Abstract
G protein-coupled receptor (GPCR) kinases (GRKs) phosphorylate activated heptahelical receptors, leading to their uncoupling from G proteins. Here we report six crystal structures of rhodopsin kinase (GRK1), revealing not only three distinct nucleotide-binding states of a GRK but also two key structural elements believed to be involved in the recognition of activated GPCRs. The first is the C-terminal extension of the kinase domain, which was observed in all nucleotide-bound GRK1 structures. The second is residues 5-30 of the N terminus, observed in one of the GRK1·(Mg2+)2·ATP structures. The N terminus was also clearly phosphorylated, leading to the identification of two novel phosphorylation sites by mass spectral analysis. Co-localization of the N terminus and the C-terminal extension near the hinge of the kinase domain suggests that activated GPCRs stimulate kinase activity by binding to this region to facilitate full closure of the kinase domain.
Rhodopsin (Rho)2 is the G protein-coupled receptor (GPCR) responsible for visual signal transduction in rod cells (1). Phosphorylation of light-activated Rho (Rho*) by rhodopsin kinase, also known as GPCR kinase 1 (GRK1), initiates a series of events that rapidly quenches signaling by the receptor (2, 3). This rapid desensitization is essential for scotopic vision and, in concert with the regeneration of visual pigment, protects rod cells from photodegeneration and permits rapid adaptation to changes in illumination. Phosphorylation of Rho* at multiple sites by GRK1 is also believed to contribute to the reproducibility of the single photon visual response (4). In humans, inactivating mutations in GRK1 are found in patients with Oguchi disease (5, 6), a stationary form of night blindness characterized by a substantial delay in dark recovery after photobleaching. In transgenic mice lacking GRK1, the single photon response is of a higher amplitude and longer duration, and the rod cells undergo photodegeneration via apoptosis, even under dim light conditions (7).
GRK1 is the founding member of the GRK kinase family, which includes β-adrenergic receptor kinase 1 (GRK2) and GRK3-7 (8, 9). All of the GRKs share an intimately associated “RH-kinase core” wherein a Ser/Thr kinase domain is inserted into a loop of a domain homologous to those found in RGS (regulator of G protein-signaling) proteins (the RH domain). The kinase domain is closely related to those of protein kinases A (PKA), G, and C (the AGC kinases) and includes the “C-terminal extension” characteristic of this kinase family. The C-terminal extension is a key interaction site for regulatory proteins and in some cases contributes residues to the active and polypeptide-binding sites (10). In all GRKs, the RH-kinase core is followed by a membrane-targeting domain (e.g. a farnesylation site in GRK1 and a pleckstrin homology domain that binds Gβγ in GRK2 and 3).
A number of observations have led to the hypothesis that Rho* engages an allosteric “docking site” on GRK1 (11, 12). GRK1 phosphorylates multiple sites in the C-terminal tail of rhodopsin, a region that is freely accessible in both the inactive and active states of the receptor. Despite this, GRK1 phosphorylates only Rho*. A single Rho* can induce transphosphorylation of hundreds of Rho molecules (13, 14), consistent with a rapidly diffusing Rho*·GRK1 complex whose catalytic site remains accessible to substrates. Furthermore, the interaction of GRK1 with Rho* stimulates kinase activity against peptide substrates by up to 160-fold, as measured by comparing the catalytic efficiency of GRK1 in the presence or absence of C-terminally truncated Rho* (11, 12).
The second and third cytoplasmic loops of Rho appear most important for binding GRK1 (12, 15), and site-directed mutants of residues in these loops inhibit GRK1 activation (16). Conversely, how GRK1 recognizes an activated receptor is far less understood. The N-terminal 30 amino acids of GRK1 appear to be critical for receptor recognition, because the in vitro binding of Ca2+-recoverin to this region (17, 18) or the binding of an antibody directed against GRK1 residues 17-34 (19) inhibits receptor phosphorylation yet has no effect on GRK1-mediated phosphorylation of peptide substrates.
The mechanism of GRK activation is also not understood. Phosphorylation at two or three sites (called the activation loop, turn motif, and hydrophobic motif) (20) is required in many AGC kinases to stabilize their kinase domains. Of these sites, GRK1 retains only the turn motif (Ser488 and Thr489), a major autophosphorylation site (21). Furthermore, the active site structures of GRK2 (22-24) and GRK6 (25) appeared to be well ordered despite the absence of activation loop and hydrophobic motif sites. However, the small and large lobes of GRK2 and GRK6 adopted comparatively “open” conformations with respect to the active, transition state complex of PKA (26). Thus, one mechanism by which GPCRs could activate GRKs would be to induce closure of the kinase domain and thereby align the catalytic machinery of the large lobe with the ATP-binding site of the small lobe. However, the binding of nucleotides may also induce kinase domain closure, as it does in PKA and protein kinase B (PKB). Because we have thus far been unable to compare apo and nucleotide-bound structures of either GRK2 or GRK6, it is not possible to determine whether nucleotide binding is sufficient to drive GRK kinase domain closure.
Here we report six different crystal forms of a soluble form of GRK1, representing three distinct nucleotide ligand states: (Mg2+)2·ATP, (Mg2+)2·ADP, and ligand-free. Critical regions of the GRK1 kinase C-terminal extension are ordered in the nucleotide-bound structures. Nucleotide binding induces a conformational change in the kinase domain, but one that still fails to fully align the catalytic machinery of the large lobe with the nucleotide-binding site. In one GRK1·(Mg2+)2·ATP structure, the extreme N terminus of the kinase is also observed. It is phosphorylated at several previously unrecognized sites and packs against the RH domain in close proximity to the hinge and the C-terminal extension of the kinase domain. Based on these structures, we propose a model for how GRK1 might interact with Rho* and how this binding in turn activates the kinase.
EXPERIMENTAL PROCEDURES
Materials—cDNA encoding bovine GRK1 in pRK5 was a gift from Dr. R. Lefkowitz (Duke University). Urea-stripped rod outer segments (ROS) were prepared as previously described (27). Dodecylmaltoside was purchased from Dojindo. The pTrec2 vector for recoverin expression was purchased from the American Type Culture Collection.
Expression, Purification, and N-terminal Sequencing of GRK1535-His6—Cloning and mutagenesis of GRK1535-His6 is described in the supplemental materials. Recombinant baculoviruses were generated using the Bac-to-Bac baculovirus expression system (Invitrogen). For protein expression, 30 ml of baculovirus was added per liter of High Five insect cells. The cells were harvested ∼40 h after viral addition. All of the purification steps were performed at 4 °C. Thawed cell pellets were resuspended in ∼100 ml of cold lysis buffer (Buffer A) containing 20 mm HEPES, pH 7.5, 300 mm NaCl, 10 mm β-mercaptoethanol, and fresh EDTA-free protease inhibitor mixture (Roche Applied Science). The cells were lysed using a C3 Avestin homogenizer (10,000 p.s.i.) and then spun in a Beckman Ti-45 rotor at 45,000 rpm for 45 min. The supernatant was filtered and diluted to 5 mg/ml final protein concentration with Buffer A. The diluted supernatant was loaded onto a 10-ml Ni2+-nitrilotriacetic acid drip column (Qiagen) pre-equilibrated with Buffer A. After washing with 10 column volumes of Buffer A and 20 column volumes of Buffer A supplemented with 20 mm imidazole, pH 8.0, the bound protein was eluted in 2-3-ml fractions with Buffer A supplemented with 150 mm imidazole, pH 8.0. Fractions containing GRK1535-His6 were diluted 6-fold in 20 mm HEPES, pH 7.5, 1 mm dithiothreitol, filtered, and then loaded onto an 8 ml Source 15 S column (Amersham Biosciences) and eluted in 0.6-ml fractions by an 80-ml gradient from 50 to 300 mm NaCl (supplemental Fig. S8). The resulting peak fractions were pooled as: peak 1 (Pool A), peaks 2 and 3 (Pool B), peak 4 (Pool C), and peak 5 (Pool D). Pool D, which eluted with the 1 m bump, appeared aggregated and was not further analyzed. Pooled peak samples were concentrated and flash-frozen in liquid nitrogen as 50-μl pellets. N-terminal sequencing of the various GRK1 pools was performed by Edman degradation using a protein sequencer (Model 494HT) from Applied Biosystems (Foster City, CA).
Crystallography—Diffraction maxima from the six crystal forms of GRK1 (supplemental methods and supplemental Fig. S11) were collected at the 19ID and 23ID-D beam lines at the Advanced Photon Source (Argonne National Laboratory) on ADSC Q315 and MAR300 CCD detectors, respectively. The crystals were cooled in a cryostream to 100 K. The data were indexed, integrated, and reduced using the HKL2000 software package (28). For crystal form I, the initial phase problem was solved by molecular replacement with PHASER in the CCP4 suite (29) using GRK6 (2ACX) as a search model. For other data sets, either crystal forms I or IV were used as search models. Coordinates were refined using REFMAC5 (30, 31), alternating with rounds of manual model building using the program O (32). NCS restraints were applied when appropriate. After Rfree converged, all of the reflections were used in the last few rounds of refinement. Stereochemistry of the refined models was validated with PROCHECK (33). The data collection and refinement statistics are summarized in supplemental Table S1, and the atomic coordinates and structure factors were deposited at the Protein Data Bank with accession codes 3C4W, 3C4X, 3C4Y, 3C4Z, 3C50, and 3C51 for crystal forms I-VI, respectively.
Purification of Wild-type GRK1 and Mutants—The High Five cell pellet was lysed similarly to GRK1535-His6. After ultracentrifugation, the membrane fraction was resuspended in Buffer A and frozen in liquid nitrogen. The soluble fraction was first purified on a Ni2+-nitrilotriacetic acid drip column at 4 °C. Fractions containing GRK1 fusion protein were pooled and dialyzed overnight against Buffer A in the presence of 2-3% (w/w) TEV protease at 4 °C. Following digestion, GRK1 no longer bound Ni2+ and thus was purified from the uncut His6-tagged fusion protein. The final purification was carried out as described above on an 8-ml Source 15S column (Amersham Biosciences). For purification of membrane-bound GRK1, the thawed membrane resuspension was stirred in an ice bath for 1 h in the presence of 1% (v/v) Triton X-100 (Amresco). After ultracentrifugation, the detergent-solubilized protein (supernatant) was purified similarly to the soluble fraction using buffers supplemented with 0.02% (w/v) dodecylmaltoside. The soluble and membrane-bound proteins also were purified as mixtures from homogenized cells.
Mass Spectrometric Analysis—Following reduction and S-alkylation, sequencing grade modified trypsin (Promega) was used for the overnight digestion of GRK1535-His6 (Pool A, preincubated with 4 mm ATP, pH 7.5, and 2 mm MgCl2) and native bovine GRK1 (in-gel) at 37 °C. MonoTip (GL Sciences Inc.) was used to enrich the phosphopeptides according to the manufacturer's protocol. Liquid chromatography-tandem mass spectrometry analysis of the tryptic digests was performed using a linear ion trap mass spectrometer (model LTQ) from Thermo-Finnigan (San Jose, CA) coupled with an Ettan MDLC system (GE Healthcare). The phosphopeptides were acquired in a positive ion mode and by an automatic data-dependent neutral loss scan method. That is, when a specific neutral loss (-49 Da and -32.7 Da for the doubly and triply charged ions) on fragment ions (MS2) was detected, MS3 was automatically triggered. The obtained data were submitted to Bioworks (Thermo Scientific) by searching the phosphorylation on Ser, Thr, and Tyr. Phosphorylation sites were confirmed by tandem MS2 and/or MS3.
Steady State Kinetics—Assay mixtures (100 μl) contained 1-40 μm urea-stripped ROS, 20 mm Bis-Tris propane, pH 7.5, 2 mm MgCl2, and 8-18 nm GRK1. The reactions were initiated by the addition of 0.1 mm [γ-32P]ATP mix (100-1000 cpm/pmol; Amersham Biosciences) and illumination at 30 °C. Two measurements were made for each concentration of urea-stripped ROS in two or three independent assays. The reactions were quenched after 10 min by the addition of 10% trichloroacetic acid (Sigma), and the samples were processed as described previously (27). Similar assays for GRK1535-His6 were carried out using 18 nm Pools A and B (10 min) and 65 nm Pool C (20 min). Soluble and membrane fractions of GRK1 were either fractionated by ultracentrifugation or purified as mixtures in detergent, each having displayed similar kinetics in these assays. Protein amounts were normalized by SDS-PAGE and A280 measurements (>95% homogenous). The data were analyzed by nonlinear analyses and curve-fitting programs of GraphPad Prism version 4.0a for Macintosh.
RESULTS
Crystal Structure of GRK1—A soluble variant of GRK1 (GRK1535-His6) was engineered based on the resolved elements of the GRK6 crystal structure (25) and was purified from baculovirus-infected insect cells. GRK1535-His6 phosphorylated Rho* with a 6-fold higher Km (12 μm) and a 2-fold lower Vmax than wild-type GRK1, possibly because of the loss of its C-terminal farnesylation site (34, 35) (Table 1). Six crystal forms of GRK1 representing (Mg2+)2·ATP, (Mg2+)2·ADP, and apo ligand states were obtained (supplemental Table S1 and supplemental Fig. S11). In all crystal forms, GRK1 uses a conserved surface on the RH domain to form a dimer interface (supplemental Fig. S1). A total of 11 unique GRK1 structures were determined, because all but one crystal form (IV) had two subunits in the asymmetric unit.
TABLE 1.
Kinetic analysis of GRK1
| GRK1535-His6 | Kma | Vmaxa |
|---|---|---|
| μm | nmol of Pi/min/mg | |
| Pool A | 12 ± 1.5 | 1100 ± 50 |
| Pool B | 5.5 ± 1.0 | 16 ± 0.8 |
| Pool C | inactiveb | inactiveb |
| GRK1 wild type | 2.1 ± 0.4 | 2300 ± 110 |
| Amino-terminal mutants | ||
| S5A | 5.1 ± 1.0 | 1600 ± 70 |
| S5D | NDc | ND |
| T8A | 1.5 ± 0.3 | 1200 ± 80 |
| T8E | 4.8 ± 1.5 | 1700 ± 130 |
| T8D | ND | ND |
| Dimer interface mutants | ||
| D164A | 3.8 ± 0.7 | 2300 ± 160 |
| L166K | 6.7 ± 0.4 | 2400 ± 60 |
| W531A | ND | ND |
| D164A/L166K | 3.1 ± 0.6 | 1100 ± 40 |
| D164A/W531A | ND | ND |
| L166K/W531A | ND | ND |
The results are listed as the means ± S.D. for two to three independent experiments, with two measurements for each concentration of ROS (1-40 μm).
These steady state kinetic parameters could not be estimated because of poor Rho* phosphorylation. However, the initial rate of Pool C was estimated to be ~20-fold lower than Pool A under similar assay conditions.
ND, not determined. These mutants either did not express or were unstable.
As in GRK2 and GRK6, the RH-kinase core of GRK1 features bipartite interactions between the RH and kinase domains (Fig. 1a and supplemental Fig. S2). The largest interface is formed between the α0, α9, and α10 helices of the RH domain and the β1, β2-β3, and αC-β4 loops of the small lobe of the kinase domain. A smaller interface is formed between the α4-α5 loop of the RH domain and the αJ helix, the first element of the kinase C-terminal extension (supplemental Fig. S2). The structure of this latter interface is maintained in all the crystal forms except in one chain of apo-GRK1, where it is disrupted as a consequence of a distinct kinase domain conformation (supplemental Table S2 and supplemental Fig. S3). Otherwise, the RH-kinase core of the nine nucleotide-bound structures of GRK1 are similar (0.37-0.85 Å root mean square deviation for 305 Cα atoms), with the regions of greatest conformational variability being the α6-α7 loops of the RH domain and portions of the kinase C-terminal extension, in agreement with the relatively high mobility of these regions (supplemental Fig. S4).
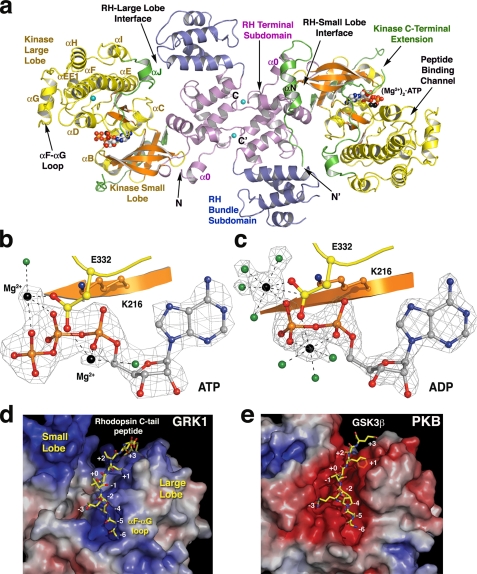
FIGURE 1.
Overview of GRK1 and its active site. a, GRK1535-His6 crystallized as a homodimer using a conserved interface of the RH domain in all the crystal forms. Shown is the most complete structure, that of crystal form I. The RH terminal subdomain is colored magenta (helices α0-3 and α8-11), and the bundle subdomain (helices α4-α7) is slate blue. The small lobe of the kinase domain (yellow) is composed of six β-strands (orange) and two α-helices (αB and αC), whereas the large lobe is primarily α-helical. The ligand (Mg2+)2·ATP is drawn as spheres. Magnesium atoms are colored black, carbons are white, nitrogens are blue, oxygens are red, phosphates are orange, and chloride ions are cyan. The extreme N-terminal region and the C-terminal extension of the kinase domain are green. b, substrate complex of GRK1. Shown is a σA-weighted |Fo| - |Fc| omit map contoured at 4 σ, wherein ATP, Mg2+, and associated waters (green) were excluded from refinement (crystal form I and chain B). Lys216 (β1 sheet, orange carbons) coordinates the α- and β-phosphates. Glu332 (yellow carbons) coordinates both Mg2+ atoms. c, product complex of GRK1. Shown is a σA-weighted |Fo| - |Fc| omit map contoured at 5 σ, wherein ADP, Mg2+, and associated waters were excluded from refinement (crystal form IV). d, the peptide-binding channel of GRK1. The molecular surface of GRK1 is colored by its electrostatic potential from -7 (red, acidic) to +7 (blue, basic) kT/e-. The channel has a strikingly basic character, explaining why GRK1 prefers acidic substrates (48, 49) and how it can phosphorylate multiple closely spaced Ser and Thr residues at the C terminus of Rho*. The channel is also wider in GRK1 than in nucleotide-bound PKB (e), reflecting the more open conformation of the GRK1 kinase domain of GRK1. As a result, the phosphoacceptor oxygen of the modeled peptide is >4 Å from the γ-phosphate of ATP, which is too far for covalent chemistry to occur. A model of residues 332-345 from the C terminus of Rho*, is shown as a stick model docked to the large lobe with Ser338 in position to be phosphorylated (position “+0”). Residues in the αF-αG loop of the large lobe appear to obstruct the N-terminal end of the peptide-binding site. e, the GSK3β peptide bound to PKB. The PKB kinase domain (Protein Data Bank code 1O6L) is in its closed conformation. The channel is markedly acidic, in line with a preference for basic substrates.
Nucleotide Binding and Conformational Change—The GRK1 kinase domain is comprised of a “small lobe” (residues 181-268) and a “large lobe” (residues 269-454), with the active site situated in a cleft formed between them. Unlike in prior GRK structures, co-crystallized nucleotides are well ordered in the GRK1 active site (Fig. 1, b and c, and supplemental Fig. S5a). Both ATP and ADP bind GRK1 concomitantly with two Mg2+ ions. In the ATP complex, the first metal ion coordinates the β and γ phosphates, and the second, less well-resolved metal ion coordinates the α phosphate. In the ADP complex, the first metal ion coordinates the β phosphate, and the second metal ion coordinates the α and β phosphates. In the ADP complex, each metal exhibits full octahedral coordination. Thus, the active site of the (Mg2+)2·ATP complex of GRK1 resembles that of the PKA·(Mn2+)2·ATP complex (36), whereas the ADP complex resembles the Aurora-A·(Mg2+)2·ADP complex (37). By analogy to structures of PKA and PKB, the peptide-binding channel of GRK1 lies on the surface of the large lobe adjacent to the nucleotide-binding site and is basic in character (Fig. 1, d and e).
The phosphate-binding loop (P-loop) of the small lobe is formed by the Gly-rich β1-β2 turn and directly interacts with the triphosphate tail of ATP and helps stabilize the phosphorylation transition state (38). In GRK1, the backbone of this loop is shifted by up to 2.9 Å away from the nucleotide-binding site relative to those of nucleotide-bound PKA or PKB and most closely resembles the structure of the P-loop in apo-PKA (39). The P-loops of GRK2 and GRK6 were both observed to adopt a similar conformation. The loop may assume a more “active” conformation (that is, like that of the transition state complex of PKA) upon receptor binding and domain closure. Indeed, our structures indicate that the kinase C-terminal extension of GRK1 packs directly on this loop (see below).
In other AGC kinases, the small and the large lobes are observed to “close” upon binding adenine nucleotides, thereby coalescing the appropriate catalytic machinery around the γ-phosphate position. Although our apo-GRK1 structure is of low resolution (∼8 Å), we can still assess changes in the disposition of the small and large lobes of GRK1. Whereas all the nucleotide-bound structures of GRK1 have kinase domains that retain essentially the same conformation (0.21-0.58 Å root mean square deviation for 229 Cα atoms; supplemental Table S2), the two unique kinase domains in the apo-GRK1 structure have distinct conformations from each other as well as from nucleotide-bound GRK1. These unique conformations are at least in part induced by unique crystal contacts and demonstrate that the GRK1 kinase domain has a high degree of flexibility in the absence of nucleotides. Thus, nucleotide binding appears to lock the GRK1 kinase domain in a relatively rigid conformational state, consistent with the relative stability conferred to native GRK1 by nucleotides in vitro (supplemental Fig. S6).
Despite the nucleotide-induced conformational change, the catalytic residues donated by the large lobe are still misaligned with the γ-phosphate of ATP. For example, the catalytic base Asp314 is ∼3 Å removed from its analogous position in the structure of the PKA·(Mg2+)2·ADP·AlF3 complex (26). To adopt a similar conformation, the large lobe of GRK1 needs to rotate an additional ∼13-15° around an axis running roughly parallel to and between the αD and αE helices (Fig. 2a and supplemental Fig. S3). This axis of rotation is similar to that required for GRK2 and GRK6 to adopt a closed conformation and similar in orientation but nonintersecting with that observed for the transition from apo- to nucleotide-bound PKA (supplemental Table S2). We propose that full closure of the kinase domain and a conformational change in the P-loop, and hence activation, is facilitated by the binding of Rho* to the allosteric docking site of GRK1.
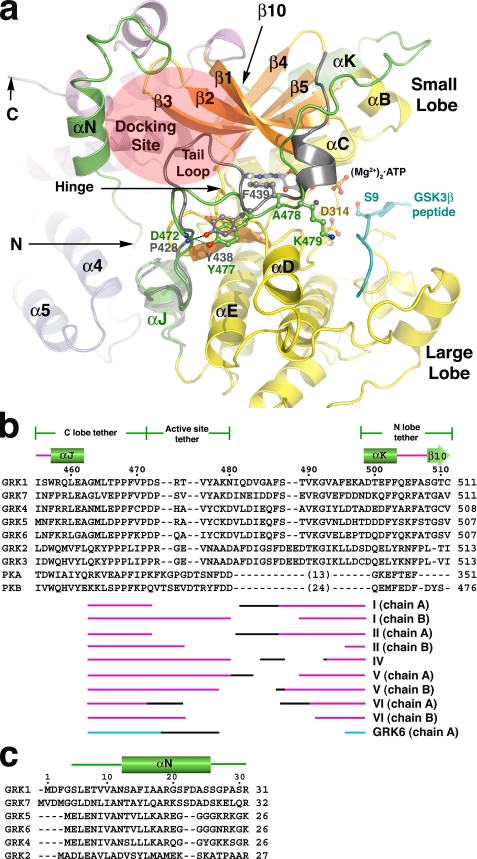
FIGURE 2.
The N terminus and kinase C-terminal extension of GRK1. a, the active site regions of GRK1 and PKB. For this comparison, the coordinates of the various GRK1 crystal forms were merged to generate a composite structure of GRK1 that spans residues 5-533 (of 558), and the large lobe was rotated to generate the expected closed, active state. The kinase C-terminal extension of PKB is gray, and the GSK3β peptide bound to PKB is cyan. The tail-loop of GRKs is four residues shorter than those of either PKB or PKA, resulting in formation of a shallow canyon with the hinge of the kinase domain at the bottom. This region forms a putative receptor-docking site (transparent ellipse). b, structural alignment of the kinase C-terminal extension of GRKs with PKA and PKB. The region immediately following the active site tether is structurally divergent among GRKs, PKA, and PKB, and thus no alignment is attempted. The numbers above the alignment refer to the sequence of bovine GRK1. The different segments of the C-terminal extension that were observed in each of our crystal forms are indicated below the alignment, with the black regions corresponding to structurally heterogeneous portions in each structure that appear influenced by crystal contacts. The accession codes for the protein sequences are: GRK1, P28327; GRK2, NP_777135.1; GRK3, P26818; GRK4, AAI17321.1; GRK5, P43249; GRK6, P43250; GRK7, NP_776757.1; PKA, 1L3R; PKB, 1O6L. c, structural alignment of GRK N-terminal regions.
The Kinase C-terminal Extension—GRK1 residues 455-511 form the C-terminal extension of its kinase domain. Three critical regions of the kinase extension have been defined (10): the C-terminal (large) lobe tether (residues 455-471 in GRK1), an active site tether (AST; residues 472-480), and N-terminal (small) lobe tether (residues 498-511). Although structures corresponding to the C-terminal lobe tether and the N-terminal lobe tether have been described previously for GRK2 and GRK6, the GRK AST has not. This region is highly variable in sequence between GRKs and other AGC kinases, precluding reliable homology modeling (Fig. 2b).
The AST of PKA contributes residues directly to the nucleotide- and peptide-binding sites and, along with the other elements of the C-terminal extension, is thought to help coordinate nucleotide and polypeptide binding with domain closure. The AST is typically disordered in structures of nucleotide-free, open PKA. Consistent with this, the AST of GRK1 is observed only in the nucleotide-bound structures (Fig. 2b), except when displaced by a crystal contact (in one chain of crystal forms I, II, and VI). The GRK1 AST begins with the “tail loop” (residues Asp472-Tyr477), which is four residues shorter than the analogous loops of PKA and PKB (Fig. 2b). The tail loop lies in close proximity to the active site and packs adjacent to the hinge of the kinase domain. The side chain of GRK1-Tyr477 (Tyr473 in GRK6, Tyr438 in PKB, Asn478 in GRK2, and Asn326 in PKA) forms a characteristic hydrogen bond with the backbone nitrogen of the first residue of the tail loop (Fig. 2a). In all non-GRK AGC kinases, the next residue is a Phe, whose side chain contributes to the adenine-binding pocket. Mutation of this residue to alanine in PKA (F327A) renders the kinase catalytically inactive (40). However, in GRKs this residue is conserved as either Ala (Ala478 in GRK1) or Cys, and thus it cannot serve a similar role. The side chain of the next residue, GRK1-Lys479, extends toward the peptide-binding channel where it could help stabilize the binding of an acidic or phosphopeptide substrate.
Residues 481-489 of GRK1, which connect the AST to the N-terminal lobe tether, pack over the surface of the β1-β2 strands and the αB helix of the small lobe. Among the various crystal forms, these residues follow several distinct paths that are at least partly influenced by unique crystal contacts (Fig. 2b). In every case, the path is distinct from those of the analogous regions in PKA and PKB, consistent with the poor sequence conservation of this region. Although the backbone atoms of Ser488 and Thr489 are visible in some structures, high temperature factors (supplemental Fig. S4) prevent interpretation as to whether they are phosphorylated in the crystal, even though phosphorylation at both sites was detected by mass spectrometry in ATP-treated GRK1535-His6 (supplemental Table S4 and supplemental Fig. S7). Given their location, phosphorylation of Ser488 and Thr489 may permit domain-stabilizing electrostatic interactions with Arg222 and Lys221, respectively, in the αB helix (Fig. 3a, inset).
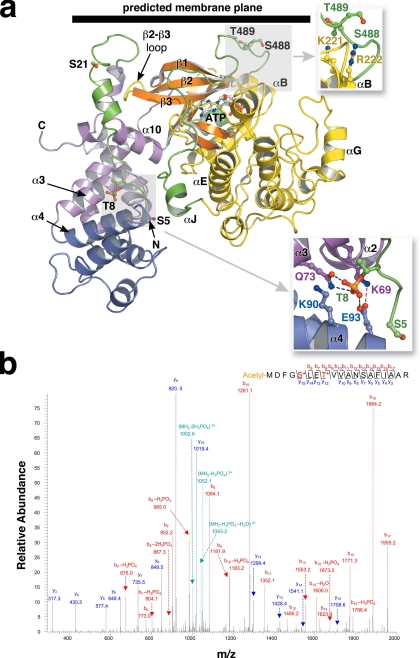
FIGURE 3.
The phosphorylation sites of GRK1. a, the RH-kinase core of GRK1. The structure corresponds to that of crystal form I (with composite C-terminal extension; see Fig. 2). The Ser5, Thr8, Ser21, Ser488, and Thr489 phosphorylation sites are drawn as stick models. The expected position of the membrane plane is indicated. Top inset, the Ser488 and Thr489 phosphorylation sites correspond to the AGC kinase turn motif. Bottom inset, interaction of Thr(P)8 with the RH domain. Gln73 and Glu93 form direct hydrogen bonds, whereas Lys69 and Lys90 complement the charge of the phosphate moiety. These crystals grew at pH 4.35, and so either Glu93 or the phosphate group could be protonated. b, tandem mass spectrometry spectra of phosphopeptides from GRK1535-His6 (Pool A, pretreated with 4 mm ATP and 2 mm MgCl2). Both Ser5 and Thr8 sites were identified in a single peptide. The Ser5 site was also readily observed in endogenous GRK1, as were the previously observed phosphorylation sites at Ser21, Ser488, and Thr489 (supplemental Fig. S7).
The N Terminus of GRK1—Residues 5-30 of the GRK1 N terminus are visible in chain B of crystal form I (Fig. 3a). This region comprises many of the residues known to be important for receptor phosphorylation by GRKs (19, 41, 42). The N terminus makes extensive contacts with both RH subdomains, burying ∼1,200 Å2 of accessible surface area. In this structure, the first six residues of GRK1 thread the hole formed between the two interfaces of the RH-kinase core, residues 7-11 interact with the cleft formed between the bundle and terminal lobes of the RH domain, and residues 12-23 form a helix (αN) whose side chains interact principally with backbone atoms in the RH α10 helix and the β2-β3 loop of the kinase domain. Finally, residues 24-32, variant among GRK subfamilies, form a loop structure engaged in an extensive 2-fold related crystal contact. The residues in the RH domain cleft that interact with the N terminus are conserved in the GRK1 subfamily, whereas those in the α10 helix are conserved in both the GRK4 and GRK1 subfamilies. Ser21, although well ordered, is not obviously phosphorylated. The structure of the GRK1 N terminus in our crystal structure is distinct from that of the GRK1 N-terminal peptide bound to Ca2+·recoverin (43), wherein residues 4-16 form an amphipathic helix.
Surprisingly, the electron density indicated that Thr8 is phosphorylated (supplemental Fig. S5b and Fig. 3a, inset). The Thr(P) side chain forms hydrogen bonds with Gln73 (α3 helix) and Glu93 (α4 helix) of the RH domain and is sandwiched between the amino groups of Lys69 (α3) and Lys90 (α4). Mass spectrometric analysis of a trypsinized N-terminal fragment (Fig. 3b and supplemental Table S4) revealed that phosphorylation at Thr8 can be readily detected in soluble GRK1 pretreated with ATP, but not in GRK1 purified directly from bovine retinas (supplemental Table S3 and supplemental Fig. S10). In contrast, an additional novel phosphorylation site was identified on Ser5 by mass spectrometry in both in vitro and native GRK1 samples (Fig. 3b, supplemental Table S4, and supplemental Fig. S7).
Phosphorylation at these novel sites, Ser5 and Thr8, taken together with Ser488 and Thr489, is consistent with the three or four phosphates predicted to be incorporated by autophosphorylation (44). We investigated the regulatory significance of these phosphorylation sites by mutating Ser5 and Thr8 to either Ala or Asp/Glu (Table 1). The S5A, T8A, and T8E mutants phosphorylated Rho* similarly to the wild-type GRK1. The mutants also exhibited similar rates of autophosphorylation, suggesting no linkage between the phosphorylation of these sites and the ability to bind ATP (data not shown). The effect of T8D and S5D mutations could not be tested because of poor protein expression. Although the effects of these mutations were not obvious in our in vitro steady state kinetic assays, it remains possible that they play more subtle regulatory roles in intact rod cells by influencing mechanisms other than innate kinase activity, e.g. by influencing membrane targeting, transport, or stability, as might be suggested by the poor expression of the S5D and T8D mutants.
N-terminal Truncations of GRK1—GRK1535-H6 purified in five chromatographic peaks (supplemental Fig. S8 and supplemental Table S5) with the active peaks corresponding to full-length (Pool A) and N-terminally truncated forms of GRK1 starting at Thr8 (Pool B) and Ala17 (Pool C). These truncations allowed us to test the importance of residues 1-16 in the context of the GRK1535 protein. Pool A was the most active against ROS membranes. Pool B retained significant activity, suggesting that the first seven residues were dispensable for, although clearly facilitate, the phosphorylation of activated receptors. The activity of Pool C was almost negligible under steady state assay conditions (Table 1). These phosphorylation defects were specific to Rho*, because autophosphorylation and phosphorylation of a soluble nonreceptor substrate, α-synuclein, were similar for all pools (data not shown). We also tested these truncated proteins for their ability to bind Ca2+·recoverin (supplemental Fig. S9). Pool B exhibited reduced recoverin binding, whereas Pool C did not retain any observable affinity. Thus, our data are consistent with residues 8-17, which span the most highly conserved region of the GRK N terminus (Fig. 2c), containing elements that are essential for both Rho* phosphorylation and recoverin binding.
GRK1 RH Domain—The RH domain of GRK1 consists of a core RGS fold of nine α-helices, with two additional GRK-specific helices (α10 and α11) contributed by sequences C-terminal to the kinase extension. As in GRK6, a third helix, α0, precedes the RH domain and interacts with both the RH and kinase domains. Although GRK1 is monomeric by size exclusion and analytical ultracentrifugation (data not shown), a dimer interface mediated by a hydrophobic surface of the RH domain is observed in all six crystal forms (Fig. 1a and supplemental Fig. S1), with a buried accessible surface area of ∼2,800 Å2. The interacting residues are conserved in all GRKs except GRK2 and GRK3.
The size and conservation of the crystalline dimer interface and the fact that a similar interface was observed in crystals of GRK6 (25) suggested that the interface could be of physiological importance. Mutation of residues in the core of the GRK6 interface did not have a significant effect on its ability to phosphorylate ROS (25). However, because GRK1 and Rho* represent a physiologically relevant enzyme-substrate pair, we believed we had a better chance of detecting a functional role for this region. We introduced single (D164A, L166K, and W531A) and double mutations (D164A/L166K, D164A/W531A, and L166K/W531A) in wild-type GRK1. Mutants containing the W531A substitution were unstable and could not be purified to homogeneity, suggesting that this residue plays a significant structural role. The kinetic properties of D164A and L166K mutants, alone and in combination, were similar to those of wild-type GRK1 on Rho*, consistent with the GRK6 data (Table 1). Autophosphorylation was also unaffected (data not shown). By choosing a different rotamer of Trp531, the indole side chain can be fit in the hydrophobic pocket occupied by the 2-fold related Trp531 with only minor adjustments to the backbone. Thus, the RH dimer interface observed in GRK1 and GRK6 may simply represent a short domain swap that occurs at the high concentrations present in protein crystals. However, the ability of this conserved region to bind other proteins in vivo remains a possibility.
DISCUSSION
Our structures of GRK1 reveal the molecular architecture of a third and vertebrate-specific subfamily of GRKs (45) in both nucleotide free- and bound-forms. As in PKA, nucleotide binding in GRK1 induces a conformational change and fixes the orientation of the small and large lobes, concomitantly with ordering of the AST of the kinase C-terminal extension. Despite this conformational change, the active site machinery of GRK1 still appears misaligned for catalysis.
As in PKA, two metal ions are observed in the active sites of ATP and ADP-bound GRK1. High concentrations of Mg2+ (>10 mm) inhibit GRK1, yet metal ions in excess of a stoichiometric Mg2+·ATP complex are necessary for full kinase activity (27). Because of the prominent structural role played by both metal ions in our structures (Fig. 1, b and c) as well as in the transition state complex of PKA (26), it is possible that the structural (second) metal ion becomes inhibitory for GRK1 at high concentrations because it traps the (Mg2+)2·ADP product complex on the enzyme. The relative ease of obtaining crystals of ADP complexes and their high resolution support the idea that GRK1·(Mg2+)2·ADP product complexes are indeed exceptionally stable.
One of our GRK1·(Mg2+)2·ATP crystal forms revealed the structure of the extreme N terminus, a structural element shown herein and by others to be important for receptor phosphorylation. This element is ordered in only one of the 11 independent GRK1 structures we determined in this study, and so the functional relevance of the observed structure is not clear. The N-terminal structure is not dependent on phosphorylation of Thr8 or even the low pH of the crystallization process because it is not ordered in the A chain of the same crystal form or in crystal form II, which was grown at the same pH. Rather, it seems dependent on a 2-fold related crystal contact. A loop from this contact “docks” in a canyon with the walls formed by αN and the tail-loop region of the kinase C-terminal extension and the floor by the hinge and adjacent surface of the small lobe. This crystalline interaction may fortuitously mimic the interactions between activated receptor and GRK1 or simply the molecular crowding that may occur when GRK1 associates with a membrane surface, leading to the ordering of the N terminus. Regardless of the actual structure of the N terminus in a Rho* complex, our structure clearly suggests that it will occupy a region close to the hinge and AST of the kinase domain.
Based on our structures, we propose that the receptor-docking site of GRKs is formed at the hinge of the kinase domain, adjacent to two key structural elements expected to be required for receptor phosphorylation: the AST of the kinase C-terminal extension and the N terminus (Fig. 2a). In the proposed docking site, a flank of the small lobe is relatively solvent-exposed compared with other AGC kinases because the tail loop is four residues shorter in the GRK1 AST. The exposed residues on this flank are highly conserved in GRKs, but not in other AGC kinases (Fig. 2, a and b). We hypothesize that the binding of Rho* to this site will not only induce the additional degree of domain closure required to bring the small and large lobes into alignment and but also induce the P-loop to adopt an active conformation similar to that of nucleotide-bound PKA.
We predict that the C terminus of Rho binds to the large lobe of the kinase domain in a manner similar to the way peptides bind PKB and that GRK1 orients itself with respect to the membrane in a manner similar to that predicted for GRK2 (Fig. 3a) (24). Because the third cytosolic loop of Rho* is the most strongly implicated in binding GRK1, we hypothesize that this loop is in close proximity to the receptor-docking site. A conceptual model that fulfills all of these expectations is shown in Fig. 4. In this model, the GRK1 active site is accessible to other molecules of Rho or Rho* in the same membrane plane, allowing high gain phosphorylation (13, 14). The active site is also accessible to soluble substrates and would allow for multiple rounds of nucleotide exchange and phosphorylation while Rho* is engaged in the docking site.
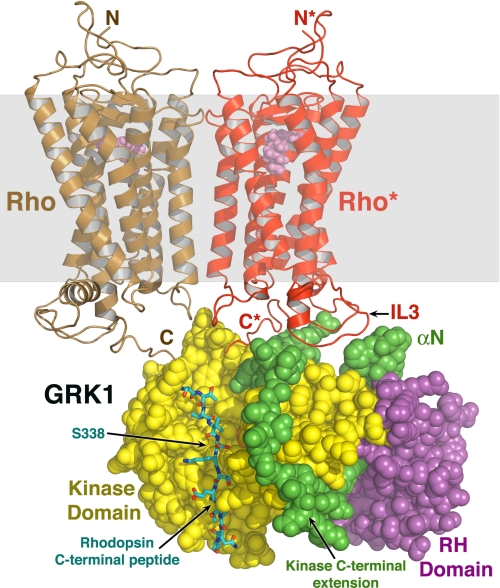
FIGURE 4.
Conceptual model of GRK1 docked to Rho*. The closed composite model of GRK1 (Fig. 2a) was docked with a model of an array of Rho molecules (Protein Data Bank code 1N3M) (50), of which two molecules are shown here for clarity. GRK1 is rendered as spheres, and the expected lipid bilayer plane is shown as a transparent gray box. A monomer of Rho* (red) was modeled such that its third cytoplasmic loop (IL3) lies close to the proposed receptor-docking site. Using the PKB-GSK3β structure (1O6L) as a guide, the C-terminal peptide of Rho* (carbons are colored cyan, oxygens are red, and nitrogens are blue) was modeled docked to the large lobe, as in Fig. 1d. The GRK1 active site would have easy access to the C-tail of Rho* or of a neighboring unactivated Rho (brown) in the same membrane plane, allowing high gain phosphorylation of ROS.
Finally, our structures of GRK1 reveal the molecular basis for some of the mutations that lead to Oguchi disease in humans. One Oguchi patient bears the V380D mutation in one allele (Val377 in bovine GRK1) and a mutation that generates a protein frame shift after Ser536 in the other allele (Pro533 in bovine GRK1) that results in a truncation of the last 22 amino acids of GRK1 (5). The side chain of Val377 contributes to the hydrophobic core of the large lobe, and so a V377D substitution is likely destabilizing (46). Interestingly, the Ser536 Oguchi allele and our bovine GRK1535 are similarly truncated at the C terminus and thus both lack the C-terminal farnesylation site. Despite this, our protein exhibits only a modest reduction in catalytic activity in vitro (Table 1), contrary to what was observed for the Oguchi Ser536 variant expressed in COS7 cells (46). Because we observed degradation of the critical N-terminal region in our GRK1535-soluble mutant but not of the wild-type protein, the Oguchi Ser536 allele may likewise suffer proteolysis at its N terminus that results in a loss of activity (Table 1). Recently, the P391H mutation in GRK1 was also reported to cause Oguchi disease (47). Pro391 (Pro388 in bovine GRK1) is a highly conserved residue in the αF-αG loop of GRKs, and the P391H mutation likely destabilizes the large lobe of the kinase domain by introducing steric clashes with neighboring hydrophobic residues.
In summary, we have characterized the structure of GRK1 and its nucleotide substrate and product complexes, providing the highest resolution and most complete structures of any GRK to date. Importantly, these studies have also revealed structural elements expected to be critical for recognition and phosphorylation of activated GPCRs, the N terminus and kinase C-terminal extension, thereby providing a testable model for how GPCRs might engage and activate GRKs (Fig. 4).
Supplementary Material
Acknowledgments
We thank Dr. Chih-chin Huang for assistance at many stages and Dr. Masaru Miyagi for assistance with mass spectrometric analysis. We thank Dr. Ute Kent for matrix-assisted laser desorption ionization time-of-flight analysis of GRK1535-His6 samples and Dr. Anthony Ludlum (both at University of Michigan, Ann Arbor) for assistance with sedimentation equilibrium analyses.
The atomic coordinates and structure factors (codes 3C4W, 3C4X, 3C4Y, 3C4Z, 3C50, and 3C51) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grants HL071818 (to J. J. G. T.) and EY 08061 and P30-EY11373 (Case Western Reserve University) (to K. P.). The General Medicine and Cancer Institutes Collaborative Access Team has been funded in whole or in part by National Cancer Institute Grant Y1-CO-1020 and Institute of General Medical Sciences Grant Y1-GM-1104. This work was also supported in part by the United States Department of Energy Office of Biological and Environmental Research under Contract DE-AC02-06CH11357. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental text, supplemental Tables S1-S5, and supplemental Figs. S1-S11.
Footnotes
The abbreviations used are: Rho, rhodopsin; Rho*, light-activated Rho; GPCR, G protein-coupled receptor; GRK, GPCR kinase; PKA, protein kinase A; PKB, protein kinase B; ROS, rod outer segment; AST, active site tether.
References
- 1.Palczewski, K. (2006) Annu. Rev. Biochem. 75 743-767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maeda, T., Imanishi, Y., and Palczewski, K. (2003) Prog. Retin Eye Res. 22 417-434 [DOI] [PubMed] [Google Scholar]
- 3.Arshavsky, V. Y., Lamb, T. D., and Pugh, E. N., Jr. (2002) Annu. Rev. Physiol. 64 153-187 [DOI] [PubMed] [Google Scholar]
- 4.Doan, T., Mendez, A., Detwiler, P. B., Chen, J., and Rieke, F. (2006) Science 313 530-533 [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto, S., Sippel, K. C., Berson, E. L., and Dryja, T. P. (1997) Nat. Genet. 15 175-178 [DOI] [PubMed] [Google Scholar]
- 6.Cideciyan, A. V., Zhao, X., Nielsen, L., Khani, S. C., Jacobson, S. G., and Palczewski, K. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 328-333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, C. K., Burns, M. E., Spencer, M., Niemi, G. A., Chen, J., Hurley, J. B., Baylor, D. A., and Simon, M. I. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 3718-3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palczewski, K., and Benovic, J. L. (1991) Trends Biochem. Sci. 16 387-391 [DOI] [PubMed] [Google Scholar]
- 9.Pitcher, J. A., Freedman, N. J., and Lefkowitz, R. J. (1998) Annu. Rev. Biochem. 67 653-692 [DOI] [PubMed] [Google Scholar]
- 10.Kannan, N., Haste, N., Taylor, S. S., and Neuwald, A. F. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 1272-1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fowles, C., Sharma, R., and Akhtar, M. (1988) FEBS Lett. 238 56-60 [DOI] [PubMed] [Google Scholar]
- 12.Palczewski, K., Buczylko, J., Kaplan, M. W., Polans, A. S., and Crabb, J. W. (1991) J. Biol. Chem. 266 12949-12955 [PubMed] [Google Scholar]
- 13.Binder, B. M., Biernbaum, M. S., and Bownds, M. D. (1990) J. Biol. Chem. 265 15333-15340 [PubMed] [Google Scholar]
- 14.Shi, G. W., Chen, J., Concepcion, F., Motamedchaboki, K., Marjoram, P., Langen, R., and Chen, J. (2005) J. Biol. Chem. 280 41184-41191 [DOI] [PubMed] [Google Scholar]
- 15.Kelleher, D. J., and Johnson, G. L. (1990) J. Biol. Chem. 265 2632-2639 [PubMed] [Google Scholar]
- 16.Shi, W., Osawa, S., Dickerson, C. D., and Weiss, E. R. (1995) J. Biol. Chem. 270 2112-2119 [DOI] [PubMed] [Google Scholar]
- 17.Levay, K., Satpaev, D. K., Pronin, A. N., Benovic, J. L., and Slepak, V. Z. (1998) Biochemistry 37 13650-13659 [DOI] [PubMed] [Google Scholar]
- 18.Higgins, M. K., Oprian, D. D., and Schertler, G. F. (2006) J. Biol. Chem. 281 19426-19432 [DOI] [PubMed] [Google Scholar]
- 19.Palczewski, K., Buczylko, J., Lebioda, L., Crabb, J. W., and Polans, A. S. (1993) J. Biol. Chem. 268 6004-6013 [PubMed] [Google Scholar]
- 20.Newton, A. C. (2002) Biochem. J. 370 361-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palczewski, K., Buczylko, J., Van Hooser, P., Carr, S. A., Huddleston, M. J., and Crabb, J. W. (1992) J. Biol. Chem. 267 18991-18998 [PubMed] [Google Scholar]
- 22.Lodowski, D. T., Pitcher, J. A., Capel, W. D., Lefkowitz, R. J., and Tesmer, J. J. (2003) Science 300 1256-1262 [DOI] [PubMed] [Google Scholar]
- 23.Lodowski, D. T., Barnhill, J. F., Pyskadlo, R. M., Ghirlando, R., Sterne-Marr, R., and Tesmer, J. J. (2005) Biochemistry 44 6958-6970 [DOI] [PubMed] [Google Scholar]
- 24.Tesmer, V. M., Kawano, T., Shankaranarayanan, A., Kozasa, T., and Tesmer, J. J. (2005) Science 310 1686-1690 [DOI] [PubMed] [Google Scholar]
- 25.Lodowski, D. T., Tesmer, V. M., Benovic, J. L., and Tesmer, J. J. (2006) J. Biol. Chem. 281 16785-16793 [DOI] [PubMed] [Google Scholar]
- 26.Madhusudan, Akamine, P., Xuong, N. H., and Taylor, S. S. (2002) Nat. Struct. Biol. 9 273-277 [DOI] [PubMed] [Google Scholar]
- 27.Palczewski, K., McDowell, J. H., and Hargrave, P. A. (1988) Biochemistry 27 2306-2313 [DOI] [PubMed] [Google Scholar]
- 28.Otwinoski, Z. (1993) in Proceedings of the CCP4 Study Weekend: Data Collection and Processing (Sawyer, L., Isaacs, N., and Bailey, S., eds) pp. 55-62, SERC Daresbury Laboratory, Warrington, UK
- 29.Winn, M. D. (2003) J. Synchrotron. Radiat. 10 23-25 [DOI] [PubMed] [Google Scholar]
- 30.Winn, M. D., Isupov, M. N., and Murshudov, G. N. (2001) Acta Crystallogr. Sect. D Biol. Crystallogr. 57 122-133 [DOI] [PubMed] [Google Scholar]
- 31.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53 240-255 [DOI] [PubMed] [Google Scholar]
- 32.Jones, T. A., Zou, J. Y., Cowan, S. W., and Kjeldgaard, M. (1991) Acta Crystallogr. Sect. A 47, 110-119 [DOI] [PubMed] [Google Scholar]
- 33.Laskowski, R. A., MacArthur, M. W., Moss, D. S., and Thornton, J. M. (1993) J. Appl. Crystallogr. 26 283-291 [Google Scholar]
- 34.Inglese, J., Glickman, J. F., Lorenz, W., Caron, M. G., and Lefkowitz, R. J. (1992) J. Biol. Chem. 267 1422-1425 [PubMed] [Google Scholar]
- 35.Inglese, J., Koch, W. J., Caron, M. G., and Lefkowitz, R. J. (1992) Nature 359 147-150 [DOI] [PubMed] [Google Scholar]
- 36.Zheng, J., Trafny, E. A., Knighton, D. R., Xuong, N. H., Taylor, S. S., Ten Eyck, L. F., and Sowadski, J. M. (1993) Acta Crystallogr. Sect. D Biol. Crystallogr. 49 362-365 [DOI] [PubMed] [Google Scholar]
- 37.Nowakowski, J., Cronin, C. N., McRee, D. E., Knuth, M. W., Nelson, C. G., Pavletich, N. P., Rogers, J., Sang, B. C., Scheibe, D. N., Swanson, R. V., and Thompson, D. A. (2002) Structure 10 1659-1667 [DOI] [PubMed] [Google Scholar]
- 38.Hemmer, W., McGlone, M., Tsigelny, I., and Taylor, S. S. (1997) J. Biol. Chem. 272 16946-16954 [DOI] [PubMed] [Google Scholar]
- 39.Akamine, P., Madhusudan, Wu, J., Xuong, N. H., Ten Eyck, L. F., and Taylor, S. S. (2003) J. Mol. Biol. 327 159-171 [DOI] [PubMed] [Google Scholar]
- 40.Batkin, M., Schvartz, I., and Shaltiel, S. (2000) Biochemistry 39 5366-5373 [DOI] [PubMed] [Google Scholar]
- 41.Noble, B., Kallal, L. A., Pausch, M. H., and Benovic, J. L. (2003) J. Biol. Chem. 278 47466-47476 [DOI] [PubMed] [Google Scholar]
- 42.Yu, Q. M., Cheng, Z. J., Gan, X. Q., Bao, G. B., Li, L., and Pei, G. (1999) J. Neurochem. 73 1222-1227 [DOI] [PubMed] [Google Scholar]
- 43.Ames, J. B., Levay, K., Wingard, J. N., Lusin, J. D., and Slepak, V. Z. (2006) J. Biol. Chem. 281 37237-37245 [DOI] [PubMed] [Google Scholar]
- 44.Buczylko, J., Gutmann, C., and Palczewski, K. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 2568-2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Premont, R. T., Macrae, A. D., Aparicio, S. A., Kendall, H. E., Welch, J. E., and Lefkowitz, R. J. (1999) J. Biol. Chem. 274 29381-29389 [DOI] [PubMed] [Google Scholar]
- 46.Khani, S. C., Nielsen, L., and Vogt, T. M. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 2824-2827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayashi, T., Gekka, T., Takeuchi, T., Goto-Omoto, S., and Kitahara, K. (2007) Ophthalmology 114 134-141 [DOI] [PubMed] [Google Scholar]
- 48.Palczewski, K., Arendt, A., McDowell, J. H., and Hargrave, P. A. (1989) Biochemistry 28 8764-8770 [DOI] [PubMed] [Google Scholar]
- 49.Onorato, J. J., Palczewski, K., Regan, J. W., Caron, M. G., Lefkowitz, R. J., and Benovic, J. L. (1991) Biochemistry 30 5118-5125 [DOI] [PubMed] [Google Scholar]
- 50.Fotiadis, D., Liang, Y., Filipek, S., Saperstein, D. A., Engel, A., and Palczewski, K. (2003) Nature 421 127-128 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.