Abstract
The PhoP-PhoQ two-component system is a well studied bacterial signaling system that regulates virulence and stress response. Catalytic activity of the histidine kinase sensor protein PhoQ is activated by low extracellular concentrations of divalent cations such as Mg2+, and subsequently the response regulator PhoP is activated in turn through a classic phosphotransfer pathway that is typical in such systems. The PhoQ sensor domains of enteric bacteria contain an acidic cluster of residues (EDDDDAE) that has been implicated in direct binding to divalent cations. We have determined crystal structures of the wild-type Escherichia coli PhoQ periplasmic sensor domain and of a mutant variant in which the acidic cluster was neutralized to conservative uncharged residues (QNNNNAQ). The PhoQ domain structure is similar to that of DcuS and CitA sensor domains, and this PhoQ-DcuS-CitA (PDC) sensor fold is seen to be distinct from the superficially similar PAS domain fold. Analysis of the wild-type structure reveals a dimer that allows for the formation of a salt bridge across the dimer interface between Arg-50′ and Asp-179 and with nickel ions bound to aspartate residues in the acidic cluster. The physiological importance of the salt bridge to in vivo PhoQ function has been confirmed by mutagenesis. The mutant structure has an alternative, non-physiological dimeric association.
Cell survival in a changing environment requires constant adaptation. In bacteria and some lower eukaryotes, two-component signaling systems couple extracellular stimuli to an intercellular adaptive response (1–3). Such systems allow a signal to be transduced across the cell membrane by employing a set of two proteins, a histidine kinase sensor and a response regulator, which interact in tandem. The typical histidine kinase is a transmembrane protein containing an N-terminal extracellular sensor domain and a C-terminal cytoplasmic kinase transmitter domain. The activity of the cytoplasmic catalytic domain is modulated by a signal transduced across the membrane from the sensor domain. In its active state, the histidine kinase sensor phosphorylates a conserved histidine residue in the cytoplasmic domain, and subsequently the phosphoryl group is transferred from the histidine kinase to a conserved aspartate residue in the N-terminal receiver domain of the cytoplasmic response regulator (4, 5). Receiver domain phosphorylation, in turn, activates the C-terminal effector domain, which mediates an adaptive response, usually by either modulating transcription or flagella rotation. In some cases, sensor stimulation promotes dephosphorylation.
The PhoP-PhoQ regulatory pair is a widely studied two-component system that has been implicated in the virulence of several Gram-negative plant and animal pathogens, including Salmonella typhimurium, Pseudomonas aeruginosa, Neisseria meningitidis, and Erwinia chrysanthemi. It appears that PhoQ may be a sensor for periplasmic concentrations of divalent cations. Both in vivo and in vitro experiments have shown that the PhoQ sensor is modulated by extracellular levels of Mg2+ and Ca2+, where low concentrations promote activation and high concentrations result in repression (6–8). The role of divalents as a signal for PhoQ is also supported by crystal structure of the S. typhimurium PhoQ sensor domain, which show Ca2+ binding sites on an acidic surface of the protein (9). It has also been suggested that host antimicrobial peptides may also serve as a signal for PhoQ, and that the competition between divalent ions and such peptides may be a means by which the invading cell can determine its subcellular environment during the infection cycle (10). Transcription of specific virulence factors is then regulated accordingly.
PhoQ is also found in non-pathogenic Gram-negative bacteria such as Escherichia coli where it controls a regulon that includes several of the same genes that are regulated in S. typhimurium and many that are not (10, 11). The role of PhoQ in E. coli appears to be to regulate a physiological response to Mg2+ starvation and is involved in acid resistance but not virulence (8). Previous experiments on soluble sensor domain constructs of E. coli PhoQ have suggested a possible direct binding of divalent cations to a cluster of acidic amino acid residues in the sensor domain. In the presence of divalents, higher thermal stability is conferred in wild-type constructs but not in mutant constructs that lack the acidic cluster (12).
We have crystallized the E. coli PhoQ periplasmic sensor domain, and we present the structures of a wild-type and a response-impaired acidic cluster mutant at atomic level resolution. Analysis of the structures reveals potential ligand binding sites and dimeric arrangements of subunits. The biological relevance and importance of the dimer interface in the wild-type structure is supported by mutational analysis and provides clues toward understanding transmembrane signaling by this molecule.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids—pLPQ2 (12) is a pSC101-derived plasmid in which expression of the E. coli phoP-phoQ operon is driven by the lacUV5 promoter. pNL3 (12) is a pBR322-derived reporter plasmid for assaying PhoP-mediated transcriptional activation that contains the phoN promoter fused to lacZ. pAED4Q (12) is a pUC19-derived plasmid in which the PhoQ sensor domain (residues 43–190) is expressed from the phage T7 φ10 promoter (13). E. coli strain CSH26ΔQ has an internal deletion in the chromosomal phoQ gene, which results in a null phenotype (12). E. coli strains BL21(DE3) and B834(DE3) were used to express PhoQ for purification.
β-Galactosidase Assays—β-Galactosidase assays were performed as described (14) in N-media with additions of MgCl2 as indicated using strain CSH26ΔQ/F′ lacIQkan/pNL3 containing derivatives of pLPQ2 with wild-type phoQ or mutant variants.
Construction of phoQ Mutations—Variants of PhoQ in which single amino acids at residues 50, 54, or 179 were individually substituted were constructed by site-directed PCR mutagenesis. Primers were synthesized in which the appropriate codons were substituted to produce mutant phoQ variants as indicated, and the resulting amplified phoQ fragments were then cloned into a plasmid vector using nearby restriction enzyme sites. Details are given as supplemental data.
Preparation of Cell Membranes and Western Blotting—E. coli CSH26ΔQ/F′ lacIQkan/pNL3 strains containing derivatives of pLPQ2 with wild-type phoQ or mutant variants were grown to mid-log phase in N-media with appropriate antibiotics as described previously (14) and then induced for 2 h at 37 °C with the addition of 1 mm isopropyl-β-d-thiogalactopyranoside. Cells were harvested by centrifugation, resuspended in resuspension buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl), and lysed by sonication on ice. Cell debris was removed by centrifugation at 14,000 × g for 5 min. Cell membranes were recovered from the supernatant by centrifugation at 100,000 × g for 15 min and were washed once with resuspension buffer. Proteins from equal amounts of membrane were separated by SDS-PAGE and were subsequently transferred to Immobilon-P transfer membrane (Millipore). Western analysis was performed using an antibody with specificity for the E. coli PhoQ cytoplasmic domain as described previously (14).
Protein Purification and Crystallization—The E. coli PhoQ wild-type sensor domain construct (residues 43–190), and the acid mutant construct (residues 43–190) in which residues 148–154 were changed from EDDDDAE to QNNNNAQ were expressed and purified from E. coli strain X90(DE3) as previously described (12). Selenomethionyl (SeMet)5 PhoQ 43–190 (wild type) was expressed from cells grown in selenomethionine minimal media, and native PhoQ 43–190 (acid mutant) was expressed from cells grown in Luria-Bertani (LB) media. Crystals of SeMet PhoQ 43–190 (wild-type) protein were grown by the hanging drop, vapor diffusion method in a buffer containing polyethylene glycol 3400, Tris (pH 8.5), sodium acetate, and 5 mm NiCl2. Crystals of the native PhoQ 43–190 (acid mutant) protein were grown by the same method in a buffer containing 1.8 m ammonium sulfate, polyethylene glycol 4000, and sodium acetate. The ratio of protein mixed to reservoir buffer was 1:1.
Data Collection and Structure Determination—Data from a four-wavelength multiwavelength anomalous dispersion experiment at the selenium K-edge were collected from a single frozen SeMet wild-type PhoQ 43–190 crystal at beamline X4A of the National Synchrotron Light Source at Brookhaven National Laboratory. Diffraction to Bragg spacings of 2.5 Å (280-mm detector distance) was collected using 1.5° rotations. Data from a single frozen crystal of native acid mutant PhoQ 43–190 were collected at a home CuKα radiation source with a Raxis-IV detector. The native crystals diffracted to Bragg spacings of 2.0 Å (140-mm detector distance), and data were collected using 1° rotations. Denzo and Scalepack of the HKL program suite (15) were used to index and merge all data sets.
Selenium sites were found from the multiwavelength anomalous dispersion data after inspection of Harker sections of Bijvoet- and dispersive-difference Patterson maps generated using CCP4 (16), and they were refined in SHARP (17) in space group P21. A partial model of SeMet PhoQ 43–190 (wild type) was generated by automatic model building using Arp/wArp (18), and this model was used in a molecular replacement search to determine phases of the native PhoQ 43–190 (acid mutant) in space group P41. The rest of the native PhoQ 43–190 (acid mutant) model was built manually in O (19), and the completed model was refined to a resolution of 2.0 Å using CNS (20) for iterative cycles of simulated annealing, conjugate gradient energy minimization, temperature-factor refinement, and manual rebuilding. An additional density feature was found to be best fit with a sulfate ion, and this was included in the model and refined as well.
The complete native PhoQ 43–190 (acid mutant) model aided the manual rebuilding of the SeMet PhoQ 43–190 (wild type) model, which was then refined by the same methods as before to a resolution of 2.5 Å against the low energy remote dataset. Three locations of additional electron density could be best fit with one acetate and two nickel ions, and these were added to the model and included in the refinement. Data collection and refinement statistics for both structures are listed in Tables 1 and 2. The atomic coordinates and structure factors for both wild-type and acid mutant E. coli PhoQ sensor domains have been deposited with the Protein Data Bank as accession codes 3BQ8 and 3BQA, respectively.
TABLE 1.
PhoQ 43–190 diffraction data
| Derivative | dmin | Wavelength | No. of reflections | Average redundancy | 〈I〉/(σ) | Completenessa | Rmergeb |
|---|---|---|---|---|---|---|---|
| Å | % | % | |||||
| SePhoQ wt λ1 | 2.5 | 0.9879 (low remote) | 10480 | 3.8 | 18.6 | 95.2 (93.2) | 6.1 (26.5) |
| SePhoQ wt λ2 | 2.5 | 0.9793 (edge) | 10401 | 3.7 | 19.4 | 94.6 (92.6) | 6.2 (25.4) |
| SePhoQ wt λ3 | 2.5 | 0.9790 (peak) | 10460 | 3.7 | 18.6 | 95.0 (93.3) | 7.0 (27.9) |
| SePhoQ wt λ4 | 2.5 | 0.9641 (high remote) | 10539 | 3.7 | 19.7 | 95.4 (94.3) | 6.5 (23.4) |
| PhoQ mutant | 2.0 | 1.5418 | 19615 | 3.3 | 22.4 | 99.0 (99.3) | 3.5 (8.9) |
Values in outermost shell are given in parentheses
Rmerge = (Σ|Ii – 〈Ii〈|)/Σ|Ii|, where Ii is the integrated intensity of a given reflection
TABLE 2.
PhoQ 43–190 refinement statistics
| Parameter | Wild-type PhoQ | Acidic mutant PhoQ |
|---|---|---|
| Bragg spacings (Å) | 20-2.5 | 20-2.0 |
| Space group | P21 | P41 |
| Cell parameters | 33.59, 113.96, 45.53 | 44.42, 44.42, 151.86 |
| a, b, c (Å)/α, β, γ(°) | 90, 109.46, 90 | 90, 90, 90 |
| Ra | 20.3% | 16.8% |
| Rfreeb | 30.1% | 23.0% |
| Number of reflections | 10,255 | 19,573 |
| Number of total atoms (non-hydrogen) | 2,421 | 2,829 |
| Number of protein atoms | 2,233 | 2,488 |
| Number of ligand atoms | 6 (1 acetate molecule, 2 nickel ions) | 5 (1 sulfate ion) |
| Number of waters | 184 | 336 |
| Average B factor | 44.8 | 27.3 |
| r.m.s. bonds (Å) | 0.009 | 0.007 |
| r.m.s. angles (°) | 1.4 | 1.3 |
R = (Σ||Fo| – |Fc||)/Σ|Fo|, where Fo and Fc denote observed and calculated structure factors, respectively
Rfree was calculated using 5% of data excluded from refinement
RESULTS
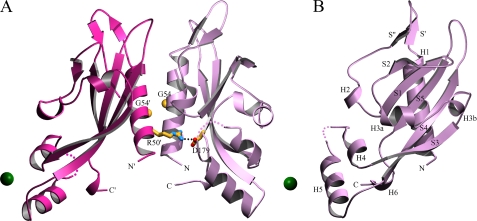
Overall Structure of PhoQ 43–190—The structure of wild-type PhoQ 43–190 (Fig. 1) was determined by multiwavelength anomalous dispersion from a single SeMet crystal at the selenium K-edge. Two molecules were found forming an apparent dimer in the asymmetric unit in space group P21. Clear electron density allowed residues to be traced in for residues 45–134 and 137–188 of molecule A and for residues 45–75, 83–135, and 138–186 of molecule B. Missing residues correspond to N- and C-terminal segments and to loops. A total of 275 ordered residues, 184 water molecules, 2 nickel ions, and 1 acetate ion was refined against the low remote data to a resolution of 2.5 Å with R and Rfree values of 20.3% and 30.1%, respectively (Tables 1 and 2).
FIGURE 1.
Structure of the wild-type PhoQ 43–190 dimer in complex with nickel. A, the overall structure in complex with nickel is shown as a ribbon diagram. Nickel ions are shown as green spheres. The Arg-50′ → Asp-179 salt bridge is shown with residue side chains depicted in a stick representation (carbon colored yellow, nitrogen colored blue, and oxygen colored red). A line of black dots marks interacting atoms of the salt bridge. The Cα atoms of Gly-54 residues are shown as yellow spheres. Residues are identified by one-letter-code labels. Colored dots are drawn to represent a hypothetical path of the protein backbone through disordered regions corresponding to residues 135 and 136 of molecule A (light purple), and residues 136 and 137 of molecule B (light red). B, the structure of one subunit (molecule A) of the wild-type PhoQ 43–190 dimer is shown from a 90° rotation about the vertical axis of the dimer as shown in A. Secondary structure elements are labeled in black. The diagram was created using MolScript (43).
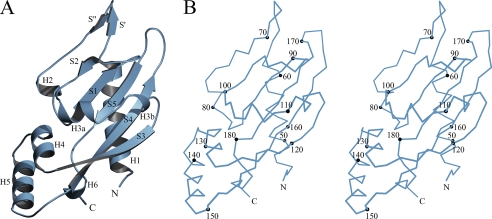
The structure of acidic cluster mutant PhoQ 43–190 (Fig. 2) was determined by molecular replacement starting from a partial model of wild-type PhoQ 43–190. Two molecules were found in the asymmetric unit in space group P41. As for the wild-type structure, these molecules are related by a quasi-diad axis of symmetry, but the arrangement is very different in this case (see below). Residues at positions 43–188 and 45–190 of molecule A and B, respectively, were found to be ordered. A total of 292 residues, 336 water molecules, and 1 sulfate ion was refined to R and Rfree values of 16.8% and 23.0%, respectively, at a resolution of 2.0 Å against data collected from a single native crystal (Tables 1 and 2).
FIGURE 2.
Structure of an acid mutant PhoQ 43–190 subunit. A, the overall structure of the acid mutant PhoQ 43–190 (molecule A) is shown as a ribbon diagram with secondary structure elements labeled in black. B, a stereo plot of the Cα trace of the same acid mutant PhoQ 43–190 molecule. Every tenth Cα atom is depicted as a black sphere and labeled accordingly. The diagrams were created using MolScript (43) and BobScript (44).
Both the wild-type and acidic cluster mutant PhoQ 43–190 structures adopt a mixed α/β-fold containing a central five-stranded anti-parallel β-sheet flanked by a long N-terminal α-helix and additional loops and helices on each side. This type of fold has been reported previously in the structures of other histidine kinase sensor domains such as DcuS (21) and CitA (22) in addition to the S. typhimurium PhoQ sensor domain (9). All four wild-type and acidic cluster mutant PhoQ 43–190 molecules are similar. Pairwise superimposition of all molecules using LSQMAN (23), based on a structurally invariant core of 47 Cα positions determined at the 5σ level of ESCET (24, 25) comparison, yields r.m.s.d. values ranging from 0.25 to 0.42 Å. Small differences in the rest of the structure may be attributed to differences in crystal packing and a certain amount of intrinsic structural lability that may be related to protein function. The E. coli and S. typhimurium sensor domains are very similar in sequence (81.3% identity), and their structures are also similar. Pairwise superimposition of the wild-type E. coli molecules with those of the S. typhimurium PhoQ yield r.m.s.d. values ranging from 0.89 to 0.97 Å over an average of 129 Cα positions, and matches for acidic cluster mutant PhoQ likewise yield r.m.s.d. values from 0.85 to 1.07 Å over an average of 131 Cα positions. Because the acidic cluster mutant PhoQ 43–190 was refined to a higher resolution and contains a greater number of ordered residues, we define the secondary structure elements based on DSSP (26) assignments of molecule A of this structure.
Structural elements of the PhoQ sensor domain are shown in Fig. 2, taking the acidic cluster mutant PhoQ 43–190 structure for reference, and secondary structure assignments are specified in Fig. 3. The PhoQ sensor domain begins with N-terminal helix H1 (residues 46–62), located on the backside of the central β-sheet. Helix H1 connects up to a pair of short antiparallel β-strands S′ (residues 64–66) and S″ (residues 69–71) that loop above the sheet. The peptide backbone then runs back down through helix H2 (residues 76–79) and connects into the first strand S1 (residues 85–89) of the sheet. Edge strand S2 (residues 95–98) follows S1 and connects into helices H3a (residues 103–108) and H3b (residues 111–113), which cross over the front of the central sheet to join the other edge strand, S3 (residues 118–125). Strand S3 runs down the far side of the sheet and leads to helices H4 (residues 126–133) and H5 (residues 137–148), which are angled below the sheet. Strand S4 (residues 154–164), the longest strand in the structure, follows H5, and runs up the length of the central sheet. The peptide backbone then loops back down into the middle strand S5 (residues 173–178) of the sheet, and the structure ends with helix H6 (residues 183–185), which follows S5 and continues to the C-terminal end.
FIGURE 3.
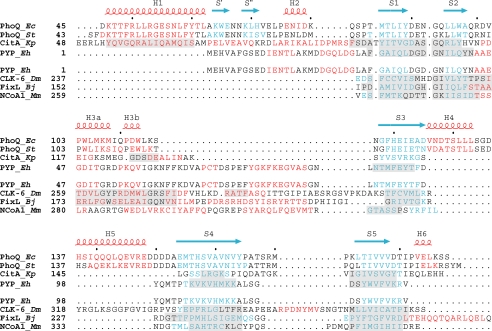
Structure-based sequence alignments. Structure-based sequence alignments comparing PDC to PAS domains. Residues in helices are colored red, and residues in strands are colored blue. Secondary structure elements of E. coli PhoQ are also shown and labeled above the alignments, with helices in red and strands in blue.Inthe top set, the PDC sensor domains PhoQ and CitA are aligned with the PYP PAS domain. Regions in CitA and PYP having Cα positions structurally aligned with those in E. coli PhoQ are shaded in light gray.Inthe bottom set, the PAS domains CLK-6, FixL, and NcoA-1 are aligned with PYP. Regions in CLK-6, FixL, and NcoA-1 having Cα positions structurally aligned with those of PYP are shaded in light gray. Organism names are abbreviated in italics (Ec for E. coli, St for S. typhimurium, Kp for Klebsiella pneumoniae, Eh for Ectothiorhodospira halophila, Dm for Drosophila melanogaster, Bj for Bradyrhizobium japonicum, and Mm for Mus musculus).
Electron-density features corresponding to non-protein moieties were found in both the wild-type and acidic cluster mutant PhoQ 43–190 structures. In the wild-type structure, nickel ions were found associated in a similar manner with each subunit. One nickel ion is coordinated by carboxylate oxygen atoms of Asp-151 and Asp-152 from the acidic cluster of subunit A and by Asp-128 from symmetry mate B′, and the other nickel ion is coordinated by Asp-151 and Asp-152 of B and Asp-128 of A′. Asp-128 is located on H4, and Asp-151 and Asp-152 are located in the “EDDDDAE” acidic cluster in the loop between H5 and S4, which had previously been implicated in divalent cation binding (12). The presence of nickel at the two sites was confirmed in a Bijvoet difference Fourier synthesis generated from a dataset collected above the nickel K-edge but below the selenium K-edge. An additional feature of nonprotein density was found in a pocket formed between the two molecules of the asymmetric unit; this was modeled as an acetate ion coordinated by side chains of residues Gln-81 of molecule A, and Arg-53 and Asn-117 of molecule B. Arg-53, Gln-81, and Asn-117 are found in H1, in the loop between H2 and S1, and in the loop between H3b and S3, respectively. Nickel was a necessary component in the crystallization buffer to obtain crystals of wild-type PhoQ 43–190, and the addition of acetate was found to improve crystal size and appearance. In the acidic cluster mutant structure, a single region of unexpected electron density was modeled as a sulfate ion, which is complexed with water molecules coordinated by guanidinium groups of Arg-50 and Arg-53 from H1 of molecule A, and Arg-169 from the loop between S4 to S5 of molecule B. Ammonium sulfate was the main precipitant in the crystal buffer used to grow the acidic cluster mutant PhoQ 43–190 crystals.
Comparison between PDC Sensor Domains and PAS Domains—A DALI (27) search comparing PhoQ 43–190 to proteins in the PDB structural data base reveals great similarity to other histidine kinase sensor domains such as S. typhimurium PhoQ (9), LuxQ (28, 29), and DcuS (21) (Z scores = 22.3, 6.3, and 6.2, respectively; r.m.s.d. values = 1.3, 3.6, and 3.1 Å, respectively). These periplasmic sensor domains have been described by others as belonging to the PAS (Per-Arnt-Sim) domain superfamily (30), and DcuS and CitA have been directly compared with PYP (31). PAS domains are found in a wide range of organisms and include a diversity of intracellular signaling domains, including FixL, NifL, NtrY, YntC, NtrB, and NifU (32). DALI (27) also finds similarities between E. coli PhoQ 43–190 and PAS domains such as the transcriptional coactivator NcoA-1 (33) and the Drosophila clock protein CLK-6 (34), although these matches score lower than those with other sensor domains (Z scores = 5.2 and 2.5, respectively; r.m.s.d. values = 2.6 and 4.4 Å, respectively), and PYP is not listed.
A structure-based alignment of the PhoQ and CitA sensor domains with the PYP, CLK-6, FixL, and NcoA-1 PAS domains illustrates the similarities and several important differences between these classes of proteins (Fig. 3). We have defined the criteria for the alignments such that corresponding structurally aligned segments have at least three contiguous Cα positions lying within 2.5 Å of each other. Pairwise sequence identities between PhoQ and these PAS domains range from 6% to 8%, and structural alignments are very restricted (30–39 Cα positions, Table 3). In all cases, more residues can be superimposed in alignments between different sensor domains or between different PAS domains than in alignments between sensor domains and PAS domains. Only the β-sheets are in common between PAS domains and the α/β sensor domain; differences primarily lie in residues between the second and third β-strands and in the N-terminal helix, present only in sensor domains. These structural differences distinguish these sensor domains from PAS domains and have led us to term this the PDC (PhoQ-DcuS-CitA) domain. Thus, PDC proteins are distinguishable both from PAS domains and from sensor domains that form four helical bundles such as Tar (35) and NarX.6 PhoQ is distinguished from DcuS and CitA in having the H4-H5 helix pair and acid cluster as an insertion.
TABLE 3.
r.m.s.d. values of PDC and PAS domain superimpositions
| PYP | PhoQ | |
|---|---|---|
| Å | Å | |
| CitA | 1.46 over 49 Cα | |
| PYP | 1.02 over 39 Cα | |
| CLK-6 | 1.34 over 65 Cα | 1.21 over 37 Cα |
| FixL | 1.07 over 66 Cα | 1.01 over 36 Cα |
| NcoA1 | 1.22 over 41 Cα | 1.28 over 30 Cα |
Dimerization of PhoQ 43–190—The wild-type PhoQ 43–190 dimer (rotation χ = 158.6°, screw translation tχ =-3.11 Å) (36) buries 1528 Å2 of total accessible surface area between the two subunits. The dimer is quasi-symmetric about an interface formed primarily by interactions between the H1 helices of each protomer that include hydrophobic contacts, hydrogen bonding, and electrostatic interactions. There are also additional interactions between residues on helix H1 and residues in the loops between S4 and S5, H2 and S1, H3b and S3, and H2 and S1. The structure reveals a salt bridge formed between Asp-179 of molecule A and Arg-50 of molecule B in which the corresponding Cα positions are separated by 11.3 Å. A symmetrically related salt bridge between Arg-50 of molecule A and Asp-179 of molecule B is not formed due to the asymmetric nature of the dimer, which places those Cα positions 16.9 Å apart.
The orientation of the wild-type PhoQ monomers is such that both the N-terminal H1 and C-terminal H6 helices are aligned in the same direction in a manner where the ends would be poised to enter into the transmembrane segments TM1 and TM2 of a full-length molecule. The roughly parallel arrangement of the N- and C-terminal helices in the wild-type PhoQ 43–190 dimer is similar to that observed in the dimeric structure of the aspartate receptor Tar (35). Two acid mutant molecules interact in their crystal environment through similar surfaces as those in the wild-type dimer, and they bury a substantial interface (2350 Å2), but orientations are radically different. This seems to be a non-physiological arrangement, because N- and C-terminal helices point in opposite directions and the Arg-50′ → Asp-179 salt bridge, seen below to be functionally important, does not form.
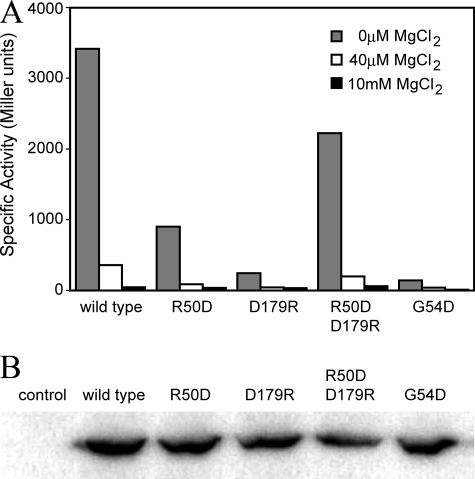
Measurements of Activity in Vivo—To examine the importance of the dimer interface, and in particular the Arg-50′ → Asp-179 salt bridge, to the function of PhoQ, R50D and D179R single mutants, which would disrupt the salt bridge, and an R50D/D179R double mutant, which could recreate the salt bridge in the opposite direction, were constructed. An additional G54D mutant was constructed in which we reasoned the insertion of a charged residue in the hydrophobic interface would be disruptive. The abilities of such mutants to activate PhoP-mediated transcription of a phoN-lacZ reporter gene in vivo were compared with the wild-type protein. As shown in Fig. 4A, phoN-lacZ expression is induced to roughly 3400 units by MgCl2 limitation when wild-type PhoQ protein is present, but it is only induced to only ∼900 and 245 units, respectively, when the PhoQ-R50D and PhoQ-D179R mutants are present. The effect of the double mutant is indeed compensatory rather than additive; reporter expression is activated to 2225 units for PhoQ-R50D/D179R, >50% of the wild-type level. The effect of the dimer-disrupting PhoQ-G54D mutant results in reporter gene induction to roughly 142 units in the magnesium limiting condition, an effect that most closely matches that of the PhoQ-D179R mutant. The mutations do not significantly affect the relative expression levels of PhoQ (Fig. 4B). These results indicate that the salt bridge observed in the crystal structure is physiologically important in the function of the protein and suggest that the integrity of the interface is crucial for the formation of the active state.
FIGURE 4.
In vivo activity and relative expression levels of PhoQ. A, E. coli strain CSH26ΔQ/F′lacIQkan, containing the pNL3 reporter plasmid and pLPQ2 for expressing the wild-type and mutant PhoQ proteins, were assayed for β-galactosidase activity following growth in N-media (0 μm MgCl2) or as supplemented with 40 μm or 10 mm MgCl2. Specific activity is shown in Miller units. Standard deviations are <10% of the mean, calculated from at least three individual experiments. B, the relative expression levels of the wild-type and mutant PhoQ proteins are shown by Western blotting, probed using an antibody specific for the cytoplasmic domain of PhoQ. The PhoQ variants are labeled above each lane accordingly. The control is the E. coli strain containing the “empty” pLPQ2 vector lacking the phoP-phoQ operon.
DISCUSSION
The sensor domain of PhoQ is clearly similar in structure to DcuS (21) and CitA (22) sensor domains despite being very dissimilar in sequence (7.4% and 4.5% identity, respectively). This PhoQ-DcuS-CitA (PDC) fold represents a different structural class (α/β) of sensor domains from the all-helical sensor domain classes such as Tar (35) and NarX.6 The PDC domain fold characteristically begins with a long N-terminal α-helix that leads into a central β-sheet core flanked by short helices on both sides. The central five-stranded antiparallel β-sheet has the same topology as that of PAS domains, and this feature dominates in structure similarity searches. This, in turn, has led others to identify PDC sensor domains as PAS domains; however, there are no actual similarities outside the β-sheet, whereas PDC and PAS domains each have their own distinguishing characteristics.
The functionally important N-terminal helix is unique to PDC domains, and the segment connecting the two β-strands at the edges of the sheet is distinctive for PDC and PAS domains. In PDC domains, the residues between the second and third strands transverse across the front of the sheet in a single span, often helical. PhoQ residues in this region align with those in CitA and in other x-ray structures of PDC sensor domains, such as DcuS, LisK, and DctB.6 In PAS domains, the segment between the second and third strands takes a longer and more tortuous path than in PDC domains, comprising multiple helices and typically ending in a helix running antiparallel to the third strand. Such differences between PDC domains (DcuS and CitA) and PAS domains were also noticed previously (37). Certain sequence similarities have been established among PAS domain family members (30), but PAS-conserved sequences are not found in the PhoQ sensor domain. It appears that, despite sharing topologically identical five-stranded β-sheets, PDC and PAS domains do nevertheless belong to separate protein superfamilies.
In the wild-type structure, the N-terminal helices that form the dimer interface lie in a parallel orientation, and we believe that the dimer interface observed in the wild-type PhoQ structure is biologically relevant. The orientation of the PhoQ protomers allows for the formation of the Arg-50′ → Asp-179 salt bridge, and the physiological importance of the salt bridge to function was confirmed by our mutagenesis studies (see above). Other studies characterizing PhoQ mutants (D179A or D179L) are also consistent with importance of the salt bridge to PhoQ function (38); however, the difference in phenotypes obtained between the two sets of mutational studies on the same location may be a result of differences in charge and size of the mutant residues. Such results suggest that normal PhoQ function is dependent and sensitive to the energetics of the dimer interface.
It is believed that histidine kinase transmembrane regions form four-helical bundles as observed in the structure of the Natronobacterium pharaonis phototaxis SRII-HtrII complex (39). The structures of the NarX sensor domain6 and the aspartate receptor Tar (35) have parallel N- and C-terminal helical regions that would be compatible with connection to such membrane-spanning regions in an intact molecule. The arrangement of the wild-type PhoQ protomers orients the N- and C-terminal helices in a parallel manner also suitable for connection with a four-helical bundle membrane-spanning region, and a similar orientation of terminal helices has also been observed in a DcuS dimer.6
It must be noted that, in contrast to the wild-type E. coli PhoQ sensor domain, the S. typhimurium PhoQ sensor (9) shows a different putative dimeric arrangement. Distances between the two N- and C-terminal residues within the S. typhimurium PhoQ dimer are reported to be 27 and 55 Å, respectively (9), whereas the distances between N- and C-terminal residues within the wild-type E. coli PhoQ are each 18 Å apart, as measured between Cα positions corresponding to residues 45 and 186 in each protomer. We believe that the dimeric arrangement of protomers in the E. coli PhoQ structure places the N- and C-terminal ends in a position more suitable for connection into a putative four-helical bundle arrangement of transmembrane helices.
The physiologically relevant Arg-50′ → Asp-179 salt bridge observed in the wild-type PhoQ dimer is not present in the S. typhimurium PhoQ dimer, which places the Cα positions of residues Arg-50 and Asp-179′, and Arg-50′ and Asp-179, 28.1 and 27.5 Å apart, respectively. In order for the salt bridge to form, a large rearrangement would be required at the dimer interface of the S. typhimurium PhoQ structure. Asp-179 in the structure of the S. typhimurium PhoQ (9) sensor domain participates in hydrogen bonding with the side chains of Thr-48 and Lys-186 (9), and mutations of Asp-179 and Lys-186 result in impaired PhoQ function (9). Equivalent residues in the E. coli PhoQ sensor domain are within weak van der Vaals contact distance but are not hydrogen-bonded. It is possible that these differing arrangements of residues along the interface between the C-terminal helix and the central sheet in the two PhoQ structures may reflect different conformational signaling states. Such residues may be involved in mediating conformational changes between regions of the protein involved with ligand binding to the membrane-spanning helices. Additional experiments will be required to address such possibilities.
In an attempt to learn more about the role of the acidic cluster in PhoQ function, we solved the PhoQ 43–190 structure containing the EDDDDAE to QNNNNAQ mutations (amino acid residues 148–154), which render the protein defective in divalent cation binding and activation. These crystals were obtained in a buffer that lacked divalent cations, and nothing is bound to the replacement sequence, although it adopts essentially the same conformation as for the wild-type structure. The acidic cluster mutant also forms a quasi-diad dimer in the crystal; however, unlike in the wild-type dimer, here the N-terminal helices lie in an antiparallel orientation at the dimer interface and the Arg-50′ → Asp-179 salt bridge does not form. We believe this arrangement to be non-physiological, because the N- and C-terminal pairs are too far apart to connect to the putative membrane-spanning four-helix bundle.
Self-association propensities of membrane proteins are diminished by orders of magnitude when freed from membrane tethering (40). Thus, even though sensor domains are expected to be functionally dimerized in situ on the membrane, they may exhibit quite low dimer affinity when isolated in solution. The E. coli PhoQ sensor domain has been shown to be monomeric in solution by analytical ultracentrifugation with a lower limit of 600 μm measured for the Kd of dimerization (12), and likewise both the CitA and DcuS sensor domains have also been reported to be mostly monomeric in solution (21, 41). It follows that a certain lability may exist in the manner by which sensor domains self-associate, and therefore dimeric interactions within in a crystal-lattice environment may not necessarily be biologically relevant. Such lability has been observed in studies involving the CitA sensor domain (22) and in our unpublished studies6 of the DctB and LisK sensor domains. In such cases, non-physiological interactions between sensor domain protomers can be found in the crystal lattice, sometimes mediated by required ions in crystallization buffers. We suggest that the “dimers” formed in the E. coli acid mutant PhoQ and S. typhimurium PhoQ structures may simply be the result of crystal packing interactions that have overridden the relatively weak affinity for the formation of a physiologically relevant dimer.
The alternative subunit association of the acid mutant PhoQ “dimer” does suggest that alteration of the acidic cluster by mutagenesis, and perhaps by divalent cation binding as well, might also affect the energetics of the dimer interface. If such effects are applicable to the intact membrane-bound molecule, then signal transduction mediated by cation binding at the acidic cluster would seem to employ a mechanism that involves a dynamic dimer interface and may not require quaternary interactions with additional proteins or with the membrane.
The PhoP-PhoQ system is activated by limiting concentrations of extracellular divalent cations. Mg2+ and Ca2+ are thought to act in part as physiologically relevant signaling ligands by directly binding the PhoQ sensor domain and producing a conformational change that influences the enzymatic activities of the cytoplasmic domain (6–8). Although we were unable to obtain suitable crystals in the presence of either Mg2+ or Ca2+ ions, the crystallization buffer contained 5 mm NiCl2, which was required for the formation of crystals. The structure of wild-type PhoQ 43–190 contains two Ni2+ ions, each associated with one of the two molecules in the asymmetric unit, forming lattice contacts between the side chains of Asp-151 and Asp-152, and Asp-128′ of a symmetry mate. Asp-151 and Asp-152 are part of the acidic cluster (148EDDDDAE154) that had previously been implicated in divalent cation binding (12). Divalent cations (Ca2+) are also associated with acid clusters of two of four Salmonella PhoQ sensor domains (9), but that site is displaced from our Ni2+ site and coordinated peripherally by Asp-149. There do not seem to be ion-associated conformational distinctions in these two structures.
Divalent cations thermodynamically stabilize the sensor domain fragment to denaturation in a fashion expected for a direct binding model, and this mode of binding requires the presence of the acidic cluster. In addition, substitution of the cluster with uncharged isosteres (QNNNNAQ) results in a protein that is defective for in vivo activation when extracellular Mg2+ concentrations are low; however, substitution of the cluster with alanine residues (AAAAAAA) results in a protein that functions normally in response to extracellular Mg2+.7 One possible explanation for these paradoxical results is that the sensor domain possesses one or more additional ligand binding sites, and neutralization of the acidic cluster site by conservative substitution with non-charged isosteres mimics divalent cation binding at this site to favor formation of the repressed state. When the acidic cluster site is substituted with alanines, the side chains necessary to form the repressed state are absent, and the protein can respond normally to binding at the other site(s). Interaction between a Ni2+ ion and Asp-128 in the crystal structure suggests that this residue might be part of such an additional physiologically relevant divalent-cation binding site. But the D128A mutation, both in the context of the wild-type acidic cluster and the alanine-substituted cluster, results in a protein that still responds normally to extracellular Mg2+ and Ca2+ (data not shown). Perhaps Asp-128 is not absolutely necessary for binding at the second site, Asp-128 either may not interact with Mg2+ or Ca2+, or such interactions do not affect protein function. Close inspection of the structure does not reveal any other potential divalent cation binding sites.
The acidic cluster resides in the lobe of PhoQ formed by the connection from H5 to S4. Alignment of PhoQ from various organisms indicates that the cluster and lobe are present in the enteric versions of PhoQ but not in non-enteric versions (42), suggesting that the acidic cluster may play a specialized role in enteric bacteria. The position of the lobe with respect to the dimer interface and the rest of the structure suggests that it might lie very close to the membrane in the intact molecule. It has been suggested that from the S. typhimurium PhoQ structure that acidic regions may interact with the membrane phospholipids in a manner to allow chelation of divalent ions such as Ca2+ (9); however, such a mechanism has not been established in vivo. Further research may be required to elucidate the specific role of the acidic cluster in its relation to divalent binding and signaling, and any involvement of membrane phospholipids in biological function.
Supplementary Material
Acknowledgments
We thank Craig Ogata and Randy Abramowitz for help with synchrotron data collection. We also thank Joel Schildbach for help in preliminary crystallization trials and data collection, and Robert Sauer, in whose laboratory the work was initiated.
The atomic coordinates and structure factors (codes 3BQ8 and 3BQA) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grants AI41566 (to C. D. W.) and GM34102 (to W. A. H.). Beamline X4A of the National Synchrotron Light Source at Brookhaven National Laboratory, a Department of Energy facility, is supported by the New York Structural Biology Center. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental text and references.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: SeMet, selenomethionyl; r.m.s.d., root mean square deviation.
J. Cheung and W. A. Hendrickson, unpublished data.
C. D. Waldburger, unpublished data.
References
- 1.Stock, A. M., Robinson, V. L., and Goudreau, P. N. (2000) Annu. Rev. Biochem. 69 183-215 [DOI] [PubMed] [Google Scholar]
- 2.West, A. H., and Stock, A. M. (2001) Trends Biochem. Sci. 26 369-376 [DOI] [PubMed] [Google Scholar]
- 3.Wolanin, P. M., Thomason, P. A., and Stock, J. B. (2002) Genome Biology http://genomebiology.com/2002/3/10/reviews/3013 [DOI] [PMC free article] [PubMed]
- 4.Robinson, V. L., Buckler, D. R., and Stock, A. M. (2000) Nat. Struct. Biol. 7 626-633 [DOI] [PubMed] [Google Scholar]
- 5.Dutta, R., Qin, L., and Inouye, M. (1999) Mol. Microbiol. 34 633-640 [DOI] [PubMed] [Google Scholar]
- 6.Sanowar, S., and Le Moual, H. (2005) Biochem. J. 390 769-776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vescovi, E. G., Ayala, Y. M., DiCera, E., and Groisman, E. A. (1997) J. Biol. Chem. 272 1440-1443 [DOI] [PubMed] [Google Scholar]
- 8.Vescovi, E. G., Soncini, F. C., and Groisman, E. A. (1996) Cell 84 165-174 [DOI] [PubMed] [Google Scholar]
- 9.Cho, U. S., Bader, M. W., Amaya, M. F., Daley, M. E., Klevit, R. E., Miller, S. I., and Xu, W. Q. (2006) J. Mol. Biol. 356 1193-1206 [DOI] [PubMed] [Google Scholar]
- 10.Zwir, I., Shin, D., Kato, A., Nishino, K., Latifi, T., Solomon, F., Hare, J. M., Huang, H., and Groisman, E. A. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 2862-2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monsieurs, P., De Keersmaecker, S., Navarre, W. W., Bader, M. W., De Smet, F., McClelland, M., Fang, F. C., De Moor, B., Vanderleyden, J., and Marchal, K. (2005) J. Mol. Evol. 60 462-474 [DOI] [PubMed] [Google Scholar]
- 12.Waldburger, C. D., and Sauer, R. T. (1996) J. Biol. Chem. 271 26630-26636 [DOI] [PubMed] [Google Scholar]
- 13.Doering, D. S., and Matsudaira, P. (1996) Biochemistry 35 12677-12685 [DOI] [PubMed] [Google Scholar]
- 14.Regelmann, A. G., Lesley, J. A., Mott, C., Stokes, L., and Waldburger, C. D. (2002) J. Bacteriol. 184 5468-5478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 16.Bailey, S. (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760-763 [DOI] [PubMed] [Google Scholar]
- 17.de La Fortelle, E., and Bricogne, G. (1997) Methods Enzymol. 276 472-494 [DOI] [PubMed] [Google Scholar]
- 18.Perrakis, A., Morris, R., and Lamzin, V. S. (1999) Nat. Struct. Biol. 6 458-463 [DOI] [PubMed] [Google Scholar]
- 19.Jones, T. A., Zou, J. Y., Cowan, S. W., and Kjeldgaard, M. (1991) Acta Crystallogr. Sect. A 47 110-119 [DOI] [PubMed] [Google Scholar]
- 20.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 21.Pappalardo, L., Janausch, I. G., Vijayan, V., Zientz, E., Junker, J., Peti, W., Zweckstetter, M., Unden, G., and Griesinger, C. (2003) J. Biol. Chem. 278 39185-39188 [DOI] [PubMed] [Google Scholar]
- 22.Reinelt, S., Hofmann, E., Gerharz, T., Bott, M., and Madden, D. R. (2003) J. Biol. Chem. 278 39189-39196 [DOI] [PubMed] [Google Scholar]
- 23.Kleywegt, G. J., and Jones, T. A. (1994) CCP4/ESF-EACBM Newsletter on Protein Crystallography 31 9-14 [Google Scholar]
- 24.Schneider, T. R. (2000) Acta Crystallogr. Sect. D Biol. Crystallogr. 56 714-721 [DOI] [PubMed] [Google Scholar]
- 25.Schneider, T. R. (2002) Acta Crystallogr. Sect. D Biol. Crystallogr. 58 195-208 [DOI] [PubMed] [Google Scholar]
- 26.Kabsch, W., and Sander, C. (1983) Biopolymers 22 2577-2637 [DOI] [PubMed] [Google Scholar]
- 27.Holm, L., and Sander, C. (1993) J. Mol. Biol. 233 123-138 [DOI] [PubMed] [Google Scholar]
- 28.Neiditch, M. B., Federle, M. J., Pompeani, A. J., Kelly, R. C., Swem, D. L., Jeffrey, P. D., Bassler, B. L., and Hughson, F. M. (2006) Cell 126 1095-1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neiditch, M. B., Federle, M. J., Miller, S. T., Bassler, B. L., and Hughson, F. M. (2005) Mol. Cell 18 507-518 [DOI] [PubMed] [Google Scholar]
- 30.Gu, Y. Z., Hogenesch, J. B., and Bradfield, C. A. (2000) Annu. Rev. Pharmacol. 40 519-561 [DOI] [PubMed] [Google Scholar]
- 31.Genick, U. K., Borgstahl, G. E. O., Ng, K., Ren, Z., Pradervand, C., Burke, P. M., Srajer, V., Teng, T. Y., Schildkamp, W., McRee, D. E., Moffat, K., and Getzoff, E. D. (1997) Science 275 1471-1475 [DOI] [PubMed] [Google Scholar]
- 32.Taylor, B. L., and Zhulin, I. B. (1999) Microbiol. Mol. Biol. Rev. 63 479-506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Razeto, A., Ramakrishnan, V., Litterst, C. M., Giller, K., Griesinger, C., Carlomagno, T., Lakomek, N., Heimburg, T., Lodrini, M., Pfitzner, E., and Becker, S. (2004) J. Mol. Biol. 336 319-329 [DOI] [PubMed] [Google Scholar]
- 34.Yildiz, O., Doi, M., Yujnovsky, I., Cardone, L., Berndt, A., Hennig, S., Schulze, S., Urbanke, C., Sassone-Corsi, P., and Wolf, E. (2005) Mol. Cell 17 69-82 [DOI] [PubMed] [Google Scholar]
- 35.Yeh, J. I., Biemann, H. P., Prive, G. G., Pandit, J., Koshland, D. E., and Kim, S. H. (1996) J. Mol. Biol. 262 186-201 [DOI] [PubMed] [Google Scholar]
- 36.Hendrickson, W. A. (1979) Acta Crystallogr. Sect. A 35 158-163 [Google Scholar]
- 37.Hefti, M. H., Francoijs, K. J., de Vries, S. C., Dixon, R., and Vervoort, J. (2004) Eur. J. Biochem. 271 1198-1208 [DOI] [PubMed] [Google Scholar]
- 38.Minagawa, S., Okura, R., Tsuchitani, H., Hirao, K., Yamamoto, K., and Utsumi, R. (2005) Biosci. Biotech. Biochem. 69 1281-1287 [DOI] [PubMed] [Google Scholar]
- 39.Gordeliy, V. I., Labahn, J., Moukhametzianov, R., Efremov, R., Granzin, J., Schlesinger, R., Buldt, G., Savopol, T., Scheidig, A. J., Klare, J. P., and Engelhard, M. (2002) Nature 419 484-487 [DOI] [PubMed] [Google Scholar]
- 40.Grasberger, B., Minton, A. P., DeLisi, C., and Metzger, H. (1986) Proc. Natl. Acad. Sci. U. S. A. 83 6258-6262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaspar, S., Perozzo, R., Reinelt, S., Meyer, M., Pfister, K., Scapozza, L., and Bott, M. (1999) Mol. Microbiol. 33 858-872 [DOI] [PubMed] [Google Scholar]
- 42.Lesley, J. A., and Waldburger, C. D. (2001) J. Biol. Chem. 276 30827-30833 [DOI] [PubMed] [Google Scholar]
- 43.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24 946-950 [Google Scholar]
- 44.Esnouf, R. M. (1997) J. Mol. Graph. Model. 15 132-134 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.