ATF6 modulated NFAT-mediated gene expression and cell growth, both of which were shown to be partially dependent upon RCAN1. Taken together, these results show for the first time that ATF6 exerts growth modulating effects and the ATF6-inducible, ERSR gene, RCAN1, is responsible for a significant portion of these effects.
Although it had not previously been shown to be an ATF6-inducible gene, earlier studies of RCAN1 are consistent with the findings of the present study. For example, the RCAN1 gene was previously identified in Chinese hamster fibroblasts and named Adapt78 after its homology to GRP78 and its ability to allow fibroblasts to adapt to oxidative stress (35). Adapt78 was subsequently found to be induced by tunicamcyin (36), and overexpression of adapt78 suppressed fibroblast growth (37). Later, adapt78 was found to bind to and inhibit the calcium-activated phosphatase, calcineurin A (28), and, therefore, was renamed MCIP1 (38). Most recently, this protein has been renamed regulator of calcineurin 1, or RCAN1 (17).
By inhibiting calcineurin, RCAN1 can modulate NFAT activation and gene expression (26). NFAT is an important mediator of many processes in cardiac myocytes, including the hypertrophic growth gene program activated in response to increases in intracellular calcium (26, 27, 39). Interestingly, the regulatory region of the RCAN1 gene examined in the present study has numerous NFAT binding sites that are known to mediate induction of RCAN1 gene expression in response to activators of hypertrophic growth, including PE (38). Thus, RCAN1 has been postulated to be a negative feedback modulator of cardiomyocyte growth in this context (27). Consistent with this hypothesis was the finding that overexpression of RCAN1 in transgenic mice inhibited cardiac hypertrophy induced by activated calcineurin A, as well as hypertrophy induced by β-adrenergic receptor agonists or exercise (40). Moreover, RCAN1-overexpressing TG mice subjected to in vivo myocardial infarction exhibited less cardiac hypertrophy, reduced heart failure, and enhanced lifespan, supporting growth modulating and protective roles for RCAN1, in vivo (41). The results of the current study show that ATF6-induced RCAN1 gene expression and decreased myocyte growth, and that knockdown of RCAN1 mRNA using siRNA attenuated this ATF6-mediated reduction. This supports the possibility that RCAN1 contributes to ATF6-MER TG mouse heart protection by modulating hypertrophic growth, which reduces the load on the ER protein folding machinery. In this way, RCAN1 would also contribute to resolving the ER stress, which would avert the consequence of continued ER stress, apoptosis (29, 42, 43).
Recent studies suggest that the mechanism of RCAN1 action is more complex than originally thought. In one study, the hearts of RCAN1 knock-out mice exhibited increased hypertrophy in response to overexpression of activated calcineurin A (44). In that same study, a paradoxical reduction of hypertrophy was observed, in RCAN1 knock-out mice subjected to pressure overload, indicating that RCAN1 can facilitate, or suppress calcineurin A signaling, depending on the growth stimulus. In another study, disrupting RCAN1 gene expression in the mouse heart impaired hypertrophy in response to overload, neuroendocrine stimulation, or exercise, suggesting that RCAN1 serves permissive roles for calcineurin/NFAT signaling in the heart (45). Thus, RCAN1 may function as either an inhibitor, or a facilitator of calcineurin/NFAT signaling, depending on the context. In support of such dual functions is a report describing a mathematical model that takes into account all of the published data on RCAN1 in the heart, and delineates physiologically feasible circumstances under which RCAN1 might actually switch from being a facilitator to an inhibitor of calcineurin/NFAT signaling (46).
In summary, the present study provides evidence that RCAN1 is an ATF6-inducible, ERSR gene, in vivo, which may contribute to the protective effects of ATF6 by modulating NFAT-regulated growth and reducing the protein synthesis load in the ER during ER stress. Moreover, whereas this study was carried out in the cardiac context, the results have potential importance in many other tissues that depend on NFAT for proper development and growth.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants HL-075573 and HL-085577. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
Footnotes
The abbreviations used are: ERSR, endoplasmic reticulum stress response; ER, endoplasmic reticulum; AdV, adenovirus; AdV-ATF6, adenovirus encoding ATF6; AdV-DN-ATF6, adenovirus encoding dominant-negative ATF6; ANP, atrial natriuretic peptide; ATF6, activating transcription factor 6; BNP, brain natriuretic peptide; ERSE, endoplasmic reticulum stress response element; GO, Gene Ontology; GRP78, glucose-regulated protein 78; Hsp90, heat shock protein 90; IRE-1, inositol-requiring protein-1; RCAN1, regulator of calcineurin 1; MER, mutant mouse estrogen receptor; NFAT, nuclear factor of activated T cells; NRVMC, neonatal rat ventricular myocyte cultures; NTG, non-transgenic; PERK, protein kinase R (PKR)-like ER kinase; PE, phenylephrine; sI, simulated ischemia; sI/R, simulated ischemia/reperfusion; siRNA, small interfering RNA; RT-qPCR, real time-quantitative PCR; TG, transgenic; MCIP1, modulatory calcineurin interacting protein-1.
References
- 1.Blobel, G. (2000) Biosci. Rep. 20 303-344 [DOI] [PubMed] [Google Scholar]
- 2.Lee, A. (2001) Trends Biochem. Sci. 26 504-510 [DOI] [PubMed] [Google Scholar]
- 3.Glembotski, C. C. (2007) Circ. Res. 101 975-984 [DOI] [PubMed] [Google Scholar]
- 4.Wu, J., and Kaufman, R. J. (2006) Cell Death Differ. 13 374-384 [DOI] [PubMed] [Google Scholar]
- 5.Bailly-Maitre, B., Fondevila, C., Kaldas, F., Droin, N., Luciano, F., Ricci, J. E., Croxton, R., Krajewska, M., Zapata, J. M., Kupiec-Weglinski, J. W., Farmer, D., and Reed, J. C. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 2809-2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thuerauf, D. J., Marcinko, M., Gude, N., Rubio, M., Sussman, M. A., and Glembotski, C. C. (2006) Circ. Res. 99 275-282 [DOI] [PubMed] [Google Scholar]
- 7.Paschen, W., Aufenberg, C., Hotop, S., and Mengesdorf, T. (2003) J. Cereb. Blood Flow Metab. 23 449-461 [DOI] [PubMed] [Google Scholar]
- 8.Romero-Ramirez, L., Cao, H., Nelson, D., Hammond, E., Lee, A. H., Yoshida, H., Mori, K., Glimcher, L. H., Denko, N. C., Giaccia, A. J., Le, Q. T., and Koong, A. C. (2004) Cancer Res. 64 5943-5947 [DOI] [PubMed] [Google Scholar]
- 9.Rzymski, T., and Harris, A. L. (2007) Clin. Cancer Res. 13 2537-2540 [DOI] [PubMed] [Google Scholar]
- 10.Xu, C., Bailly-Maitre, B., and Reed, J. C. (2005) J. Clin. Investig. 115 2656-2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshida, H., Haze, K., Yanagi, H., Yura, T., and Mori, K. (1998) J. Biol. Chem. 273 33741-33749 [DOI] [PubMed] [Google Scholar]
- 12.Ye, J., Rawson, R. B., Komuro, R., Chen, X., Dave, U. P., Prywes, R., Brown, M. S., and Goldstein, J. L. (2000) Mol. Cell 6 1355-1364 [DOI] [PubMed] [Google Scholar]
- 13.Haze, K., Yoshida, H., Yanagi, H., Yura, T., and Mori, K. (1999) Mol. Biol. Cell 10 3787-3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, Y., Shen, J., Arenzana, N., Tirasophon, W., Kaufman, R. J., and Prywes, R. (2000) J. Biol. Chem. 275 27013-27020 [DOI] [PubMed] [Google Scholar]
- 15.Szegezdi, E., Logue, S. E., Gorman, A. M., and Samali, A. (2006) EMBO Rep. 7 880-885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martindale, J. J., Fernandez, R., Thuerauf, D., Whittaker, R., Gude, N., Sussman, M. A., and Glembotski, C. C. (2006) Circ. Res. 98 1186-1193 [DOI] [PubMed] [Google Scholar]
- 17.Davies, K. J., Ermak, G., Rothermel, B. A., Pritchard, M., Heitman, J., Ahnn, J., Henrique-Silva, F., Crawford, D., Canaider, S., Strippoli, P., Carinci, P., Min, K. T., Fox, D. S., Cunningham, K. W., Bassel-Duby, R., Olson, E. N., Zhang, Z., Williams, R. S., Gerber, H. P., Perez-Riba, M., Seo, H., Cao, X., Klee, C. B., Redondo, J. M., Maltais, L. J., Bruford, E. A., Povey, S., Molkentin, J. D., McKeon, F. D., Duh, E. J., Crabtree, G. R., Cyert, M. S., de la Luna, S., and Estivill, X. (2007) FASEB J. 21 3023-3028 [DOI] [PubMed] [Google Scholar]
- 18.Wu, H., Peisley, A., Graef, I. A., and Crabtree, G. R. (2007) Trends Cell Biol. 17 251-260 [DOI] [PubMed] [Google Scholar]
- 19.Irizarry, R. A., Hobbs, B., Collin, F., Beazer-Barclay, Y. D., Antonellis, K. J., Scherf, U., and Speed, T. P. (2003) Biostatistics 4 249-264 [DOI] [PubMed] [Google Scholar]
- 20.Jain, N., Thatte, J., Braciale, T., Ley, K., O'Connell, M., and Lee, J. K. (2003) Bioinformatics 19 1945-1951 [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto, K., Yoshida, H., Kokame, K., Kaufman, R. J., and Mori, K. (2004) J. Biochem. (Tokyo) 136 343-350 [DOI] [PubMed] [Google Scholar]
- 22.Zechner, D., Thuerauf, D. J., Hanford, D. S., McDonough, P. M., and Glembotski, C. C. (1997) J. Cell Biol. 139 115-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadoshima, J., Jahn, L., Takahashi, T., Kulik, T. J., and Izumo, S. (1992) J. Biol. Chem. 267 10551-10560 [PubMed] [Google Scholar]
- 24.Taigen, T., De Windt, L. J., Lim, H. W., and Molkentin, J. D. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 1196-1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., Harris, M. A., Hill, D. P., Issel-Tarver, L., Kasarskis, A., Lewis, S., Matese, J. C., Richardson, J. E., Ringwald, M., Rubin, G. M., and Sherlock, G. (2000) Nat. Genet. 25 25-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vega, R. B., Bassel-Duby, R., and Olson, E. N. (2003) J. Biol. Chem. 278 36981-36984 [DOI] [PubMed] [Google Scholar]
- 27.Rothermel, B. A., Vega, R. B., and Williams, R. S. (2003) Trends Cardiovasc. Med. 13 15-21 [DOI] [PubMed] [Google Scholar]
- 28.Fuentes, J. J., Genesca, L., Kingsbury, T. J., Cunningham, K. W., Perez-Riba, M., Estivill, X., and de la Luna, S. (2000) Hum. Mol. Genet. 9 1681-1690 [DOI] [PubMed] [Google Scholar]
- 29.Terai, K., Hiramoto, Y., Masaki, M., Sugiyama, S., Kuroda, T., Hori, M., Kawase, I., and Hirota, H. (2005) Mol. Cell. Biol. 25 9554-9575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pu, W. T., Ma, Q., and Izumo, S. (2003) Circ. Res. 92 725-731 [DOI] [PubMed] [Google Scholar]
- 31.Thuerauf, D. J., Hoover, H., Meller, J., Hernandez, J., Su, L., Andrews, C., Dillmann, W. H., McDonough, P. M., and Glembotski, C. C. (2001) J. Biol. Chem. 276 48309-48317 [DOI] [PubMed] [Google Scholar]
- 32.Ardehali, H., O'Rourke, B., and Marban, E. (2005) Circ. Res. 97 740-742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanford, D. S., Thuerauf, D. J., Murray, S. F., and Glembotski, C. C. (1994) J. Biol. Chem. 269 26227-26233 [PubMed] [Google Scholar]
- 34.Molkentin, J. D., and Dorn, G. W. (2001) Annu. Rev. Physiol. 63 391-426 [DOI] [PubMed] [Google Scholar]
- 35.Crawford, D. R., Leahy, K. P., Abramova, N., Lan, L., Wang, Y., and Davies, K. J. (1997) Arch. Biochem. Biophys. 342 6-12 [DOI] [PubMed] [Google Scholar]
- 36.Leahy, K. P., Davies, K. J., Dull, M., Kort, J. J., Lawrence, K. W., and Crawford, D. R. (1999) Arch. Biochem. Biophys. 368 67-74 [DOI] [PubMed] [Google Scholar]
- 37.Leahy, K. P., and Crawford, D. R. (2000) Arch. Biochem. Biophys. 379 221-228 [DOI] [PubMed] [Google Scholar]
- 38.Yang, J., Rothermel, B., Vega, R. B., Frey, N., McKinsey, T. A., Olson, E. N., Bassel-Duby, R., and Williams, R. S. (2000) Circ. Res. 87 E61-E68 [DOI] [PubMed] [Google Scholar]
- 39.Molkentin, J. D., Lu, J. R., Antos, C. L., Markham, B., Richardson, J., Robbins, J., Grant, S. R., and Olson, E. N. (1998) Cell 93 215-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rothermel, B. A., McKinsey, T. A., Vega, R. B., Nicol, R. L., Mammen, P., Yang, J., Antos, C. L., Shelton, J. M., Bassel-Duby, R., Olson, E. N., and Williams, R. S. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 3328-3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Rooij, E., Doevendans, P. A., Crijns, H. J., Heeneman, S., Lips, D. J., van Bilsen, M., Williams, R. S., Olson, E. N., Bassel-Duby, R., Rothermel, B. A., and De Windt, L. J. (2004) Circ. Res. 94 e18-e26 [DOI] [PubMed] [Google Scholar]
- 42.Nickson, P., Toth, A., and Erhardt, P. (2007) Cardiovasc. Res. 73 48-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szegezdi, E., Duffy, A., O'Mahoney, M. E., Logue, S. E., Mylotte, L. A., O'Brien, T., and Samali, A. (2006) Biochem. Biophys. Res. Commun. 349 1406-1411 [DOI] [PubMed] [Google Scholar]
- 44.Vega, R. B., Rothermel, B. A., Weinheimer, C. J., Kovacs, A., Naseem, R. H., Bassel-Duby, R., Williams, R. S., and Olson, E. N. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 669-674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanna, B., Brandt, E. B., Kaiser, R. A., Pfluger, P., Witt, S. A., Kimball, T. R., van Rooij, E., De Windt, L. J., Rothenberg, M. E., Tschop, M. H., Benoit, S. C., and Molkentin, J. D. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 7327-7332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shin, S. Y., Choo, S. M., Kim, D., Baek, S. J., Wolkenhauer, O., and Cho, K. H. (2006) FEBS Lett. 580 5965-5973 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.