Abstract
The present study demonstrated that invadopodia are associated with invasion by degradation of matrix in prostate cancer cells PC3. To find out the presence of invadopodia in PC3 cells, we performed a few comparative analyses with osteoclasts, which utilize podosomes for migration. Our investigations indeed demonstrated that invadopodia are comparable to podosomes in the localization of Wiskott-Aldrich syndrome protein (WASP)/matrix metalloproteinase-9 and the degradation of matrix. Invadopodia are different from podosomes in the localization of actin/vinculin, distribution during migration, and the mode of degradation of extracellular matrix. Invadopodia enable polarized invasion of PC3 cells into the gelatin matrix in a time-dependent manner. Gelatin degradation was confined within the periphery of the cell. Osteoclasts demonstrated directional migration with extensive degradation of matrix underneath and around the osteoclasts. A pathway of degradation of matrix representing a migratory track was observed due to the rearrangement of podosomes as rosettes or clusters at the leading edge. Reducing the matrix metalloproteinase-9 levels by RNA interference inhibited the degradation of matrix but not the formation of podosomes or invadopodia. Competition experiments with TAT-fused WASP peptides suggest that actin polymerization and formation of invadopodia involve the WASP-Arp2/3 complex pathway. Moreover, PC3 cells overexpressing osteopontin (OPN) displayed an increase in the number of invadopodia and gelatinolytic activity as compared with PC3 cells and PC3 cells expressing mutant OPN in integrin-binding domain and null for OPN. Thus, we conclude that OPN/integrin αvβ3 signaling participates in the process of migration and invasion of PC3 cells through regulating processes essential for the formation and function of invadopodia.
Cell migration plays a key role in several cellular processes. Cell migration is a multistep process, primarily initiated by an external stimulus that leads to the formation of cytoskeletal changes and membrane protrusions. Changes in cell shape and formation of adhesive structures are regulated by the dynamic regulation of the actin cytoskeleton. Temporal and spatial localization of actin-binding proteins is likely to play a vital role in the dynamic regulation of the actin cytoskeleton along with the specialized structures involved in cell migration (1, 2). Although actin structures such as lamellipodia or filopodia-like extensions play a role in cell migration, they are connected to the extracellular matrix (ECM)2 via adhesive structures, including focal adhesions (3), podosomes (1), and invadopodia (4, 5).
Focal adhesions are structural links on the cell membrane where the actin cytoskeleton inside the cell is connected to the ECM on the outside in eukaryotic cells. Cells expressing focal adhesions are characterized by lower rates of motility due to different and slower mechanisms of structural assembly/disassembly. Podosomes are found in highly invasive cancer cells, macrophages, Rous sarcoma virus-transformed chick embryo chondrocytes, endothelial cells, certain transformed fibroblasts, and osteoclasts (2, reviewed in 5). Podosomes are F-actin-containing dot-like adhesion structures. Podosomes form clusters (6) and rosettes (7) during cell migration. These are highly dynamic structure that contained actin, WASP, Arp2/3, gelsolin, and MMP-9 (1, 6, 8). Consistent with their function as adhesion sites, podosomes contain many of the same proteins found in focal adhesions, such as F-actin, vinculin, talin, fimbrin, and α-actinin (9, 10). Podosomes contain proteins that regulate actin polymerization such as gelsolin, the Arp2/3 complex, N-WASP/WASP, and fimbrin, whereas focal adhesion does not contain these proteins (reviewed in Ref. 5). Osteoclasts are unique in that their mechanism of cell motility is podosome-based. The speed of podosome assembly and disassembly generates high rates of motility in osteoclasts (11). Also, release of MMPs from podosomes facilitates migration of osteoclasts through degradation of ECM (6).
Invadopodia are membrane protrusions formed by invasive carcinoma cells (4, 5). Podosomes and invadopodia share ultrastructures and molecular components. Invadopodia are enriched with actin and actin-binding proteins, proteases, seprase, and signaling proteins. Actin-binding and -signaling proteins present in invadopodia have roles in the regulation of membrane remodeling and actin cytoskeleton. Due to the presence of proteases, invadopodia have a matrix degradation property (reviewed in 5 and 12). A significant development in our understanding of the role of WASP (Wiskott-Aldrich Syndrome protein) family proteins in actin dynamics has been achieved in recent years from a number of studies on a variety of cell systems (8, 13, 14). WASP has been shown to control podosome assembly and sealing ring formation in cooperation with Cdc42Hs in primary macrophages and osteoclasts (13, 15). Similarly, N-WASP, Arp2/3 complex, Nck, Cdc42, and WIP regulate invadopodium formation in highly invasive cancer cells (4, 16, 17).
Although focal adhesions, invadopodia, and podosomes mediate cell adhesions (3, 5), focal adhesions are spatially, morphologically, and compositionally distinct from the other two structures (18). Focal adhesions are determined by localization of focal adhesion kinase, which is not present in invadopodial complexes (18). Although both podosomes and invadopodia release proteases at the cell-to-substratum adhesion sites and are involved in cell adhesion and migration, some morphological and functional differences exist. Podosomes are invaginations from the ventral membrane toward cytoplasm, whereas invadopodia are protrusions (evaginations) from ventral and peripheral membrane surfaces into the matrix (4). Podosomes are highly dynamic structures with a half-life of 2–12 min (11), and invadopodia are less dynamic but highly stable structures. Their lifetime varies from minutes to several hours (16).
It is not known whether prostate cancer cells use podosomes or invadopodia as adhesive and migratory structures. Pericellular activation of proteases has been shown to play a role in prostate cancer cell proliferation and invasion (19). Because invadopodia are implicated in migration and invasion of metastatic tumor cells to surrounding tissues (5, 12, 20, 21), we hypothesized that prostate cancer cells (PC3) would also use invadopodia for migration and invasion. To determine the adhesive structure involved in the migration of prostate cancer cells, we performed a few comparative analyses in cells that express podosomes (e.g. osteoclasts) and focal adhesions (e.g. melanoma cells). As hypothesized, invadopodia-like adhesive structures were found inside and at the periphery of prostate cancer cells; colocalization of both F-actin and vinculin was found in these structures. These structures are different from podosomes and focal adhesions in the localization of vinculin. Localization of WASP in these structures suggests that WASP may have a role in the formation of invadopodia. Nevertheless, these structures display localization of MMP-9, which is capable of degrading the matrix.
EXPERIMENTAL PROCEDURES
Reagents—Antibodies to actin, α-actinin, vinculin, WASP, and MMP-9 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Cy2- and Cy3-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA). Glyceraldehyde-3-phosphate dehydrogenase antibody was purchased from Abcam, Inc. (Cambridge, MA). An MMP-9 activity detection kit was purchased from Biomol (Plymouth Meeting, PA). Rhodamine phalloidin was purchased from Sigma-Aldrich. Nickel-nitrilotriacetic acid-Sepharose beads to purify TAT proteins were purchased from Amersham Biosciences. Polyacrylamide solution and protein estimation reagents were purchased from Bio-Rad.
cDNA Constructs, Cell Lines, and Culture—Mutation in the integrin-binding motif (Arg-Gly-Asp (RGD) ΔArg-Gly-Ala (RGA)) of human osteopontin (OPN (22)) was generated using altered sites II in vitro mutagenesis system (Promega, Madison WI). Prostate cancer epithelial cells (PC3 and CRL-1435, ATCC, Manassas, VA) were transfected with full-length (PC3/OPN), mutant OPN (PC3/OPN (RGA)) in pCEP4 vector, and vector without insert (pCEP4) with use of Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. The OPN siRNA expression vector was generated using Seqwright DNA Technology Services (Houston, TX) as described previously (23). PC3 cells were transfected with the small interfering RNA (siRNA) constructs and pSilencer 4.1-CMV neo vector (as vector control) using a silencer siRNA transfection kit (Ambion, Austin, TX). Individual clones that exhibited maximum reduction in endogenous OPN levels were generated as described previously (23). PC3 clones that express the highest levels of OPN (full-length and mutant) and a maximum reduction in the endogenous levels of OPN were used for all the experiments described here. These clones were designated as PC3/OPN, PC3/OPN (RGA), and PC3/siRNA. PC3 cell lines were cultured in RPMI 1640 media containing 5% fetal bovine serum at 37 °C (Invitrogen).
Preparation of Osteoclast Precursor from Mice—C57/BL6 mice were used for in vitro osteoclast generation. Bone marrow cells isolated from five mice were cultured into 100-mm dishes with 20 ml of α-minimal essential medium, supplemented with 10% fetal bovine serum. After culturing for 24 h, non-adhered cells were layered on histopaque-1077 (Sigma) and processed as described previously (1). Cells were cultured with the appropriate concentrations of macrophage colony stimulating factor-1 (10 ng/ml, R & D Systems) and receptor activator for nuclear factor κB-ligand (55–75 ng/ml). After 3 days in culture, media were replaced with fresh cytokines. The multinucleated osteoclasts were seen from day 4 onward.
Cloning of WASP Fragments—Bacterial expression constructs coding various HIV-TAT fusion peptides of WASP were generated by the PCR method. WASP constructs were generated from human cDNA library by using the following primers: Full length (FL): 5′-catgccatgggcatgagtgggggcccaatgggaggaag-3′ and 5′-acatgcatgcacttatcagtcatcccattcatcatcttcatc-3′; Verpolin-like, central, and acidic domain (VCA): 5′-catgccatgggcgtgcctgccgggggcctg-3′ and 5′-acatgcatgcacttatcagtcatcccattcatcatcttcatc-3′. The PCR product was digested with NcoI and SphI restriction enzymes and inserted into the HA-TAT fusion vector, which was cut with the same restriction enzymes. The sequences of all the clones were confirmed for reading frame by DNA sequencing.
Purification of TAT-fused Proteins—Escherichia coli BL-21 strain (Invitrogen) was used to transform HA-TAT vector containing the above-mentioned WASP constructs. Protein was purified using nickel-nitrilotriacetic acid column as described previously (24). SDS-PAGE and Western analysis with a hemagglutinin (HA) antibody as well as Coomassie Blue staining were performed to test the purity of the purified proteins (data not shown).
Transduction of TAT-fused Proteins into PC3 Cells—Transduction of TAT-fused peptides into PC3 cells was performed as described previously (24). Based on the dose-dependent uptake experiment (data not shown), a concentration of 100 nm TAT-fused WASP protein was decided for transduction experiments shown in Figs. 7 and 8. The uptake of TAT-fused WASP proteins was determined by immunoblotting with an antibody to HA tag present in the transduced protein. The uptake reaches maximum levels at 60–90 min and decreases after 3 h. Proteins transduced into PC3 cells were stable for 8–10 h. Based on the time-course experiment, cells were transduced with TAT-proteins for 60 min in serum-free medium, and the F-actin content was determined as described previously (25). Untransduced cells as well as cells transduced with HA-TAT vector (8–10 kDa) and herpes simplex virus-thymidine kinase (Hsv-TK, 42kDa) proteins were used as controls for transduction experiments.
FIGURE 7.
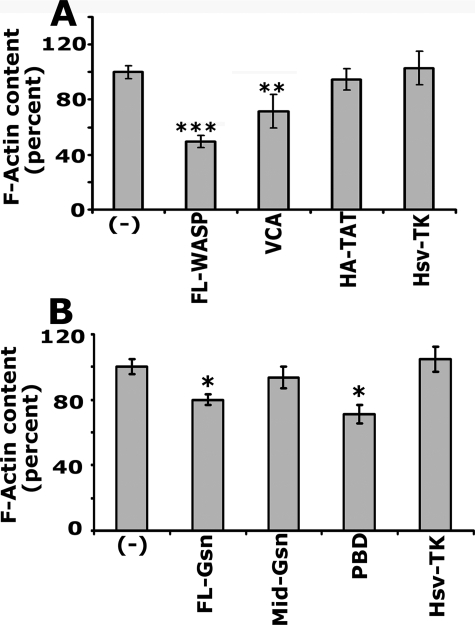
Measurement of F-actin content in PC3 cells transduced with TAT-fused WASP and gelsolin peptides. F-actin content was measured by rhodamine phalloidin binding in PC3 cells transduced with peptides indicated in A and B. Cells were grown in 24-well tissue culture dishes, and 3 to 4 wells were used for each peptide. The results represented are mean ± S.E. for three experiments. ***, p < 0.001; **, p < 0.01; *, p < 0.05 versus untransduced (-) and control proteins (HA-TAT or Hsv-TK)-transduced PC3 cells.
FIGURE 8.
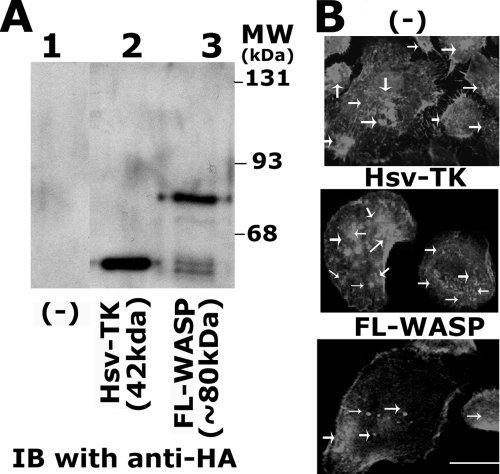
The effects of transduction of WASP peptide on actin distribution in PC3 cells. A, immunoblotting analysis of the levels of transduced proteins in PC3 cells with a HA-antibody is shown. PC3 cells were transduced with TAT-fused Hsv-TK (lane 2) and FL-WASP peptides. Lysates (∼200 μg) were immunoblotted with an antibody to HA to determine the levels of transduced proteins in PC3 cells. B, confocal analysis of distribution of actin (red) is shown in PC3 cells transduced with FL-WASP and Hsv-TK proteins. Untransduced PC3 cells are indicated as (-) in A and B. Scale bar, 50 μm. The results shown are representative of three independent experiments.
Measurement of F-actin Content Using Rhodamine Phalloidin Binding—Prostate cancer cells were cultured in 24-well culture plates for 12–18 h. After various treatments, cells were rinsed thrice with phosphate-buffered saline. Subsequently, cells were fixed, permeabilized, and incubated with rhodamine phalloidin (1:200) as described earlier (25). Cells were extracted with absolute methanol, and the fluorescence of each sample was measured with fluorometry (Bio-Rad spectrofluorometer) (25).
Immunohistochemistry and Actin Staining—Cells were fixed and permeabilized with 3.7% paraformaldehyde and 0.1% Triton X-100 for 10 min. Cells were washed with phosphate-buffered saline and immunostained with WASP or MMP-9 (1:100 dilution) for 60 min, followed by labeling with secondary antibody (CY2; 1:100) for 60 min. Cells stained for MMP-9 were stained for actin with rhodamine phalloidin as described previously (24). The immunostained cells were viewed and photographed on a Bio-Rad confocal laser-scanning microscope. Images were stored in TIF format and processed by using Photoshop (Adobe Systems, Inc., Mountain View, CA).
siRNA for MMP-9 and Immunoblotting Analysis—Two different siRNA were designed and synthesized by Ambion as follows: the first target sequences (sense and antisense) were 5′-ggcauacuuguaccgcuautt-3′ and 5′-auagcgguacaaguaugcctc-3′ (cat. no. 16806); the second sequences were 5′-ggcauacuuguaccgcuautt-3′ and 5′-auagcgguacaaguaugcctc-3′ (cat. no. 16706). MMP-9 levels were reduced to a greater extent by the second target sequence (cat. no. 16706) in osteoclasts. Hence, we used the second target sequences to reduce MMP-9 levels in osteoclasts and PC3 cells. Control RNAi (cRNAi) sequence consisting of a scrambled sequence, which does not have specificity to any known cellular mRNAs was used as control. Cy3-conjugated luciferase siRNA (cat. no. D-001110, Upstate Cell Signaling Solutions, Charlottesville, VA) was also used to optimize the condition to obtain the greatest amount of target-specific knockdown of MMP-9 protein in osteoclasts and PC3 cells. Transfection of siRNA and cRNAi sequences was performed as described previously (26). Following incubation with siRNA to MMP-9 (0.25 and 0.5 μm) and cRNAi (0.5 μm) for 48 h, cells (osteoclasts and PC3 cells) were washed three times with cold phosphate-buffered saline and lysed in RIPA buffer (10 mm Tris-HCl, pH 7.2, 150 mm NaCl, 1% deoxycholate, 1% Triton X-100, 0.1% SDS, 1% aprotinin, and 2 mm phenylmethylsulfonyl fluoride) and centrifuged at 15,000 rpm for 15 min at 4 °C. Protein contents were measured using the Bio-Rad protein assay reagent. Equal amounts of lysate proteins were used for immunoblotting as described previously (26).
Fluorescent Gelatin Degradation Assay—Cross-linked-fluorophore (FITC)-conjugated gelatin matrix-coated coverslips were prepared as described (18). Gelatin-coated coverslips were quenched with RPMI containing 10% fetal bovine serum at 37 °C for 60 min prior to plating cells. To assess the formation of podosomes or invadopodia and degradation of FITC-gelatin matrix, cells (osteoclasts and PC3 cells) were cultured on FITC-gelatin-coated coverslips for different time periods (8–16 h). Cells were fixed and stained for actin with rhodamine phalloidin. Degraded matrix was noticed as foci after 4–6 h. Degradation was more after incubation for 12–14 h. Gelatin matrix and actin-stained cells were viewed and photographed with a Bio-Rad confocal laser-scanning microscope. Images were stored in TIF image format and processed by using Photoshop.
MMP-9 Activity Assay—Equal amounts of lysate proteins (200 μg) were used for immunoprecipitation with antibodies to WASP, actin, vinculin, talin, and α-actinin (24). MMP-9 activity present in the immune complexes was pulled down with protein-A Sepharose and measured according to the manufacturer's guidelines provided in the MMP-9 assay kit (BIOMOL). After the reaction, optical density was measured at 450 nm to obtain the relative gelatinase activity. Standard MMP-9 protein provided in the kit was used to generate a standard graph. MMP-9 activity coprecipitated with the immune complexes was calculated and plotted.
Cell Migration Assay (Invasion Assay)—Invasion assays were performed in Transwell chambers with 8-μm pore size membranes as described previously (1). Transwell inserts were layered with an ECM solution (i.e. collagen), which functions as an in vitro basement membrane (Millipore-QCM cell invasion assays). Cells were incubated for 12–14 h at 37 °C. PC3 cells migrated through the collagen layer attached to the bottom of the Transwell membrane. Cells were stained with acid hematoxylin and counted with a light microscope. Statistical comparisons were performed as described below.
Data Analysis—All values presented are expressed as the means ± S.E. of three or more experiments done at different times normalized to intra-experimental control values. Asterisks were used to graphically indicate the statistical significance. A value of p < 0.05 was considered significant. For statistical comparisons, analysis of variance was used with the Bonferroni corrections. GraphPad Instat (GraphPad Software, San Diego, CA) was used to perform the statistical tests.
RESULTS
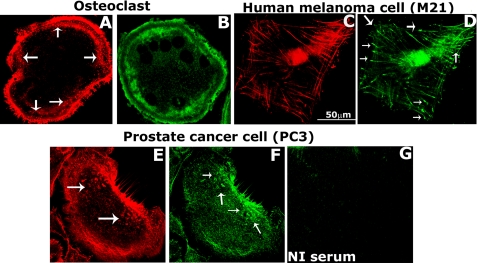
Characterization of Adhesive Structure in PC3 Cells, a Comparative Analysis—Staining of prostate cancer cells (PC3/OPN) for actin with rhodamine phalloidin (red, Fig. 1E) displayed punctate F-actin-enriched structures at the center and periphery (Fig. 1, E and F, indicated by arrows). Vinculin was also observed in these structures (green, Fig. 1F). We have previously demonstrated focal adhesions and podosomes as adhesive/migratory structures in human melanoma cells (M21) and osteoclasts, respectively (6, 15). To exhibit that the structures observed in prostate cancer cells are unique and different from these adhesive/migratory structures, we performed actin (red) and vinculin (green) staining in M21 and osteoclasts. F-actin-enriched dot-like podosomes were observed at the periphery in the clear zone area of osteoclasts plated on coverslip (Fig. 1A, indicated by arrows). Vinculin distribution was observed as a double ring and circumferences the clear-zone area and podosomes (Fig. 1B). Human melanoma cells displayed spear-shaped elongated focal adhesion structures enriched in F-actin and vinculin (Fig. 1, C and D, indicated by arrows in D). F-actin-enriched adhesive structures in PC3 cells are diverse from those of podosomes and focal adhesions. It is possible that the adhesive structures enriched in actin and vinculin are invadopodia in prostate cancer cells, because cancer cells use invadopodia for adhesion and degradation of the matrix (4, 27).
FIGURE 1.
A comparative analysis of adhesive structures in different cells. Distribution of vinculin (green) and actin (red) in osteoclasts (A and B), human melanoma (C and D) and prostate cancer (PC3, E and F) cells is shown. Immunostaining with corresponding non-immune (NI) serum for vinculin antibody is shown in panel E. The results shown are representative of three independent experiments.
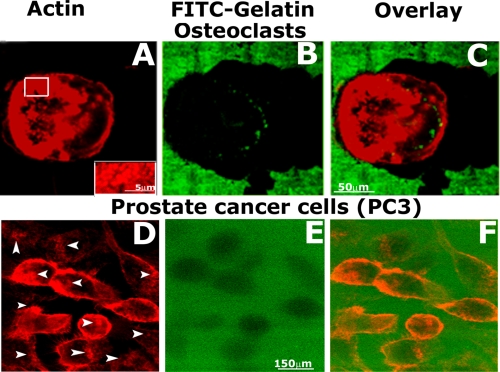
Comparative Analysis of the Matrix Degradation Property of Prostate Cancer Cells and Osteoclasts—The ability of osteoclasts and prostate cancer cells to degrade the matrix was compared by culturing cells on a cross-linked fluorophore (FITC)-conjugated-gelatin matrix-coated coverslip for 12 h (18). Cells were stained with rhodamine phalloidin for actin. Actin staining of osteoclasts revealed rosettes (Fig. 2A) or clusters (Fig. 3C) of podosomes. In higher magnification, ring-like rosettes at the migration front or leading edge demonstrated individual podosomes enriched in F-actin (inset in Fig. 2A). Manifestation of rosettes or clusters of podosomes during migration is different from the sealing ring, which is formed during bone resorption (26). Localization of the rosettes of podosomes at the leading edge and a track of cleared area of gelatin matrix indicates that osteoclasts degrade matrix during migration, and podosomes play a key role in this process. PC3 cells displayed areas of actin enrichment in the cell peripheries as well as within the cell body (Fig. 2D, indicated by arrows). Gelatin degradation was present underneath the cell body (Fig. 2, E and F). Degradation was confined within the cell boundary, and cells were found within the degraded matrix. These observations suggest that actin-enriched invadopodia-like structures may perhaps assist in the polarized invasion of cells into matrix boundaries.
FIGURE 2.
Comparative analysis of the gelatin matrix degradation property of osteoclasts and prostate cancer cells (PC3). The ability of osteoclasts (A–C) and prostate cancer cells (D–F) to degrade the matrix were compared by culturing cells on a cross-linked fluorophore (FITC)-conjugated-gelatin matrix. Cells were stained with rhodamine phalloidin for actin as shown in A and D. Merged images (overlay) show FITC-gelatin matrix (green) and staining for actin in an osteoclast (C) and PC3 cells (F). Images at the matrix level that exhibit the dark (non-fluorescent) areas of degradation are shown in B and E. Larger areas of degradation by osteoclast were observed (B). Inset, white box in panel A is shown at a higher magnification in the inset. Podosomes enriched in F-actin were observed in the enlarged view shown in the inset. These results represent one of three separate experiments performed with the same results.
FIGURE 3.
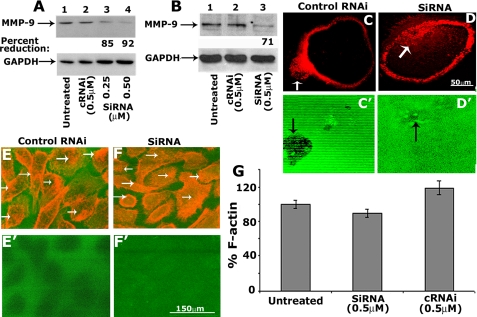
The effects of siRNA to MMP-9 on the degradation of FITC-gelatin matrix. A and B, the effect of siRNA to MMP-9 on protein levels of MMP-9 in osteoclasts (A) and PC3 cells (B). Osteoclasts and PC3 cells were treated with the indicated concentration of siRNA (lanes 3 and 4 in A; lane 3 in B) or cRNAi (lane 2 in A and B) sequences. Untreated cells (lane 1 in A and B) were used as controls. Equal amounts of lysate proteins were used for the immunoblot analysis with an antibody to MMP-9. Loading was normalized by an immunoblotting analysis with a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody after stripping the MMP-9 blot. Quantitation and statistical analysis of the levels of MMP-9 in cells treated with siRNA is provided at the bottom of subpanel (A and B). A significant reduction in the levels of MMP-9 was observed in siRNA-treated cells. C–F, immunostaining of osteoclasts (C and D) and PC3 cells (E and F) with rhodamine phalloidin cultured on FITC-coated gelatin coverslips is shown. Merged images (overlay) show FITC-gelatin matrix (green) and staining for actin in an osteoclast (C and D) and PC3 cells (E and F). Images at the FITC-gelatin matrix level are shown in C′, D′, E′, and F′. The dark (non-fluorescent) areas of degradation were observed in cells treated with cRNAi (C′ and E′). Degradation was reduced (D′) or not observed (F′) in cells treated with siRNA to MMP-9. G, measurement of F-actin content. F-actin content was measured by rhodamine phalloidin binding to PC3 cells treated as given below the figure. About 3–5 wells in 24-well tissue culture plates were used for each treatment. siRNA to MMP-9 had no effect on the F-actin content of PC3 cells. The data presented are mean ± S.E. of three different experiments.
MMP-9 in Podosome Clusters and Invadopodia Have Matrix Degradation Property Effect of MMP-9 Silencing—We have previously shown that MMP-9 has a significant role in migration of PC3 cells and osteoclasts (6, 23). To determine the role of MMP-9 in cell migration/invasion, we used siRNA strategy to reduce endogenous levels of MMP-9 in osteoclasts (Fig. 3, A, C, and D) and PC3 cells (Fig. 3, B, E, and F). Cells were incubated with siRNA for 36 h at 37 °C, at doses of 0.25 μm (Fig. 3A, lane 3) and 0.5 μm (Fig. 3A, lane 4, and Fig. 3B, lane 3). The MMP-9 protein levels were reduced to a significant level in osteoclasts (by 92%) and in PC3 cells (by 71%) at a final concentration of 0.5 μm siRNA as compared with the levels of protein in cRNAi transfected cells (lane 2 in A and B) at the same concentration. Mock transfected (untreated) cells were also used as controls (lane 1 in A and B). We used 0.5 μm siRNA and cRNAi for the experiments shown in panels C–G.
To elucidate the role of MMP-9 in gelatin degradation, cells incubated with siRNA or cRNAi for 24 h were plated on FITC-gelatin matrix, and the incubation was continued for another 12 h. Cells were stained with rhodamine phalloidin for actin (Fig. 3, C–F). Osteoclasts plated on gelatin matrix for 12 h displayed podosome clusters at the membrane projection and peripheral podosomes in the clear zone area (Fig. 3C). Gelatin degradation (indicated by a black arrow in C′) was observed in the area exactly below the membrane protrusion where a cluster of podosomes (indicated by a white arrow in C) was observed. Degradation was not observed at the periphery where podosomes are present at the clear zone area. In osteoclasts treated with siRNA to MMP-9, a significant decrease in the degradation of gelatin matrix was observed under the area (Fig. 3D′, indicated by a black arrow) where a cluster of podosomes (Fig. 3D, indicated by a white arrow) is located. These osteoclasts neither show failure of formation of peripheral podosomes in the clear zone nor cluster of podosome at the membrane projection (Fig. 3D). It appears that MMP-9 present in the podosomes at the clear zone (6) is not required for cell adhesion. Nevertheless, MMP-9 is required for migration, because osteoclasts treated with siRNA to MMP-9 (data not shown) or a broad spectrum MMP inhibitor GM6001 are hypomotile (6).
Similar to the finding shown in osteoclasts, siRNA to MMP-9 considerably reduced the amount of matrix degradation (Fig. 3F′) as compared with cRNAi-transfected PC3 cells (Fig. 3E′). Apparently, prostate cancer cells treated with siRNA to MMP-9 had no changes in the formation of invadopodia (indicated by arrows) or F-actin content as compared with untreated or cRNAi-treated PC3 cells (Fig. 3G). These observations corroborate the possible role of MMP-9 activity in the migration of osteoclasts and prostate cancer cells (28, 29), although formation of invadopodia or podosomes does not require MMP-9 activity.
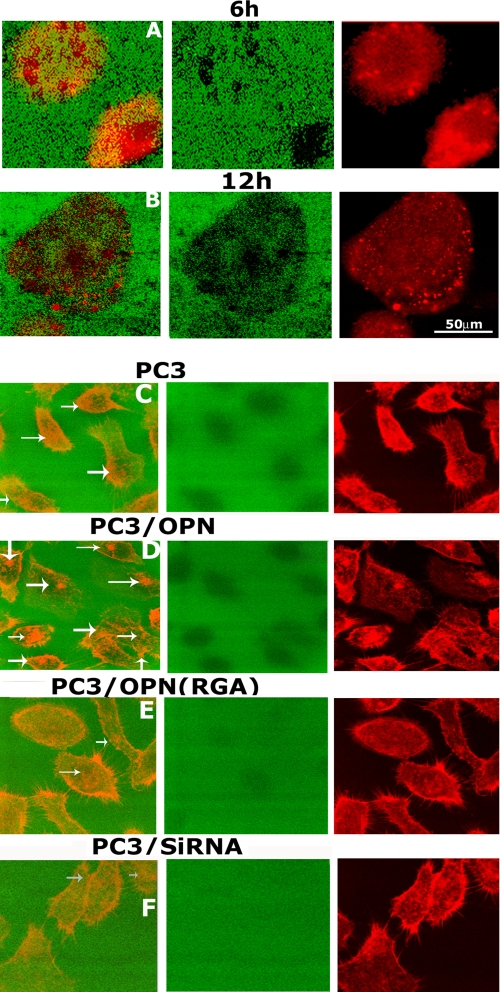
Modulation of Gelatin Degradation and Invasion by OPN/αvβ3 Signaling—Invadopodia have been shown to form a focal degradation on the gelatin matrix (reviewed in Refs. 5, 12). We have observed a time-dependent focal degradation of FITC-gelatin matrix incubated with PC3 cells (Fig. 4). Higher magnification images of PC3 cells revealed several distinctive invadopodia-like adhesive structures enriched in F-actin (Fig. 4, A and B, red). Focal degradation was observed at 4–6 h (Fig. 4, A and B, green) and continued incubation for 12–14 h increased the number and depth of degradation and not the area. Degradation is restricted within the dimension of a cell (Fig. 4B).
FIGURE 4.
Analysis of the effect of OPN expression in PC3 cells on the degradation of FITC-conjugated gelatin matrix. A and B, time-dependent degradation of FITC-gelatin matrix was observed in PC3 cells (A and B). Cells cultured on FITC-gelatin matrix for 6 h (A) and 12 h (B) were stained for actin with rhodamine phalloidin (red). Higher magnification view of images at the matrix level exhibits a time-dependent increase in the dark (non-fluorescent) areas of focal degradation of gelatin matrix below the cell (green panels) and not around the cells. Actin staining displayed actin-enriched punctate invadopodia-like structures at the center and periphery (redpanels). Merged images (A and B) demonstrate that focal degradation areas on the gelatin-matrix correspond with the actin-enriched invadopodia-like structures in PC3 cells. C–F, the effect of OPN expression on the degradation of FITC-gelatin matrix was analyzed in the following PC3 cell lines: PC3 (C), PC3/OPN (D), PC3/OPN (RGA) (E), and PC3/siRNA (F). The effects of OPN expression in the regulation of gelatin-degradation is shown in multiple cells at lower magnification. Cells were incubated for 12 h on the gelatin matrix. Merged images show FITC-gelatin matrix (green) and PC3 cells stained for actin (C–F). The degree of gelatin degradation in different PC3 cells lines are shown at the matrix level in green panels. Actin staining (red panels) displayed actin-enriched punctate invadopodia-like structures at the center and periphery (indicated by arrows in the merged panels). The results represent one of three experiments performed.
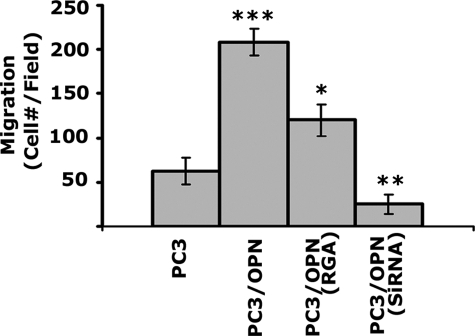
OPN plays a key role in the bone tropism and growth of prostate cancer cells. Integrin αvβ3 signaling pathway mediates OPN-associated androgen-independent growth of prostate cancer cells, a common characteristic feature found in advanced prostate cancer cells (30, 31). Despite the clinical importance of integrin αvβ3 and OPN, little is known about the mechanisms mediating prostate cancer cell progression and invasion. We have previously demonstrated the possible role of OPN/αvβ3 signaling in prostate cancer cell migration by using the following PC3 cell lines (23): PC3 cells lines that overexpress osteopontin (PC3/OPN), mutant OPN in the integrin binding-site (PC3/OPN (RGD ΔRGA)), and null for OPN (PC3/siRNA). These PC3 cell lines were used in the present study to investigate the role of OPN/integrin αvβ3 signaling in the regulation of degradation of matrix (Fig. 4, C–F) and invasion (Fig. 5). Untransfected PC3 cells were used as control. The effects of OPN expression on gelatin degradation was shown at lower magnification with multiple cells in Fig. 4 (C–F). Cells were stained with rhodamine phalloidin for actin. A higher magnification view of PC3 cell lines stained for WASP and MMP-9 displaying the number and location of invadopodia is shown in Fig. 6 (A–D). A decrease in the number of invadopodia in PC3/OPN (RGA) and PC3/siRNA cells corresponds with the substantial reduced level of degradation of gelatin matrix in these cells (Fig. 4, green panels of E and F). PC3 and PC3/OPN cells look smaller in the overlay images (Fig. 4, C and D), because they are present within the excavated gelatin matrix. Gelatin degradation and the number of invadopodia were observed in the following order: PC3/OPN > PC3 > PC3/OPN (RGA) > PC3/siRNA.
FIGURE 5.
The effects of OPN expression on the invasion of PC3 cells. Cell invasion was assessed in vitro using PC3 cell lines indicated at the bottom. Invasion assay with each cell line was performed in triplicates and counted. The data are mean ± S.E. of three separate experiments. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus PC3 cells.
FIGURE 6.
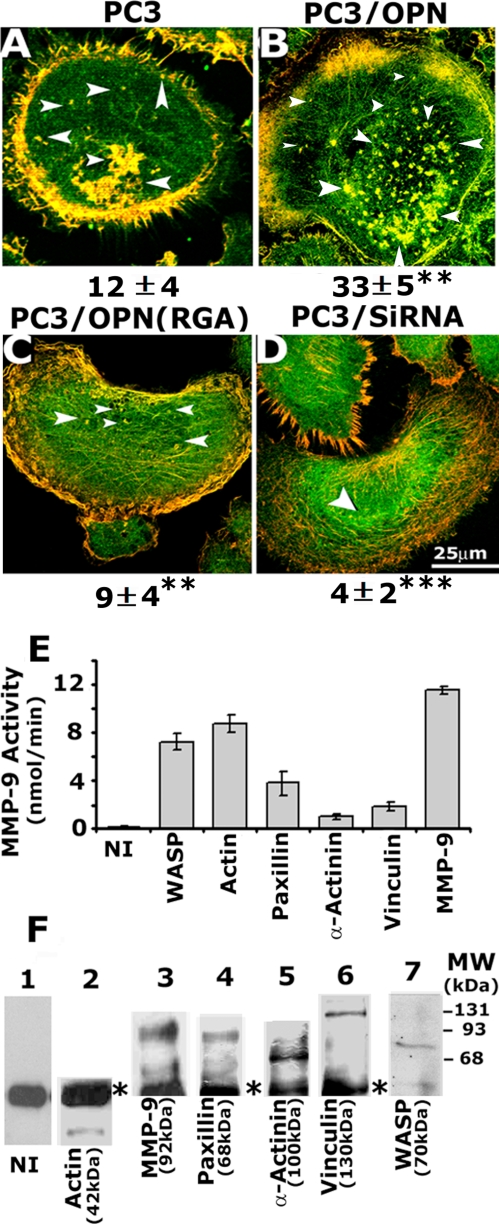
Analysis of association of WASP with MMP-9 in PC3 cell lines. A–D, confocal analysis of distribution of WASP (red) and MMP-9 (green) in PC3 cell lines. The results shown are representative of three independent experiments. The number of invadopodia is provided at the bottom. **, p < 0.01 versus PC3/cells; ***, p < 0.001 versus PC3 and PC3/OPN cells. The data are mean ± S.E. of >100 cells of two experiments. E and F, analysis of MMP-9 activity by colorimetric assay. Proteins indicated at the bottom were immunoprecipitated with the respective antibodies. The immune complexes were subjected to MMP-9 activity assay as described under “Experimental Procedures.” Assay was performed in triplicates, and the data presented as mean ± S.E. These results represent one of three separate experiments performed with the same results. To determine the precipitated protein levels in each immunoprecipitate, immunoblotting was performed with the same antibody indicated below each lane in F. Molecular weight of the proteins is indicated in parentheses. Asterisks indicate IgG heavy chain.
The ability of PC3 and PC3/OPN to degrade the gelatin matrix directly connects the invasive nature of these cells. Invasion into the ECM of the basement membrane is an important step in the process of metastasis. Because invasion involves degradation of and migration through ECM, we have used the standard invasion assay as described under “Experimental Procedures.” Invasion of PC3/OPN was significantly greater than that of PC3, PC3/OPN (RGA), and PC3/siRNA cells (Fig. 5). The above observations suggest that the OPN/αvβ3 signaling pathway is involved in the formation of invadopodia. It also corroborates with the previously described regulatory role of OPN/αvβ3 signaling pathway in the activation and secretion of MMP-9 (6, 23).
Analysis of Association of WASP with MMP-9 in PC3 Cell Lines—The WASP-Arp2/3 pathway has been shown to be necessary for the stabilization and maturation of invadopodia (4). Membrane-associated proteases, separase, and gelatinase A have been shown to localize in invadopodia (5, 12). To identify the components of invadopodia, PC3 cell lines were immunostained with antibodies to WASP and MMP-9. Immunostaining analyses revealed colocalization (yellow) of WASP (red) and MMP-9 (green) in punctate spots and clusters at the center and periphery. Colocalization was also observed at the periphery of the membrane (Fig. 6, A–C). The number of invadopodia per cell is provided at the bottom of each panel. The number of invadopodia exhibiting colocalization of WASP/MMP-9 was more in PC3/OPN cells (Fig. 6B), and a considerable reduction in the number of invadopodia is apparent in the immunostaining analysis of PC3/OPN (RGA) and PC3/siRNA cells.
We proceeded to determine whether WASP association with MMP-9 in immunostaining analysis (Fig. 6) construes as WASP-bound MMP-9 activity. We performed MMP-9 activity assay using immunoprecipitates made with a WASP-antibody. For comparison, the MMP-9 activity associated with other proteins including actin, paxillin, vinculin, and α-actinin were analyzed (Fig. 6E). Equal amount of lysate protein (200 μg) from PC3 cells were immunoprecipitated with antibodies to the proteins indicated above. MMP-9 activity was determined in the immunocomplexes pulled down with protein-A Sepharose beads as described under “Experimental Procedures.” An equal amount of protein when used for MMP-9 immunoprecipitation gave significantly higher levels of activity. Hence, an MMP-9 activity assay following MMP-9 immunoprecipitation was performed with a 10% (20 μg) protein amount compared with the other proteins. Among all the proteins examined, the highest MMP-9 activity was associated with actin, followed by WASP. Paxillin immunoprecipitates exhibited about half as much as WASP-associated MMP-9 activity. Vinculin and α-actinin demonstrated negligible MMP-9 activity (Fig. 6E). Western blots in Fig. 6F demonstrate the amount of proteins immunoprecipitated with the indicated antibodies. Because proteins such as MMP-9 and actin are abundant, blots exposed to x-ray films for shorter time (<30 s) are shown (Fig. 6F, lanes 2 and 3). Blots exposed for >5 min are shown for other proteins (lanes 1 and 4–7). Immunoblotting analysis with the indicated antibodies was provided to demonstrate the presence of proteins in the indicated immunoprecipitates. It is not used as a quantitative measure to compare the activity to the level of each protein. The MMP-9 activity seen may not be a result of differences in levels of the different proteins that were immunoprecipitated. It may depend on the specific interactions with other proteins and localization in a given cell. However, our observations indeed demonstrate that WASP associates with active MMP-9 either directly or through some adapter protein(s), thereby providing a functional aspect to its role in the degradation of matrix by means of invadopodia. These results suggest that WASP integrates MMP-9 activity in invadopodia downstream of integrin αvβ3 signaling, although the possible association has to be determined.
Effects of WASP and Gelsolin Peptides on the F-actin Content of PC3 Cells—Actin polymerization involves severing of preexisting actin filaments, uncapping of barbed ends, and actin nucleation. Gelsolin and WASP-Arp2/3 complex were shown to have a role in actin polymerization by uncapping and nucleation of actin filaments in osteoclasts (15, 25). Because localization of WASP was observed in invadopodia of PC3 cells (Fig. 6, A–D), we addressed the question whether WASP would have a role in actin polymerization in PC3 cells.
Central (C) and acidic (A) domains of VCA have a binding site for Arp2/3 complex (13). Therefore, we cloned FL-WASP and VCA domain of WASP in TAT-fused expression vector and purified the proteins as described previously (24). Gelsolin is a high affinity barbed end capping protein in osteoclasts (24). Transduction of TAT-fused gelsolin peptides, which contain full-length gelsolin (FL-Gsn, 98.5 kDa), and phosphoinositide binding domain (PBD, 13.5kDa) disrupted podosome organization in osteoclasts (32). PC3 cells were transduced with TAT-fused WASP and gelsolin peptides as indicated in Fig. 7. The Mid-GSN fragment did not contain any PBDs. Mid-GSN (36 kDa), HA-TAT (6–8 kDa, vector protein), and Hsv-TK (42 kDa, a nonspecific TAT-fused protein) were used as controls (32). F-actin content was determined as described previously (25). A decrease in the F-actin content was found in PC3 cells transduced with FL-WASP, VCA domain of WASP, FL-Gsn, and PBD of Gsn as compared with untransduced (-) or control peptides transduced cells (Fig. 7, A and B). These experiments suggest that transduced peptides can competitively inhibit the endogenous function of respective proteins. However, a significant decrease in the F-actin content with FL-WASP and VCA domain suggests the possible role of WASP in cytoskeletal structural organization in PC3 cells.
The Effects of Transduction of WASP Peptide on Actin Distribution in PC3 Cells—A significant decrease in the levels of F-actin in PC3 cells transduced with FL-WASP (Fig. 7A) prompted subsequent analysis of the effects of this peptide on the distribution of invadopodia (Fig. 8). The uptake of TAT-fused WASP proteins was determined by immunoblotting with an antibody to HA (Fig. 8A). ∼200 μg of osteoclast lysate was used for immunoblotting analysis. Immunoblotting with an antibody to HA demonstrates the levels of transduced peptides such as Hsv-TK (42 kDa, lane 2) and FL-WASP (∼80 kDa, lane 3). Untransduced (-) and Hsv-TK protein-transduced cells were used as controls. Cells stained for actin with rhodamine phalloidin (Fig. 8B) exhibited a decrease in the number of invadopodia in cells transduced with FL-WASP peptide for 12 h. Clusters of invadopodia were observed in untransduced (-) or Hsv-TK-transduced cells (Fig. 8B, indicated by arrows). A decrease in the number of invadopodia corresponds with a considerable reduced level of degradation of gelatin matrix despite no changes in the total MMP-9 activity was observed in cells transduced with FL-WASP as compared with control cells (data not shown). The ability of FL-WASP to reduce the number of invadopodia and F-actin content suggests a possible role for WASP in the regulation of actin dynamics and invadopodia formation in prostate cancer cells.
DISCUSSION
Cell migration through the ECM is necessary for cancer cells to invade adjacent tissues and metastasize to an organ distant from primary tumors. Because podosomes and invadopodia have been linked with invasion of cancer cells, we evaluated the adhesive structure involved in the invasion of prostate cancer cells. To determine the composition of these structures, cells were stained for actin and vinculin. PC3 cells displayed distribution of actin and vinculin in punctate structures at the center and periphery in the filopodia-like extensions (Fig. 1). Actin-enriched adhesive structures present in PC3 cells neither look like nor function like enlarged podosomes in migration or matrix degradation. Both general structure and function of podosomes and invadopodia draw our attention to clarify some issues related to cell migration/invasion process. The function of podosomes was also speculated to provide local anchorage and thus stabilize membrane protrusions, which might allow more efficient directional migration (12). Our observations in osteoclasts concur with this hypothesis that podosomes, which are present as clusters or rosettes at the membrane projections, may provide directional movement as well as degradation of the underlying matrix during migration (Figs. 2 and 3). This type of podosome organization in osteoclasts corroborates with the previously described rosettes of podosome arrangement in endothelial and mouse NIH3T3 cells (33, 34). The rosettes of podosomes seem to emerge from the clusters (34). We have observed both rosettes and clusters of podosomes at the migration front. Podosomes present in these clusters or rosettes are capable of degrading gelatin matrix (Figs. 2 and 3). Degradation of matrix by podosomes was shown to be confined to areas corresponding to both size and location of podosomes (reviewed in Refs. 5, 12, 35, 36). However, our observations in osteoclasts exhibited cell migrations with a track formed by the degradation of matrix (Fig. 2).
PC3 cells also displayed punctate (Figs. 1 and 4) and clusters (Figs. 2 and 8) of actin enriched invadopodia-like structures. These structures neither present at the migration front nor function in the directed migration of cells (Fig. 2). Gelatin degradation assay indeed demonstrated polarized and diffused degradation of matrix below the cell (Figs. 2, 3, 4). There are no sign of directed migration in PC3 cells as observed in osteoclasts (Fig. 2). PC3 cells subjected to gelatin degradation assay demonstrated that actin-rich adhesive structures correspond with the focal degradation area of matrix (Fig. 4B). Degradation was confined within the cell boundary, and cells were found within the degraded matrix (Figs. 2, 3, 4). In the gelatin degradation assay, we have shown the ability of PC3 cells to invade the matrix, which is in large part dependent on the formation of invadopodia. This is different from the migratory phenotype observed in osteoclasts, which utilize podosomes for migration.
Subsequently, we carried out experiments to identify the nature of the adhesive structures present in PC3 cells. PC3 cells cultured on the FITC-gelatin matrix for 12–14 h were stained for actin and subjected to XZ scanning by confocal microscopy. In the lateral XZ scan view of the gelatin matrix, actin staining was observed in the membrane protrusions. Periodic membrane protrusions were observed from the ventral surface of the cell into the matrix.3 These observations corroborate the findings by others on the specifications of invadopodia (4, 5). Invadopodia are considered to be important for cell invasion and metastasis in the majority of studies performed in cancer cell lines (20, 37, 38). Based on the gelatin degradation assay (Figs. 2, 3, 4) and the distribution of actin/vinculin (Fig. 1), we consider that the adhesive structures present in PC3 cells (Figs. 1 and 6) are invadopodia and have a role in the process of cell invasion and metastasis processes. We were unable to detect focal adhesion sites in PC3 cells. It is possible that these cells may use focal adhesions for migration. It will be of interest to determine the role of focal adhesions in prostate cancer cell migration.
Invadopodia have been shown to exhibit an association of integrins along with membrane type-1 matrix metalloproteinase (MT1-MMP) (20), and not just active enzymes like MMP-2 and MMP-9 alone (39). We have shown here localization of MMP-9 in invadopodia of PC3 cells. Although both MMP-9 and MMP-2 are secreted by PC3 cells, only the secretion of MMP-9 is regulated by the integrin αvβ3 signaling mechanism. Inhibition of MMP-9 activity with a broad spectrum MMP-9 inhibitor, GM6001, reduced the ability of PC3 cells to migrate (23). Reducing the proteolytic activity in invadopodia with a MMP-9 and MMP-2 inhibitor, batimastat (BB-94 (40)) decreases invasion indicating that breast cancer cell invasion is dependent upon proteolytically active invadopodia (38). Therefore, to further elucidate the regulation of matrix degradation and the role of MMP-9 in this process, we used the siRNA strategy to reduce the endogenous levels of MMP-9. Reducing the endogenous levels of MMP-9 significantly lowered the ability of PC3 cells and osteoclast to degrade the gelatin matrix (Fig. 3). These observations also provide evidence for the non-redundant function of MMP-9 in ECM degradation.
Osteoclasts treated with a MMP inhibitor, KB-R7785, demonstrated an increase in the life span of podosomes (41). Similarly, knockdown of MT1-MMP in transforming growth factor-β-treated endothelial cells reduced matrix degradation, whereas the number of podosome-containing cells was increased (reviewed in 12, 33). Formation of podosomes does not necessarily seem to be dependent on the expression or activity of MT1-MMP and MMP-9 (33). Our data corroborate the finding by others that inhibition of MMP-9 activity had no effect in the formation, localization, or number of podosomes in osteoclasts or invadopodia in PC3 cells. It is possible that the presence of invadopodia and podosomes in siRNA-treated cells may be due to an increase in the in the stability and decrease in the turnover rate of podosomes and invadopodia (Fig. 3). Although the above-mentioned studies have provided critical information about the localization and function of MMPs in matrix degradation and migration/invasion processes, further studies will determine the exact mechanism by which MMP-9 regulates the function of podosomes and invadopodia.
WASP is known to have a regulatory role in actin polymerization. However, the findings of WASP-associated MMP-9 activity (Fig. 6E) and colocalization of WASP with MMP-9 (Fig. 6) are interesting and not expected. A study by Sossey-Alaoui (42) provided a link between WAVE-3 (a member of the WASP/WAVE family) expression and the regulation of expression of MMPs (MMP-1, -3, and -9) in human neuroblastoma cells. They reported that knockdown of WAVE3 expression inhibited the expression levels of MMP-1, -3, and -9 but not MMP-2 (42). Because experiments with siRNA to MMP-9 suggest the non-redundant role of MMP-9 in gelatin degradation assay (Fig. 3), we envision that inhibition of interaction of WASP and MMP-9 would have the potential to modulate invadopodia function. The above findings provide interesting insights into the role of WASP in the regulation of MMP-9 expression, localization, or activity in invadopodia.
Phosphoinositide-binding proteins such as WASP and gelsolin have been shown to play key roles in the regulation of formation of podosomes and invadopodia (8, 13, 15, 16). Given that podosomes and invadopodia share many of the properties, some of the facts in the formation of podosomes could be related to invadopodia formation. We found that transduction of gelsolin and WASP peptides reduced the F-actin content in PC3 cells (Fig. 7). Transduction of PBD of gelsolin not only blocked the interaction of phosphatidylinositol 4,5-bisphosphate (PIP2) with gelsolin but also with ezrin and WASP in osteoclasts (32). A decrease in the F-actin content in cells transduced with PBD suggests a role for PIP2-binding proteins in actin polymerization in PC3 cells. One possible reason for a decrease in F-actin content in PC3 cells transduced with FL-Gsn and PBD may be due to their ability to bind PIP2 and competitively block the binding of PIP2 with WASP as shown previously (32). Because FL-WASP peptide reduced the F-actin content, experiments were performed to determine the number of invadopodia by rhodamine phalloidin staining for actin (Fig. 8). Reduced actin staining and the number of invadopodia accentuate the potential role of WASP in the dynamics of actin filament during invadopodia formation.
Our observations in PC3/OPN (RGA) and PC3/siRNA cells suggest that the various signaling steps involved in the regulation of formation of invadopodia, localization, and activity of MMP-9 are down-regulated due to reduced αvβ3 signaling. Several studies show that integrins play a role in signal transduction that promotes invadopodia activities in cancer cells (37, 43, 44). Although integrin αvβ3 is critical in the activation of signaling pathways that regulate actin cytoskeletal remodeling toward invadopodia formation, it is not known at this time how it regulates this process. The findings from this study are our first step to identify the type of invasive structures present in PC3 cells and the possible regulatory mechanism involved in the regulation of invasion. These results should advance experiments aimed at testing the role of other potential actin-binding/regulatory proteins in actin dynamics besides WASP-Arp2/3 complex.
Conclusions—The main conclusions of this study are 1) activation of αvβ3 integrin stimulates the formation of invadopodia and invasion of PC3 cells; 2) the WASP-Arp2/3-mediated pathway may contribute to the formation of invadopodia as the number of invadopodia is decreased in cells transduced with FL-WASP peptide; and 3) formation of invadopodia is coordinated with the localization of MMP-9 and focal degradation of matrix during invasion process.
Acknowledgments
We thank Dr. Michael Kiefer for OPN cDNA and Dr. Stefan Linder (Institut fuer Prophylaxe und Epidemiologie der Kreislaufkrankheiten, Ludwig-Maximilians-Universitaet, Muenchen, Germany) for WASP cDNA and VCA domain of WASP. We also thankfully acknowledged the technical assistance of Dr. Samanna Venkatesababa (Gazes Cardiac Research Institute at Medical University of South Carolina, Charleston, SC) in the preparation of FITCgelatin matrix and Drs. Chandra M. Khatwal and Peter Swaan (University of Maryland, Baltimore, MD) in the measurement of MMP-9 activity using the enzyme-linked immunosorbent assay reader.
This work was supported, in whole or in part, by National Institutes of Health Grant AR46292 (to M. A. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ECM, extracellular matrix; WASP, Wiskott-Aldrich syndrome protein; MMP-9, matrix metalloproteinase-9; RGA, arginine-glycine-alanine amino acid sequences (this is a mutated version of integrin binding sequences RGD; D is aspartic acid); PC3, prostate cancer cell; OPN, osteopontin; siRNA, small interference RNA; PTP, protein-tyrosine phosphatase; cRNAi, control RNA interference; PIP2, phosphatidylinositol 4,5-disphosphate; F-actin, filamentous actin; VCA, verpolin, cofilin, acidic domain; FL, full-length; HA, hemagglutinin; GM6001, a broad spectrum inhibitor to MMPs; Gsn, gelsolin; PBD, phosphoinositide binding domain; FITC, fluorescein isothiocyanate; Hsv-TK, herpes simplex virus-thymidine kinase.
M. A. Chellaiah, unpublished observations.
References
- 1.Chellaiah, M., Kizer, N., Silva, M., Alvarez, U., Kwiatkowski, D., and Hruska, K. A. (2000) J. Cell Biol. 148 665-678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Linder, S., and Aepfelbacher, M. (2003) Trends Cell Biol. 13 376-385 [DOI] [PubMed] [Google Scholar]
- 3.Burridge, K., and Chrzanowska-Wodnicka, M. (1996) Annu. Rev. Cell Dev. Biol. 12 463-518 [DOI] [PubMed] [Google Scholar]
- 4.Yamaguchi, H., Pixley, F., and Condeelis, J. (2006) Eur. J. Cell Biol. 85 213-218 [DOI] [PubMed] [Google Scholar]
- 5.Buccione, R., Orth, J. D., and McNiven, M. A. (2004) Nat. Rev. Mol. Cell. Biol. 5 647-657 [DOI] [PubMed] [Google Scholar]
- 6.Samanna, V., Ma, T., Mak, T. W., Rogers, M., and Chellaiah, M. A. (2007) J. Cell. Physiol. 213 710-720 [DOI] [PubMed] [Google Scholar]
- 7.Moreau, V., Tatin, F., Varon, C., Anies, G., Savona-Baron, C., and Genot, E. (2006) Eur. J. Cell Biol. 85 319-325 [DOI] [PubMed] [Google Scholar]
- 8.Calle, Y., Jones, G. E., Jagger, C., Fuller, K., Blundell, M. P., Chow, J., Chambers, T., and Thrasher, A. J. (2004) Blood 103 3552-3561 [DOI] [PubMed] [Google Scholar]
- 9.Marchisio, P. C., Cirillo, D., Teti, A., Zambonin-Zallone, A., and Tarone, G. (1987) Exp. Cell Res. 169 202-214 [DOI] [PubMed] [Google Scholar]
- 10.Zambonin, Z. A., Teti, A., Gaboli, M., and Marchisio, P. C. (1989) Connect. Tissue Res. 20 143-149 [DOI] [PubMed] [Google Scholar]
- 11.Kanehisa, J., Yamanaka, T., Doi, S., Turksen, K., Heersche, J. N., Aubin, J. E., and Takeuchi, H. (1990) Bone 11 287-293 [DOI] [PubMed] [Google Scholar]
- 12.Linder, S. (2007) Trends Cell Biol. 17 107-117 [DOI] [PubMed] [Google Scholar]
- 13.Linder, S., Nelson, D., Weiss, M., and Aepfelbacher, M. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 9648-9653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rohatgi, R., Nollau, P., Ho, H. Y., Kirschner, M. W., and Mayer, B. J. (2001) J. Biol. Chem. 276 26448-26452 [DOI] [PubMed] [Google Scholar]
- 15.Chellaiah, M. A. (2005) J. Biol. Chem. 280 32930-32943 [DOI] [PubMed] [Google Scholar]
- 16.Yamaguchi, H., Lorenz, M., Kempiak, S., Sarmiento, C., Coniglio, S., Symons, M., Segall, J., Eddy, R., Miki, H., Takenawa, T., and Condeelis, J. (2005) J. Cell Biol. 168 441-452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gimona, M., and Buccione, R. (2006) Int. J. Biochem. Cell Biol. 38 1875-1892 [DOI] [PubMed] [Google Scholar]
- 18.Bowden, E. T., Coopman, P. J., and Mueller, S. C. (2001) Methods Cell Biol. 63 613-627 [DOI] [PubMed] [Google Scholar]
- 19.Festuccia, C., Guerra, F., D'Ascenzo, S., Giunciuglio, D., Albini, A., and Bologna, M. (1998) Int. J. Cancer 75 418-431 [DOI] [PubMed] [Google Scholar]
- 20.Artym, V. V., Zhang, Y., Seillier-Moiseiwitsch, F., Yamada, K. M., and Mueller, S. C. (2006) Cancer Res. 66 3034-3043 [DOI] [PubMed] [Google Scholar]
- 21.Courtneidge, S. A., Azucena, E. F., Pass, I., Seals, D. F., and Tesfay, L. (2005) Cold Spring Harbor Symp. Quant. Biol. 70 167-171 [DOI] [PubMed] [Google Scholar]
- 22.Kiefer, M. C., Bauer, D. M., and Barr, P. J. (1989) Nucleic Acids Res. 17 3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desai, B., Rogers, M. J., and Chellaiah, M. A. (2007) Mol. Cancer 6 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chellaiah, M. A., Soga, N., Swanson, S., McAllister, S., Alvarez, U., Wang, D., Dowdy, S. F., and Hruska, K. A. (2000) J. Biol. Chem. 275 11993-12002 [DOI] [PubMed] [Google Scholar]
- 25.Chellaiah, M., and Hruska, K. (1996) Mol. Biol. Cell 7 743-753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chellaiah, M. A., Kuppuswamy, D., Lasky, L., and Linder, S. (2007) J. Biol. Chem. 282 10104-10116 [DOI] [PubMed] [Google Scholar]
- 27.Chen, W. T. (1996) Enzyme Protein 49 59-71 [DOI] [PubMed] [Google Scholar]
- 28.Ishibashi, O., Niwa, S., Kadoyama, K., and Inui, T. (2006) Life Sci. 79 1657-1660 [DOI] [PubMed] [Google Scholar]
- 29.Incorvaia, L., Badalamenti, G., Rini, G., Arcara, C., Fricano, S., Sferrazza, C., Di, T. D., Gebbia, N., and Leto, G. (2007) Anticancer Res. 27 1519-1525 [PubMed] [Google Scholar]
- 30.Thalmann, G. N., Sikes, R. A., Devoll, R. E., Kiefer, J. A., Markwalder, R., Klima, I., Farach-Carson, C. M., Studer, U. E., and Chung, L. W. (1999) Clin. Cancer Res. 5 2271-2277 [PubMed] [Google Scholar]
- 31.Cooper, C. R., Chay, C. H., and Pienta, K. J. (2002) Neoplasia 4 191-194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biswas, R. S., Baker, D., Hruska, K. A., and Chellaiah, M. A. (2004) BMC Cell Biol. 5 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varon, C., Tatin, F., Moreau, V., Van Obberghen-Schilling, E., Fernandez-Sauze, S., Reuzeau, E., Kramer, I., and Genot, E. (2006) Mol. Cell. Biol. 26 3582-3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collin, O., Tracqui, P., Stephanou, A., Usson, Y., Clement-Lacroix, J., and Planus, E. (2006) J. Cell Sci. 119 1914-1925 [DOI] [PubMed] [Google Scholar]
- 35.Burgstaller, G., and Gimona, M. (2005) Am. J. Physiol. 288 H3001-H3005 [DOI] [PubMed] [Google Scholar]
- 36.Mizutani, K., Miki, H., He, H., Maruta, H., and Takenawa, T. (2002) Cancer Res. 62 669-674 [PubMed] [Google Scholar]
- 37.Mueller, S. C., Ghersi, G., Akiyama, S. K., Sang, Q. X., Howard, L., Pineiro-Sanchez, M., Nakahara, H., Yeh, Y., and Chen, W. T. (1999) J. Biol. Chem. 274 24947-24952 [DOI] [PubMed] [Google Scholar]
- 38.Kelly, T., Yan, Y., Osborne, R. L., Athota, A. B., Rozypal, T. L., Colclasure, J. C., and Chu, W. S. (1998) Clin. Exp. Metastasis 16 501-512 [DOI] [PubMed] [Google Scholar]
- 39.Abecassis, I., Olofsson, B., Schmid, M., Zalcman, G., and Karniguian, A. (2003) Exp. Cell Res. 291 363-376 [DOI] [PubMed] [Google Scholar]
- 40.Low, J. A., Johnson, M. D., Bone, E. A., and Dickson, R. B. (1996) Clin. Cancer Res. 2 1207-1214 [PubMed] [Google Scholar]
- 41.Goto, T., Maeda, H., and Tanaka, T. (2002) J. Bone Miner. Metab. 20 98-105 [DOI] [PubMed] [Google Scholar]
- 42.Sossey-Alaoui, K., Ranalli, T. A., Li, X., Bakin, A. V., and Cowell, J. K. (2005) Exp. Cell Res. 308 135-145 [DOI] [PubMed] [Google Scholar]
- 43.Nakahara, H., Mueller, S. C., Nomizu, M., Yamada, Y., Yeh, Y., and Chen, W. T. (1998) J. Biol. Chem. 273 9-12 [DOI] [PubMed] [Google Scholar]
- 44.Gimona, M. (2008) Semin. Cancer Biol. 18 23-34 [DOI] [PubMed] [Google Scholar]