Abstract
To explore guinea pigs as models of chymase biology, we cloned and expressed the guinea pig ortholog of human chymase. In contrast to rats and mice, guinea pigs appear to express just one chymase, which belongs to the α clade, like primate chymases and mouse mast cell protease-5. The guinea pig enzyme autolyzes at Leu residues in the loop where human chymase autolyzes at Phe. In addition, guinea pig α-chymase selects P1 Leu in a combinatorial peptide library and cleaves Ala-Ala-Pro-Leu-4-nitroanilide but has negligible activity toward substrates with P1 Phe and does not cleave angiotensin I. This contrasts with human chymase, which cleaves after Phe or Tyr, prefers P1 Phe in peptidyl 4-nitroanilides, and avidly hydrolyzes angiotensin I at Phe8 to generate bioactive angiotensin II. The guinea pig enzyme also is inactivated more effectively by α1-antichymotrypsin, which features P1 Leu in the reactive loop. Unlike mouse, rat, and hamster α-chymases, guinea pig chymase lacks elastase-like preference for P1 Val or Ala. Partially humanized A216G guinea pig chymase acquires human-like P1 Phe- and angiotensin-cleaving capacity. Molecular models suggest that the wild type active site is crowded by the Ala216 side chain, which potentially blocks access by bulky P1 aromatic residues. On the other hand, the guinea pig pocket is deeper than in Val-selective chymases, explaining the preference for the longer aliphatic side chain of Leu. These findings are evidence that chymase-like peptidase specificity is sensitive to small changes in structure and provide the first example of a vertebrate Leu-selective peptidase.
Chymases are serine peptidases expressed and secreted mainly by mast cells. They are proposed to play a variety of roles in host defense, homeostasis and disease, including anti-parasite defense (1), blood pressure regulation (2, 3), connective tissue turnover (4), angiogenesis (5), cardiovascular remodeling (6), ischemia-reperfusion injury (7), lung fibrosis (8), airway inflammation (9, 10), and venom detoxification (11). Most chymases can cleave a variety of extracellular peptides and proteins, including endogenous targets like angiotensin I and exogenous pathogen or allergen-derived targets like profilin (12). Because the pool of cleavable host and pathogen targets is large, chymases have the potential to influence a broad range of events associated with mast cell activation and secretion.
The closest relatives of chymases are cathepsin G and granzyme B-like proteins, which are expressed in myelomonocytes and cytolytic lymphocytes, respectively, in addition to mast cells. Genes encoding these enzymes are tightly clustered in mammalian genomes. In humans, four genes in this group are expressed: chymase (CMA1), cathepsin G (CTSG), granzyme B (GZMB), and granzyme H (GZMH). In mice and rats, however, chymase and granzyme B-like genes markedly expanded and diversified by gene duplication, conversion, and point mutation (13, 14). The rat locus has an estimated 66 genes, of which 28 may be expressed (15). Although products of chymase, cathepsin G, and granzyme genes are structurally and phylogenetically related, they have a range of roles and target specificities. Granzyme B, for example, is an Aspase that cleaves specific host proteins and kills tumor and other target cells by triggering apoptosis, although humans and mice differ in this regard (16). Cathepsin G, on the other hand, has broad activity (tryptic, chymotryptic, Met-ase) (17) and regulates adhesion-dependent function of neutrophils (18). Within the chymase group, expressed proteins examined to date are either primarily chymotryptic (hydrolyzing peptides after residues with aromatic side chains) or elastolytic (hydrolyzing after residues with small, aliphatic side chains) (19, 20). Even among chymotryptic chymases, there is a range of catalytic competence and target preference. For example, mammalian chymotryptic chymases vary in their tendency to hydrolyze after one or the other of two aromatic residues in angiotensin I (21, 22). Cleavage at one site activates angiotensin I to angiotensin II, whereas cleavage at the other destroys activity. These preferences are influenced by sometimes subtle variations of amino acids in the vicinity of the substrate-binding site (22). In the case of elastolytic chymases, notably the mouse chymase MCP-5 and its rat ortholog, the switch from chymotryptic to elastolytic specificity is attributable to natural mutation of a single amino acid (19, 20). Similar elastolytic properties have recently been described for hamster α chymase (23). Thus, small changes in structure, including alteration of just one amino acid in the vicinity of the substrate-binding site, can cause large changes in function.
Guinea pigs are used to model allergic events and a variety of other phenomena. Although the properties of trypsin-like mast cell tryptases have been explored in guinea pigs, the enzymatic properties of their chymases have been an enigma, especially because unlike in primates, dogs, and hamsters, there is little evidence that chymase-like enzymes in guinea pigs contribute to angiotensin I-induced vascular contraction (24). However, guinea pigs can respond to exogenous chymotryptic peptidase, because human chymase injected into guinea pig skin increases microvascular permeability and provokes neutrophilic and eosinophilic inflammation (25). The present work shows that guinea pigs express an α-chymase-like peptidase with novel specificity for Leu in peptide and protein targets.
EXPERIMENTAL PROCEDURES
Cloning and Sequencing of Guinea Pig Chymase cDNA— Fresh guinea pig (Cavia porcellus) heart muscle was harvested into RNAlater (Qiagen). Total RNA was extracted from homogenates following the RNeasy Mini procedure (Qiagen). Single-stranded cDNA was synthesized using random hexamers. To amplify a portion of guinea pig chymase by homology PCR, rodent mast cell protease consensus sequence was used to design the following primers: 5′-GAGTCAAAGCCACACTCCCGCCCTTACATGG and 5′-KYRCACASDARDGGNCCNCCDGAGTCYCC. Amplimers were cloned into pCR2.1-TOPO-TA (Invitrogen) and sequenced. Primers for rapid amplification of cDNA ends were designed from the obtained sequence. Amplimers obtained by 3′-rapid amplification of cDNA ends using primer 5′-GAGAAAGACTGTAGTGGTTTCTTGATAC were cloned and sequenced. To obtain complete cDNA sequence, nested reverse transcriptase-PCR was performed with a 5′-rapid amplification of cDNA ends protocol (Invitrogen), using 5′-ACCCCTAGGTTGACTGTTAG and 5′-GTATCAAGAAACCACTACAGTCTTTCTC as first and second stage primer, respectively. The resulting amplimer was cloned and sequenced. To assess for potential polymorphisms, protein-coding sequence was verified by PCR cloning and DNA sequencing of a transcript from a guinea pig from an alternate supplier.
Phylogenetic Analysis—Guinea pig chymase amino acid sequence was compared with that of cathepsin G and 27 other chymase-like mammalian peptidases using Geneious software (Biomatters). Rooted dendrograms were prepared from aligned sequences using the unweighted pair group method with arithmetic mean (UPGMA)2 or neighbor-joining techniques with bootstrap resampling.
Generation of Humanized Mutant Guinea Pig Chymase cDNA—Guinea pig chymase was partially humanized using a QuikChange multisite-directed mutagenesis kit (Stratagene) with primer 5′-GCCCAGGGCATTGTATCCTATGGTCATCGGAATG. This changes the codon encoding specificity triad residue Ala216 into a codon for Gly. The resulting A216G mutant cDNA was used to express recombinant mutant enzyme, as described below.
Expression and Mutagenesis of Recombinant Chymases—For expression of guinea pig chymases in BL21(DE3) E. coli, primers 5′-CATATGGATGACGACGACAAGCTCCCATTACCGGCCGTG and 5′-GCGGCCGCTTAATTTGCTTTCAAGATCTTGTTGATC were used to PCR-clone mature peptide-coding sequence into NdeI and NotI sites of vector pET28a (Stratagene). Primers were designed so that enteropeptidase cleavage of expressed product, which has a 26-residue prosequence containing hexahistidine and an enteropeptidase cleavage recognition sequence, yields a peptide matching that of predicted wild type mature protein. Inclusion bodies containing recombinant guinea pig chymase were isolated and purified from E. coli extracts. Insoluble protein from lysed cells was dissolved in 0.1 m Tris-HCl (pH 8.0), 1 mm β-mercaptoethanol, and 6 m guanidine HCl and chromatographed in the same buffer over an Ni2+-nitrilotriacetic acid column. Purified, denatured protein then was incubated with a mixture of oxidized glutathione (150 mm) and reduced glutathione (1.5 mm) and then dialyzed overnight against H2O (pH 4–4.5) at 4 °C. Precipitated protein was recovered by centrifugation and dissolved in 6 m guanidine HCl, 20 mm EDTA (pH 4.5). Refolding was accomplished by dilution into a large volume of refolding buffer: 50 mm Tris-HCl (pH 8.0), 0.5 m arginine, 1 mm EDTA, and 0.5 mm cysteine. After incubation for 2 days at 4 °C, the preparation was concentrated and dialyzed in 50 mm Tris-HCl (pH 7.0) containing 0.3 m NaCl and 1 mm β-mercaptoethanol, purified by Ni2+-nitrilotriacetic acid chromatography, and activated by incubation with enteropeptidase. The activated preparation was further purified by gel filtration in Superdex 75 using running buffer containing 50 mm Tris-HCl (pH 8.0), 0.3 m NaCl, 1 mm tris(2-carboxyethyl) phosphine, and 10% glycerol, followed by rechromatography in 50 mm MES (pH 5.5), 150 mm NaCl, 1 mm tris(2-carboxyethyl) phosphine, and 10% glycerol. The purified protein was monomeric by gel filtration and analytical ultracentrifugation. For kinetic and inhibitor studies, recombinant human chymase expressed in insect cells was prepared, activated, and purified as described (21). Active chymase concentration in samples used in kinetic studies was estimated using the specific activity of highly purified recombinant human chymase assayed under standard conditions (21).
Protein Sequencing—Purified, self-incubated, recombinant wild type guinea pig chymase (∼100 pmol) was desalted by application to and washing of a polyvinylidene difluoride membrane. Membrane-bound protein was subjected directly to N-terminal amino acid sequencing on a Procise 494 HT sequencer (PerkinElmer Life Sciences).
Combinatorial Peptide Library Survey of Cleavage Site Preferences—Fluorescence resonance energy transfer substrate combinatorial libraries were synthesized on PEGA1900 resin via split synthesis after standard Fmoc chemistry using 2-(7-aza-1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate as the condensing reagent. Fmoc-Gly-OH, Fmoc-Glu-Lucifer Yellow, and all Fmoc-protected amino acids were coupled to amino PEGA1900 resin in the presence of diisopropylethylamine in N-methylpyrrolidone. Double coupling was performed using O-(1,2-dihydro-2-oxo-1-pyridyl)-N,N,N′,N′-tetramethyluronium tetrafluoroborate in the presence of diisopropylethylamine in N-ethylpyrrolidone. Synthesis was performed on a semiautomated shaking vessel machine. For variable positions in the peptide sequences, the resin was split into 19 portions, to each of which a different Fmoc-protected amino acid (except Fmoc-Cys-OH) was coupled. The portions then were mixed together and washed. Depending on the library, splitting was repeated, or the synthesis proceeded in a single portion. The last building block was t-butoxycarbonyl-Lys-(dabsyl)-OH. Protecting groups were removed by incubation with trifluoroacetic acid/H2O/triisopropylsilane (95:3:2 by volume). For screening of the Lys(dabsyl)-QAXXXXAQQG-Glu(Gly-PEGA)-Lucifer Yellow peptide library, ∼5000 beads were washed with reaction buffer (50 mm Tris/HCl, pH 8.0, 300 mm NaCl, 1 mm tris(2-carboxyethyl)-phosphine, 10% glycerol). Purified enzyme (50 μg/ml in 1 ml) in reaction buffer was added to the resin. After shaking for 2–3 h at room temperature, beads were filtered and washed first with reaction buffer and then with 100 mm MES (pH 4.8) containing 5% glycerol and 0.05% Triton X-100. Brightly fluorescent beads identified by microscopy were isolated and submitted for N-terminal sequencing. Screening of the SSVXAXSAPG peptide library was similar.
Analysis of Inhibitor Sensitivity—Enzymes were preincubated for 15 min at 25 °C with or without a potential inhibitor and then assayed for residual activity, as assessed with Suc-AAPL-4-NA (wild type guinea pig chymase) and Suc-AAPF-4-NA (A216G guinea pig mutant, human chymase, bovine chymotrypsin) substrates (Sigma). Concentrations of wild type and mutant guinea pig chymase, human chymase, and chymotrypsin (Sigma) were 110, 58, 29, and 8.4 nm, respectively. Concentrations of soybean trypsin inhibitor, phenylmethanesulfonyl fluoride, chymostatin (Sigma), α1-antitrypsin, α1-antichymotrypsin (EMD Chemicals), and E-64 (MP Biomedical) were 70 μm, 0.25 mm, 2.5 μm, 0.25 μm, 0.25 mm, and 0.5 mm, respectively.
Active Site Titration—To estimate active enzyme concentration, amidolytic activity of purified preparations of wild type and A216G mutant guinea pig chymase was titrated with α1-antichymotrypsin as described by Karlson et al. (26). Active site-titrated preparations were assayed under standard conditions to obtain a specific activity, which was used to determine the concentration of active enzyme in future studies.
Determination of Hydrolysis Kinetics—Kinetic parameters, including catalytic constant kcat and Michaelis constant Km, were derived from double reciprocal plots of substrate concentration versus reaction velocity for wild type and A216G mutant guinea pig chymase and human chymase cleaving a panel of peptidyl 4-NA substrates and angiotensin I. Hydrolysis of 4-NA substrates was assessed spectophotometrically at 410 nm. Assay buffers contained phosphate-buffered saline with 0.01% Triton X-100 and 0.05% dimethyl sulfoxide, with concentrations of 4-NA substrates ranging from 71 to 425 μm. Cleavage of angiotensin I was monitored by reverse phase high performance liquid chromatography of fragments generated by incubation with guinea pig and human chymases using modifications of approaches described previously (21, 22). Peptidases were incubated with angiotensin I (8–24 nm) in phosphate-buffered saline (pH 7.4) in aliquots of 50 μl at 37 °C. Reactions were stopped by the addition of 1 μl of 12 n HCl and then diluted with 60 μl of an aqueous solution of 10% acetonitrile, 0.1% trifluoroacetic acid. Outflow of samples chromatographed on a 2.1 × 250-mm Thermo Fisher Scientific BioBasic C-18 column with a linear gradient of 10–40% acetonitrile, 0.1% trifluoroacetic acid was monitored at 280 nm. Peak areas were determined using Unicorn 5.0 software (GE Healthcare).
Molecular Modeling—Models of human and hamster 2 chymases were prepared from crystal-based coordinates from Protein Data Bank 1pjp (27) and 2rdl (23), respectively. Homology models of wild type and A216G guinea pig chymase were built by Swiss-Model (available on the World Wide Web) starting with the 1pjp scaffold. Images were produced using Chimera (available on the World Wide Web) from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco and rendered with POV-Ray (available on the World Wide Web).
RESULTS
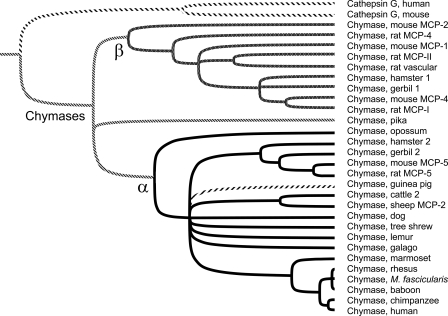
Amino Acid Sequence of Guinea Pig Preprochymase—As shown in Fig. 1, the guinea pig chymase cDNA and gene sequence predict a 337-amino acid protein identical in length to human chymase and to the mouse ortholog MCP-5, both of which are α-chymases (see Fig. 2). Overall, the guinea pig sequence is slightly more similar (77% identical) to human chymase than to mouse chymase MCP-5 (75% identical), which is its closest relative among several chymase-like proteins in rodents. The guinea pig protein features a typical 19-residue signal peptide followed by an acidic dipeptide (Gly-Glu) similar or identical to the propeptide featured in other chymases and in cathepsin G, B-type granzymes, and neutrophil elastase-related peptidases, all of which are thought to be activated by dipeptidylpeptidase I (cathepsin C) (28–30). The propeptide is followed by a 226-residue catalytic domain with an N-terminal Val, which is atypical for chymase and its relatives (which usually have an Ile at this site) but is present in some other trypsin family serine peptidases. Catalytic triad residues (His57, Asp102, and Ser195 using chymotrypsinogen numbering) essential for the amidolytic function of all serine peptidases are intact. However, “specificity triad” residues (189, 216, and 226 of chymotrypsinogen) that influence preferences for the amino acid at the site of hydrolysis in target peptides (31) are unique among enzymatically characterized serine peptidases (Table 1). The guinea pig triad differs in two residues from human chymase, which is chymotryptic, and in all triad residues in comparison with mouse MCP-5, which is elastolytic. A chymase-like peptidase we deduced from recently deposited whole genome shotgun sequence of the pika contains an identical specificity triad. However, this pika chymase is not closely related to guinea pig chymase in overall structure, as demonstrated by the phylogenetic analysis in Fig. 2.
FIGURE 1.
Multiple sequence alignment. Wild type guinea pig preprochymase (GPα) amino acid sequence is compared with that of orthologous mouse and human α-chymases (Humα and Musα, respectively). Putative signal and propeptide segments are italicized and underlined, respectively. Chymase-specific numbering begins with Met-21, which is the first residue of the signal peptide. The mature catalytic domain begins at Val1. Key residues in the catalytic domain are also numbered in italics using standard chymotrypsinogen numbering. ▾, sites of hydrolysis by signal peptidase and dipeptidylpeptidase I or of autolysis; • and #, catalytic and specificity triad residues, respectively; †, predicted sites of N-glycosylation. The human and mouse chymase sequences are from GenBank™ files M64269 and NP_034910, respectively.
FIGURE 2.
Tree of α/β chymases. Phylogenetic relationships between guinea pig chymase and other chymases are probed with this rooted dendrogram generated by UPGMA analysis of aligned mature chymase amino acid sequences using 1000 iterations of bootstrap resampling. The threshold for node assignment was 70%. Tines of peptidases belonging to α and β clades are black and gray, respectively, except for guinea pig chymase, which is forward-hatched. Examples of cathepsin G, the closest relative of chymases, are included for comparison. Accession numbers of sequences used in tree construction are as follows: human and mouse cathepsin G (NP_001902 and CAA55290), mouse MCP-1, -2, -4, and -5 (AAB23194, NP_032597, NP_034909, and NP_034910), rat MCP-I and -II (AAB48268 and P00700), rat MCP-4 and -5 (U67907 and NP_037224), rat vascular chymase (AAC16657), hamster chymase 1 and 2 (BAA19932 and BAA28615), gerbil chymase 1 and 2 (P50340 and P50341), opossum chymase (XP_001369716), guinea pig chymase (this work; AM851020), cattle chymase 2 (XP_593156), sheep MCP-2 (P79204), dog chymase (NP_001013442), baboon chymase (AAA91159), rhesus chymase (BAA22070), M. fascicularis chymase (crab-eating macaque; BAA22070), chimpanzee chymase (XP_001170224), and human chymase (M64269). We deduced the sequence of additional chymases from unannotated whole genome shotgun sequence as follows: American pika (Ochotona princeps, AAYZ01164217), tree shrew (AAPY01734039), mouse lemur (ABDC01231172), galago (AAQR01659070), and marmoset (contig 1790.4 from the Washington University Genome Sequencing Center, available on the World Wide Web).
TABLE 1.
Comparison of specificity triad residues
| Peptidase | Specificity triad | P1 preference | Source/Reference |
|---|---|---|---|
| Guinea pig chymase | SAV | Leu >> Met > Phe | This work |
| Guinea pig chymase A216G | SGV | Phe, Tyr >> Leu > Met | This work |
| Human chymase | SGA | Phe, Tyr >> Leu, Trp > Met | Ref. 43 and this work |
| Dog chymase | SGA | Phe, Tyr > Trp | Refs. 12, 22, 44, and 45 |
| Mouse chymase 4 | SGA | Phe > Tyr > Trp | Refs. 21 and 35 |
| Rat chymase 1 | SGA | Phe > Trp > Tyr >> Leu? > Met | Refs. 35, 46, and 47 |
| Hamster chymase 1 | SGA | Phe, Tyr > Leu >> Trp, His | Ref. 23 |
| Human Pancreatic Ela 2 | SGS | Met > Leu > Tyr > Phe > Ala | Ref. 48 |
| Cattle chymotrypsin | SGG | Tyr > Phe > Trp >> Met > Leu | Ref. 49 |
| Human PSA | SGS | Met > Ala > Tyr > Arg > Lys > Leu > Phe | Ref. 50 |
| Rat chymase 2 | AGA | Phe > Tyr >> Met | Ref. 46 |
| Human SCCE | NGG | Tyr > Ala = Met > Phe = Leu, Arg, Lys | Ref. 50 |
| Human granzyme H | TGG | Phe > Tyr > Met > Leu > Ala > Arg | Ref. 51 |
| Human cathepsin G | AGE | Lys = Phe > Arg = Leu > Met > Ala > Asp | Refs. 17 and 44 |
| Cattle duodenase | NGD | Arg, Lys, Phe, Leu, Tyr | Ref. 52 |
| Rat chymase 4 | LGI | Phe > Tyr | Ref. 26 |
| Rat granzyme M | AST | Met > Leu | Ref. 53 |
| Human granzyme M | ASP | Met > Leu | Ref. 54 |
| Human neutrophil elastase | GVD | Ala > Val > Ile > Thr > Met | Ref. 49 |
| Mouse MCP-5 | NVA | Val > Ile > Ala | Ref. 19 |
| Rat MCP-5 | NVA | Val, Ala, Ile | Refs. 19 and 20 |
| Hamster chymase 2 | NVS | Ala > Val | Ref. 23 |
| Rat granzyme B | AGR | Asp | Ref. 49 |
| Human β-tryptase | DGG | Arg, Lys | Ref. 55 |
| Rat trypsin | DGG | Arg > Lys | Ref. 49 |
| Human plasmin | DGG | Lys > Arg | Ref. 49 |
| Human thrombin | DGG | Arg > Lys | Ref. 49 |
| Mouse MCP-2 | SYA | Inactive | Ref. 36 |
| Mouse MCP-8 | ARR | Inactive | Ref. 37 |
Guinea pig chymase has more catalytic domain cysteines (i.e. 8) than other known chymases, including one more than α-chymases like human chymase and mouse MCP-5 and two more than most β-chymases. Cys127 (chymotrypsinogen numbering) appears to be unique among serine peptidases; in the human and mouse enzyme, Phe is at this location, which is the principal site of autolytic cleavage in human chymase. Modeling results discussed in more detail below indicate that Cys127 does not form an intramolecular disulfide bond; thus, guinea pig chymase has two unpaired cysteines, the other being Cys22, which is also unpaired in human chymase and presumably in other α-type chymases with a cysteine at this site. Guinea pig chymase contains two sites of predicted N-linked attachment to carbohydrates. The Asn95 site is conserved in humans, not mice; the Asn113 site, although not present in human chymase or mouse MCP-5, is present in a variety of other chymases, including opossum chymase, analyzed in Fig. 2.
Guinea Pig Chymase Is an α-Chymase—As revealed by the phylogenetic analysis in Fig. 2, guinea pig chymase allies strongly with the α clade of chymases. Although a number of rodents express one or more chymases belonging to the β clade, no evidence of β-chymase expression was detected in guinea pigs. Tree analysis by UPGMA does not group guinea pig α-chymase with other available rodent α-chymases (i.e. from hamsters, gerbils, mice, and rats). Similar results were obtained by neighbor-joining analysis. This is consistent with other evidence that guinea pig sequences exhibit substantial variation from other mammals classified within the order Rodentia. This high degree of divergence has been a basis for questioning the assumed monophyletic nature of this order (32).
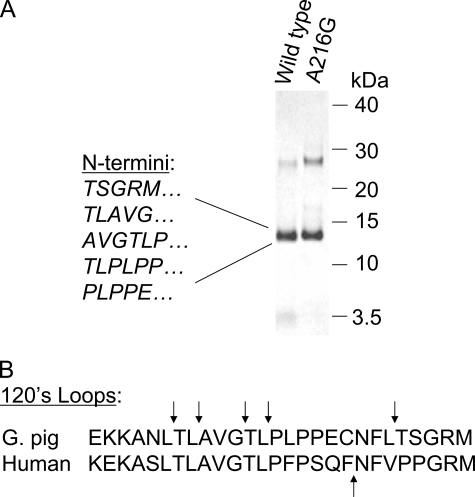
Wild Type Guinea Pig Chymase Autolyzes at Leu Residues— Self-incubation of wild type guinea pig chymase resulted in limited autolysis with main fragments of 13–14 kDa evident upon gel electrophoresis (Fig. 3), consistent with cleavage primarily at a single region. N-terminal sequence of the major fragments reveals N termini consistent with hydrolysis mainly at Leu residues in the “120s loop” of guinea pig chymase, as shown in Fig. 3. Intriguingly, purified human chymase autolyzes within the same loop at Phe127 (33, 34). Like autolyzed human chymase, self-cleaved guinea pig chymase remains active (data not shown). As shown in Fig. 3, hydrolysis at leucines in the 120s loop yields 13–14-kDa fragments under nonreducing conditions, confirming that the guinea pig enzyme's two extra cysteines are not mutually paired, because if they were, the fragments would appear only under reducing conditions. As shown in Fig. 3A, fragments generated by autolyzed A216G are similar to those generated by wild type guinea pig chymase, suggesting that cleavage probably occurs in the 120s loop. Exact autolysis sites in the A216G mutant remain to be determined.
FIGURE 3.
Autolytic cleavage. A, Coomassie Blue-stained, self-incubated wild type and A216G guinea pig chymase after nonreducing SDS-PAGE. The uncleaved parent band is at ∼28 kDa. N termini within the 13–14 kDa bands generated by autolytic hydrolysis of wild type guinea pig chymase, as identified by Edman sequencing, are shown to the left. Elution positions of marker proteins are shown to the right. B demonstrates that cleavage sites reside in the surface-exposed 120s loop, which also contains the major site of autolytic hydrolysis in human chymase, which, however, hydrolyzes after Phe, not Leu. The arrows identify sites of hydrolysis.
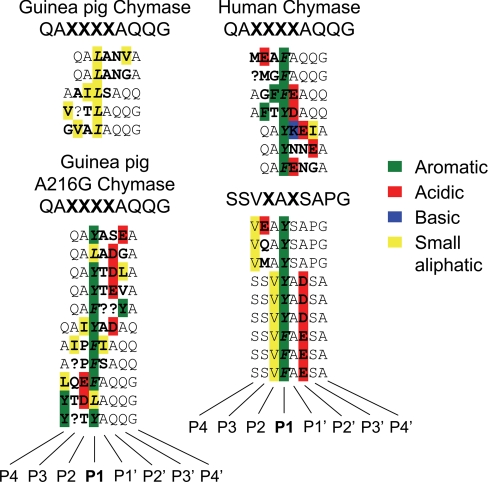
Wild Type Guinea Pig Chymase Hydrolyzes Combinatorial Library Peptides at P1 Leu—As shown in Fig. 4, wild type guinea pig chymase prefers Leu at the site of hydrolysis (i.e. P1) in four peptide sequences identified by screening a QAXXXXAQQG combinatorial peptide library and preferred Ile in one sequence. No substrates with P1 aromatic residues were identified. On the other hand, the partially humanized A216G mutant is much less Leu-selective at P1 when screened using the same library, with aromatic residues predominating (5 Tyr, 4 Phe, 2 Leu). Human chymase is even more selective for P1 aromatic residues when tested with this library (4 Phe, 3 Tyr) and also when tested with an SSVXAXSAPG library (3 Phe, 6 Tyr). At the varying positions in the library, no striking preferences at subsites other than P1 are observed, although residues with acidic side chains are somewhat preferred by the A216G mutant at P2′ and also by human chymase. A preference for acidic residues at P2′ also is a feature of mouse chymase 4 (35).
FIGURE 4.
Hydrolysis of substrates in a combinatorial peptide library. Wild type guinea pig chymase, A216G mutant guinea pig chymase, and human chymase were incubated with peptides of sequence QAXXXXAQQG (where X represents random amino acids). Human chymase also was screened with peptides displaying SSVXAXSAPG, with two randomly varying amino acids. The sequences of individual peptides hydrolyzed by the enzymes are shown, with the hydrolysis site (P1) italicized and with variable positions in boldface. Substrate residues with aromatic, acidic, basic, and small aliphatic side chains are color-coded green, red, blue, and yellow, respectively.
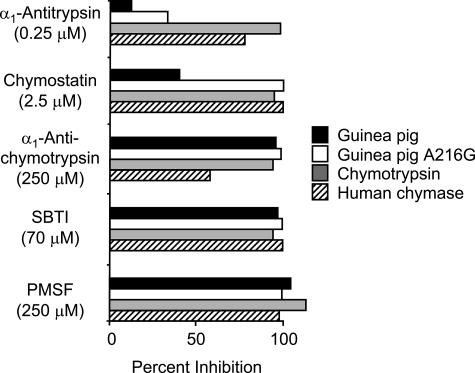
Guinea Pig Chymase Has a Novel Inhibition Profile—As shown in Fig. 5, the inhibition profile of wild type guinea pig chymase deviates substantially from that of the A216G mutant, human chymase, and bovine chymotrypsin. Most strikingly, wild type guinea pig chymase is much more resistant to the serpin α1-antitrypsin and the Phe analog chymostatin than is chymotrypsin or human chymase. On the other hand, wild type guinea pig chymase is fully inhibited by broad-spectrum inhibitors soybean trypsin inhibitor and phenylmethanesulfonyl fluoride and is more sensitive than human chymase to α1-antichymotrypsin. When wild type or mutant guinea pig chymase is incubated in molar excess with α1-antichymotrypsin, the inhibitor is degraded (not shown). All of the enzymes resist Glu64 (not shown), which mainly inhibits cysteine peptidases and some tryptic serine peptidases.
FIGURE 5.
Inhibitor susceptibility. The indicated inhibitors were preincubated in duplicate with wild type recombinant guinea pig chymase (guinea pig), partially humanized recombinant guinea pig chymase (A216G), bovine chymotrypsin, and recombinant human chymase and then assayed for residual peptidyl 4-NA-hydrolyzing activity. The graph shows mean percentage inhibition as compared with the enzymes preincubated without added inhibitor.
Wild Type and A216G Guinea Pig and Human Chymase Strongly Differ in Rates of Hydrolysis of Tetrapeptides and Angiotensin I—Active site titration with α1-antichymotrypsin was used to determine the percentage of active enzyme in preparations of wild type and mutant guinea pig chymases. These values in turn were used to calculate kcat in kinetic studies. The concentration of active enzyme in preparations of human chymase was determined using previously determined specific activity toward Suc-AAPF-4-NA under standard assay conditions (21). As shown in Tables 2 and 3, wild type guinea pig, mutant A216G, and human chymase differ strikingly with regard to kinetic parameters and selectivity when tested for amidolytic activity against Suc-AAPX-4-NA substrates, where X represents Phe, Leu, or Met. Wild type guinea pig enzyme, although weak overall, strongly prefers the substrate with P1 Leu, consistent with the results of combinatorial screening. There is slight preference for P1 Met over Phe, although Met and Phe both are weak alternatives to Leu. In contrast, the A216G mutant prefers P1 Phe over Leu and even more so over Met, but the kinetic bias of the human enzyme for P1 Phe is even more pronounced. The difference in specificity constant kcat/Km for Suc-AAPF-4-NA cleavage is especially striking: 480-fold between wild type and mutant guinea pig and 5800-fold between wild type guinea pig and human chymase. When tested against the best known natural peptide substrate of human chymase, angiotensin I, wild type guinea pig enzyme has no ability to cleave at Phe8 and thereby generate angiotensin II. However, the A216G mutant readily hydrolyzes angiotensin I at this site, suggesting that the difference between wild type guinea pig and human chymase in angiotensin-converting activity mostly is due to a single amino acid change in the vicinity of the substrate binding site. Additional substrates tested included Suc-AAA-4-NA and Suc-AAPV-4-NA, which are not hydrolyzed by guinea pig and human chymases under conditions in which P1 Phe, Leu, and Met substrates are cleaved.
TABLE 2.
Kinetic data: Hydrolysis of peptide 4-NAs and angiotensin
| Chymase | Kinetic parameter | AAPF-4-NA | AAPL-4-NA | AAPM-4-NA | Angiotensin I at Phe8 |
|---|---|---|---|---|---|
| Wild type guinea pig | kcat (s–1) | 0.028 ± 0.013 | 0.93 ± 0.15 | 0.098 ± 0.040 | NDa |
| Km (mm) | 0.53 ± 0.32 | 1.55 ± 0.32 | 0.97 ± 0.58 | ||
| kcat/Km (s–1 mm–1) | 0.063 ± 0.018 | 0.60 ± 0.03 | 0.11 ± 0.03 | ||
| A216G Guinea pig | kcat (s–1) | 16 ± 10 | 3.9 ± 1.7 | 13 ± 18 | 10 |
| Km (mm) | 0.65 ± 0.60 | 1.50 ± 0.95 | 27 ± 37 | 0.36 | |
| kcat/Km (s–1 mm–1) | 30 ± 8 | 3.8 ± 0.4 | 0.72 ± 0.30 | 28 | |
| Human | kcat (s–1) | 143 ± 74 | 19.3 ± 0.8 | 23 ± 27 | 20 |
| Km (mm) | 0.47 ± 0.41 | 1.02 ± 0.13 | 3.0 ± 4.1 | 0.028 | |
| kcat/Km (s–1 mm–1) | 363 ± 109 | 19 ± 3 | 2.4 ± 4.0 | 710 |
No cleavage detected
TABLE 3.
Selectivity based on kcat/Km for AAPX-4-NA substrates
| Chymase | P1 Phe | P1 Leu | P1 Met |
|---|---|---|---|
| Wild type guinea pig | 1 | 9.5 | 1.7 |
| A216G guinea pig | 42 | 5.3 | 1 |
| Human | 151 | 7.9 | 1 |
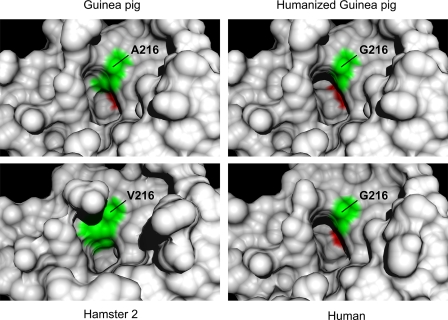
Molecular Models Reveal Basis of P1 Selectivity—Comparisons of human, hamster 2, and guinea pig chymase active sites are shown in Fig. 6. These models predict that the wild type guinea pig P1 side chain-binding primary specificity pocket is narrower and shallower than in human chymase but that the pocket in humanized A216G guinea pig chymase resembles that of the human enzyme. On the other hand, the volume of the P1 side chain pocket in hamster 2 chymase is even more dramatically constrained by the bulkier side chain of Val216. The models, along with functional comparisons of the human and two guinea pig enzymes, provide compelling evidence that the residue 216 side chain is the principal determinant of observed differences in P1 specificity between wild type guinea pig, human, and in particular hamster 2 chymase. The wild type guinea pig P1 slot no longer has the volume, shape, or depth to accommodate sterically demanding substituents, like phenyl side chains, or very long side chains, like that of Met, but is big enough and sufficiently hydrophobic to engage and attract the shorter and less bulky side chains of Leu and to some extent Ile. However, the model suggests that it has significantly more room than the hamster P1 site, which only accepts P1 residues with small compact side chains, preferring Ala and to a lesser extent Val (23).
FIGURE 6.
Models of P1 side chain binding pocket. The panels show corresponding regions of wild type guinea pig, humanized A216G guinea pig, human, and hamster 2 chymases. The human and hamster models are based on crystal-derived structures of Protein Data Bank code 1pjp and 2rdl, respectively. The guinea pig structures are homology models. Specificity triad residues 216 and 226 (chymotrypsinogen numbering) are green and red, respectively. Note that the slotlike crevice accommodating the aromatic P1 side chain in the human enzyme is partly filled in by the Ala216 side chain of wild type guinea pig chymase but is largely restored in the humanized A216G mutant. By comparison, the bulky side chain of Val216 in hamster chymase 2 markedly reduces the volume available for occupancy by a P1 side chain.
DISCUSSION
This study identifies a serine peptidase in guinea pigs that is phylogenetically similar to human and other mammalian α-chymases while differing radically in substrate specificity. Autolysis sites, peptide cleavage preferences, hydrolysis kinetics, and inhibitor susceptibility patterns consistently reveal an enzyme that is highly P1 Leu-selective. This selectivity is strong but not absolute, as suggested by the identification of one combinatorial peptide substrate with P1 Ile and a site of autolysis at Gly in the 120s loop. Leucine selectivity is unique among known chymase and granzyme-like enzymes. To our knowledge, guinea pig chymase is the first Leu-selective vertebrate peptidase described. This “Leu-ase” activity can be added to the already impressively wide spectrum of chymase/granzyme family primary specificities, including chymotryptic (e.g. human chymase and mouse MCP-4), tryptic (e.g. granzyme A and human cathepsin G), elastolytic (e.g. mouse and rat MCP-5), Met-ase (granzyme M), and Asp-ase (e.g. granzyme B). Even within the α-chymase group, which is closely related in overall primary structure and clearly monophyletic (see Fig. 2), Leu-ase activity can now be added to the chymotryptic or elastolytic activities characteristic of other enzymatically characterized members of this clade. Thus, the present work provides further evidence that phylogenetic similarity is an unreliable guide to function among chymase- and granzyme-like peptidases.
The causes and putative benefits of functional plasticity and hyperevolution in this group of peptidases are not known. However, because most if not all granzymes and chymases are expressed in immune cells and many make demonstrable contributions to host defense, guinea pig chymase also may serve in this capacity. We speculate that evolutionary pressures exerted by pathogens select for specificity-altering mutations, thereby accelerating the pace of functional change. This hypothesis implies that these peptidases probably evolved to cleave exogenous (i.e. pathogen-derived) targets rather than endogenous targets, like angiotensin I. Such a role in detoxification or killing has been suggested for some chymases and other secreted immune cell peptidases (11, 12).
Husain and co-workers (31) presented evidence that the chameleon-like capacity of chymase and granzyme-like peptidases to change specificity in response to one or a few amino acid changes was preceded 170 or more million years ago by despecializing mutations that created a nonspecific “stemzyme” from an ancestral peptidase with tryptic specificity. More sequence-selective chymases and granzyme B-like peptidases evolved from the broad-specificity stemzyme, which was unusually tolerant of amino acid changes in the substrate binding pocket. After millions of years of mutation, selection, and specialization, these enzymes now appear to have become intolerant of mutations in this region, especially those involving the so-called specificity triad residues, which play a major role in forming the P1 pocket accommodating the side chain of the amino acid at the site of target peptide hydrolysis. Chymases are prime examples of the great sensitivity of P1 target site preferences to mutations in specificity triad residues. Prior work showed that mutation of a single amino acid (specificity triad residue 216) from Gly to Val transformed rat and mouse α-chymase specificity from chymotryptic to elastolytic (19, 20). The example provided by guinea pig α-chymase in the present work is similarly dramatic, with a fundamental change from chymotryptic to Leu-ase specificity being largely attributable to a single amino acid change, in this case of Gly216 to Ala. In both examples, reversion of residue 216 in the native chymases to the Gly found in classic chymases of chymotryptic specificity (see Table 1) “restores” chymotryptic activity. Thus, this work underscores the key role of Gly216 in preserving chymotryptic activity.
As shown in Table 1, all known serine peptidases with an ability to hydrolyze targets after aromatic amino acids contain Gly216. The present work, combined with work of previous investigators, establishes that the presence at this position of any kind of side chain, including the minimal methyl group of Ala216 in guinea pig chymase, essentially abolishes chymotryptic activity. Amino acids with large side chains, such as Tyr and Arg, in the 216-position appear to be completely inactivating, as reported for mouse MCP-2 and MCP-8 (36, 37). Residues with side chains of intermediate size, such as Val216 in murine and hamster α-chymases and Ser216 in granzyme M, are associated with elastase and Met-ase activity, respectively, but have little if any ability to hydrolyze peptides after aromatic amino acids. The “gatekeeping” role of residue 216 also is supported by the structural and modeling studies (19, 20, 23, 31), including those in the present work, which suggests that the 216 side chain influences the volume, shape, and hydrophobicity of the binding pocket accommodating the P1 residue of the substrate (see Fig. 6). Interestingly, guinea pig chymase is the only serine peptidase characterized to date with Ala in position 216, although recently deposited genomic sequence suggests the presence of an α-like chymase in the American pika, which is a lagomorph with a specificity triad, including Ala216, identical to that of the guinea pig enzyme. Thus, there are likely to be other mammalian chymase-like enzymes preferring P1 Leu.
Notwithstanding the above evidence of the essential nature of Gly in the 216-position, it should be emphasized that Gly216 may be necessary but certainly is not sufficient for chymotryptic activity. Granzyme B, for example, features Gly216 but is an Asp-ase due to the presence of Arg226 in another specificity triad position. Furthermore, most if not all peptidases of tryptic specificity feature Gly216, although almost universally in combination with the acidic specificity triad residue Asp189, which attracts basic substrate side chains to the P1 binding pocket and is not present in chymotryptic peptidases. It also may be important that wild type guinea pig chymase differs from other serine peptidases in another specificity triad position, 226, which is occupied by relatively bulky Val rather than the Ala found in most if not all active chymase-like peptidases (see Table 1). This residue and potentially others in the more extended substrate binding site may need to be changed to more fully “humanize” the substrate preferences of guinea pig α-chymase.
Somewhat unexpectedly, our comparisons of overall structure in Figs. 1 and 2 suggest that guinea pig chymase is much more closely related to human chymase than are other rodent α-chymases, including mouse, rat, and hamster. Indeed, as shown in Fig. 1, the amino acid sequence of the guinea pig enzyme is more closely related to human α-chymase than to mouse α. This adds fuel to the long smoldering controversy about whether guinea pigs are rodents and whether the order Rodentia is monophyletic (32, 38, 39). From a phylogenetic standpoint, it also is interesting that opossum chymase aligns with the α clade, suggesting that α-chymases existed before marsupial and placental mammals diverged ∼185 million years ago (14) and that α may be the ancestral chymase. Origins of the β-chymases that so far appear to be expressed uniquely in myomorphic rodents (and not in guinea pigs) remain obscure. Because β-chymases are less conserved than α, as shown in Fig. 2, the molecular clocks of the clades probably differ, complicating attempts to identify their respective origins. Intriguingly, genomes of dogs (40) and guinea pigs (data not shown) contain a β-like apparent pseudogene, which may be the remnant of an ancestral β locus now active only in myomorphic rodents and which, in the case of rats and mice, not only persists but is greatly expanded by chains of duplications (14, 15).
The profile of susceptibility to a panel of inhibitors of wild type guinea pig chymase is unique compared with the A216G mutant and with human chymase. The ability to resist complete inhibition by chymostatin even when used in an ∼2000-fold molar excess is especially notable. Chymostatin is a natural product thought to block the P1 substrate binding site by one or more of its Phe-like phenyl moieties, which our models suggest are inaccessible to chymostatin in wild type guinea pig chymase. The unusually high level of resistance to human α1-antitrypsin is also notable and may be due to the shallowness of the guinea pig P1 side chain binding pocket, which will not readily accommodate the elongated side chain of the Met in the P1 position of the α1-antitrypsin reactive site loop. Similar considerations appear to explain the greater sensitivity (compared with human chymase) of wild type guinea pig chymase to an alternative major circulating serpin, α1-antichymotrypsin, because the inhibitor's P1 reactive site loop residue is Leu, which is preferred by the guinea pig enzyme in target peptides. On the other hand, P1 preferences alone do not explain why human chymase resists α1-antichymotrypsin more strongly than does guinea pig chymase, given that the human enzyme may be as active a Leu-ase in a kinetic sense (see Table 2). This apparent paradox may be explained by the reported nonsuicidal, serpin-degrading activity of human chymase, which occurs in parallel with formation of a serpin-chymase inhibitory complex (41), thereby inflating the stoichiometry of inhibition. Furthermore, swapping of serpin reactive site loops shows that the kinetics and stoichiometry of inhibition are not solely functions of the identity and position of residues in the reactive site loop (42).
Acknowledgments
We thank Katrin Schliemann and Martin Weber for technical assistance in cloning and purifying chymases, respectively.
The nucleotide sequence reported in this paper has been submitted to the DDBJ/GenBank™/EBI Data Bank with accession number(s) AM851020.
This work was supported, in whole or in part, by National Institute of Health Grants HL024136 (to G. H. C. and W. W. R.) and T32 07185 (to N. N. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: UPGMA, unweighted pair group with arithmetic mean; MES, 2-(N-morpholino)ethanesulfonic acid; NA, nitroanilide; MCP, mast cell protease; Fmoc, N-(9-fluorenyl)methoxycarbonyl; contig, group of overlapping clones.
References
- 1.Knight, P. A., Wright, S. H., Lawrence, C. E., Paterson, Y. Y., and Miller, H. R. (2000) J. Exp. Med. 192 1849-1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ju, H., Gros, R., You, X., Tsang, S., Husain, M., and Rabinovitch, M. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 7469-7474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li, M., Liu, K., Michalicek, J., Angus, J. A., Hunt, J. E., Dell'Italia, L. J., Feneley, M. P., Graham, R. M., and Husain, A. (2004) J. Clin. Invest. 114 112-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tchougounova, E., Lundequist, A., Fajardo, I., Winberg, J. O., Abrink, M., and Pejler, G. (2005) J. Biol. Chem. 280 9291-9296 [DOI] [PubMed] [Google Scholar]
- 5.Coussens, L. M., Raymond, W. W., Bergers, G., Laig-Webster, M., Behrendtsen, O., Werb, Z., Caughey, G. H., and Hanahan, D. (1999) Genes Dev. 13 1382-1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishimoto, M., Takai, S., Kim, S., Jin, D., Yuda, A., Sakaguchi, M., Yamada, M., Sawada, Y., Kondo, K., Asada, K., Iwao, H., Sasaki, S., and Miyazaki, M. (2001) Circulation 104 1274-1279 [DOI] [PubMed] [Google Scholar]
- 7.Abonia, J. P., Friend, D. S., Austen, W. G., Jr., Moore, F. D., Jr., Carroll, M. C., Chan, R., Afnan, J., Humbles, A., Gerard, C., Knight, P., Kanaoka, Y., Yasuda, S., Morokawa, N., Austen, K. F., Stevens, R. L., and Gurish, M. F. (2005) J. Immunol. 174 7285-7291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakaguchi, M., Takai, S., Jin, D., Okamoto, Y., Muramatsu, M., Kim, S., and Miyazaki, M. (2004) Eur. J. Pharmacol. 493 173-176 [DOI] [PubMed] [Google Scholar]
- 9.Greco, M. N., Hawkins, M. J., Powell, E. T., Almond, H. R., Jr., de Garavilla, L., Hall, J., Minor, L. K., Wang, Y., Corcoran, T. W., Di Cera, E., Cantwell, A. M., Savvides, S. N., Damiano, B. P., and Maryanoff, B. E. (2007) J. Med. Chem. 50 1727-1730 [DOI] [PubMed] [Google Scholar]
- 10.de Garavilla, L., Greco, M. N., Sukumar, N., Chen, Z. W., Pineda, A. O., Mathews, F. S., Di Cera, E., Giardino, E. C., Wells, G. I., Haertlein, B. J., Kauffman, J. A., Corcoran, T. W., Derian, C. K., Eckardt, A. J., Damiano, B. P., Andrade-Gordon, P., and Maryanoff, B. E. (2005) J. Biol. Chem. 280 18001-18007 [DOI] [PubMed] [Google Scholar]
- 11.Metz, M., Piliponsky, A. M., Chen, C. C., Lammel, V., Abrink, M., Pejler, G., Tsai, M., and Galli, S. J. (2006) Science 313 526-530 [DOI] [PubMed] [Google Scholar]
- 12.Mellon, M. B., Frank, B. T., and Fang, K. C. (2002) J. Immunol. 168 290-297 [DOI] [PubMed] [Google Scholar]
- 13.Caughey, G. H. (2006) Curr. Resp. Med. Rev. 2 263-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallwitz, M., Reimer, J. M., and Hellman, L. (2006) Immunogenetics 58 655-669 [DOI] [PubMed] [Google Scholar]
- 15.Puente, X. S., and Lopez-Otin, C. (2004) Genome Res. 14 609-622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaiserman, D., Bird, C. H., Sun, J., Matthews, A., Ung, K., Whisstock, J. C., Thompson, P. E., Trapani, J. A., and Bird, P. I. (2006) J. Cell Biol. 175 619-630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polanowska, J., Krokoszynska, I., Czapinska, H., Watorek, W., Dadlez, M., and Otlewski, J. (1998) Biochim. Biophys. Acta 1386 189-198 [DOI] [PubMed] [Google Scholar]
- 18.Raptis, S. Z., Shapiro, S. D., Simmons, P. M., Cheng, A. M., and Pham, C. T. (2005) Immunity 22 679-691 [DOI] [PubMed] [Google Scholar]
- 19.Kunori, Y., Koizumi, M., Masegi, T., Kasai, H., Kawabata, H., Yamazaki, Y., and Fukamizu, A. (2002) Eur. J. Biochem. 269 5921-5930 [DOI] [PubMed] [Google Scholar]
- 20.Karlson, U., Pejler, G., Tomasini-Johansson, B., and Hellman, L. (2003) J. Biol. Chem. 278 39625-39631 [DOI] [PubMed] [Google Scholar]
- 21.Caughey, G. H., Raymond, W. W., and Wolters, P. J. (2000) Biochim. Biophys. Acta 1480 245-257 [DOI] [PubMed] [Google Scholar]
- 22.Muilenburg, D. J., Raymond, W. W., Wolters, P. J., and Caughey, G. H. (2002) Biochim. Biophys. Acta 1596 346-356 [DOI] [PubMed] [Google Scholar]
- 23.Kervinen, J., Abad, M., Crysler, C., Kolpak, M., Mahan, A. D., Masucci, J. A., Bayoumy, S., Cummings, M. D., Yao, X., Olson, M., de Garavilla, L., Kuo, L., Deckman, I., and Spurlino, J. (2008) J. Biol. Chem. 283 427-436 [DOI] [PubMed] [Google Scholar]
- 24.Miyazaki, M., Takai, S., Jin, D., and Muramatsu, M. (2006) Pharmacol. Ther. 112 668-676 [DOI] [PubMed] [Google Scholar]
- 25.He, S., and Walls, A. F. (1998) Br. J. Pharmacol. 125 1491-1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karlson, U., Pejler, G., Froman, G., and Hellman, L. (2002) J. Biol. Chem. 277 18579-18585 [DOI] [PubMed] [Google Scholar]
- 27.Pereira, P. J. B., Wang, Z. M., Rubin, H., Huber, R., Bode, W., Schechter, N. M., and Strobl, S. (1999) J. Mol. Biol. 286 163-173 [DOI] [PubMed] [Google Scholar]
- 28.Wolters, P. J., Pham, C. T., Muilenburg, D. J., Ley, T. J., and Caughey, G. H. (2001) J. Biol. Chem. 276 18551-18556 [DOI] [PubMed] [Google Scholar]
- 29.Pham, C. T., and Ley, T. J. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 8627-8632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adkison, A. M., Raptis, S. Z., Kelley, D. G., and Pham, C. T. (2002) J. Clin. Invest. 109 363-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wouters, M. A., Liu, K., Riek, P., and Husain, A. (2003) Mol. Cell 12 343-354 [DOI] [PubMed] [Google Scholar]
- 32.D'Erchia, A. M., Gissi, C., Pesole, G., Saccone, C., and Arnason, U. (1996) Nature 381 597-600 [DOI] [PubMed] [Google Scholar]
- 33.Urata, H., Kinoshita, A., Misono, K. S., Bumpus, F. M., and Husain, A. (1990) J. Biol. Chem. 265 22348-22357 [PubMed] [Google Scholar]
- 34.Wang, Z., Walter, M., Selwood, T., Rubin, H., and Schechter, N. M. (1998) Biol. Chem. 379 167-174 [DOI] [PubMed] [Google Scholar]
- 35.Andersson, M. K., Karlson, U., and Hellman, L. (2008) Mol. Immunol. 45 766-775 [DOI] [PubMed] [Google Scholar]
- 36.Pemberton, A. D., Brown, J. K., Wright, S. H., Knight, P. A., McPhee, M. L., McEuen, A. R., Forse, P. A., and Miller, H. R. (2003) Clin. Exp. Allergy 33 1005-1012 [DOI] [PubMed] [Google Scholar]
- 37.Gallwitz, M., Enoksson, M., and Hellman, L. (2007) Immunogenetics 59 391-405 [DOI] [PubMed] [Google Scholar]
- 38.Graur, D., Hide, W. A., and Li, W. H. (1991) Nature 351 649-652 [DOI] [PubMed] [Google Scholar]
- 39.Huchon, D., Chevret, P., Jordan, U., Kilpatrick, C. W., Ranwez, V., Jenkins, P. D., Brosius, J., and Schmitz, J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7495-7499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallwitz, M., and Hellman, L. (2006) Immunogenetics 58 641-654 [DOI] [PubMed] [Google Scholar]
- 41.Schechter, N. M., Jordan, L. M., James, A. M., Cooperman, B. S., Mei, W. Z., and Rubin, H. (1993) J. Biol. Chem. 268 23626-23633 [PubMed] [Google Scholar]
- 42.Plotnick, M. I., Schechter, N. M., Wang, Z. M., Liu, X., and Rubin, H. (1997) Biochemistry 36 14601-14608 [DOI] [PubMed] [Google Scholar]
- 43.Raymond, W. W., Waugh Ruggles, S., Craik, C. S., and Caughey, G. H. (2003) J. Biol. Chem. 278 34517-34524 [DOI] [PubMed] [Google Scholar]
- 44.Powers, J. C., Tanaka, T., Harper, J. W., Minematsu, Y., Barker, L., Lincoln, D., Crumley, K. V., Fraki, J. E., Schechter, N. M., Lazarus, G. G., Nakajima, K., Nakashino, K., Neurath, H., and Woodbury, R. G. (1985) Biochemistry 24 2048-2058 [DOI] [PubMed] [Google Scholar]
- 45.Caughey, G. H., Leidig, F., Viro, N. F., and Nadel, J. A. (1988) J. Pharmacol. Exp. Ther. 244 133-137 [PubMed] [Google Scholar]
- 46.Yoshida, N., Everitt, M. T., Neurath, H., Woodbury, R. G., and Powers, J. C. (1980) Biochemistry 19 5799-5804 [DOI] [PubMed] [Google Scholar]
- 47.Le Trong, H., Neurath, H., and Woodbury, R. G. (1987) Proc. Natl. Acad. Sci. U. S. A. 84 364-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Del Mar, E. G., Largman, C., Brodrick, J. W., Fassett, M., and Geokas, M. C. (1980) Biochemistry 19 468-472 [DOI] [PubMed] [Google Scholar]
- 49.Harris, J. L., Backes, B. J., Leonetti, F., Mahrus, S., Ellman, J. A., and Craik, C. S. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 7754-7759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Debela, M., Magdolen, V., Schechter, N., Valachova, M., Lottspeich, F., Craik, C. S., Choe, Y., Bode, W., and Goettig, P. (2006) J. Biol. Chem. 281 25678-25688 [DOI] [PubMed] [Google Scholar]
- 51.Edwards, K. M., Kam, C. M., Powers, J. C., and Trapani, J. A. (1999) J. Biol. Chem. 274 30468-30473 [DOI] [PubMed] [Google Scholar]
- 52.Zamolodchikova, T. S., Vorotyntseva, T. I., and Antonov, V. K. (1995) Eur. J. Biochem. 227 866-872 [DOI] [PubMed] [Google Scholar]
- 53.Smyth, M. J., Wiltrout, T., Trapani, J. A., Ottaway, K. S., Sowder, R., Henderson, L. E., Kam, C. M., Powers, J. C., Young, H. A., and Sayers, T. J. (1992) J. Biol. Chem. 267 24418-24425 [PubMed] [Google Scholar]
- 54.Smyth, M. J., O'Connor, M. D., Trapani, J. A., Kershaw, M. H., and Brinkworth, R. I. (1996) J. Immunol. 156 4174-4181 [PubMed] [Google Scholar]
- 55.Harris, J. L., Niles, A., Burdick, K., Maffitt, M., Backes, B. J., Ellman, J. A., Kuntz, I., Haak-Frendscho, M., and Craik, C. S. (2001) J. Biol. Chem. 276 34941-34947 [DOI] [PubMed] [Google Scholar]