Abstract
The critical tumor suppressor p53 is mutated or functionally inactivated in nearly all cancers. We have shown previously that the MDM2-MDMX complex functions as an integral unit in targeting p53 for degradation. Here we identify the small protein 14-3-3 as a binding partner of MDMX, which binds at the C terminus (Ser367) in a phosphorylation-dependent manner. Importantly, we demonstrate that the serine/threonine kinase Akt mediates phosphorylation of MDMX at Ser367. This phosphorylation leads to stabilization of MDMX and consequent stabilization of MDM2. Previous studies have shown that Akt phosphorylates and stabilizes MDM2. Our data suggest that stabilization of MDMX by Akt may be an alternative mechanism by which Akt up-regulates MDM2 protein levels and exerts its oncogenic effects on p53 in tumor cells.
The p53 tumor suppressor is a critical regulator of cell proliferation. In response to cellular stresses, p53 elicits a response that results in cell cycle arrest or cell death. The activation of p53 occurs in a robust manner, and its proper regulation is therefore critical to maintain normal cellular functions (1). The MDM2 protein is one key regulator of p53 activity. The manner by which MDM2 inhibits p53 activity is 2-fold. First, MDM2 binds to the transactivation domain of p53, preventing transcription of downstream targets. Second, and perhaps more importantly, MDM2 functions as an E3 ligase to maintain low protein levels of p53 under non-stressed conditions and to return p53 to normal levels after a damage response is resolved (2).
More recently, a close homologue of MDM2, MDMX, has been shown to be another critical regulator of p53 activity (3, 4). MDMX also suppresses p53 transcriptional activity by binding to its transactivation domain (5–7). Studies have shown that MDMX can bind to and stabilize MDM2; however, MDMX has not been shown to possess E3 ligase activity in vivo (8, 9). Furthermore, we (10) and others (11) have shown that the MDM2-MDMX heterocomplex is more efficient at ubiquitylating p53. Of interest, MDM2 and MDMX have been found amplified in certain cancers that retain wild-type p53, indicating that these critical negative regulators of p53 contribute to its inactivation and thus promote cell proliferation (12–14).
The serine/threonine protein kinase Akt promotes cell survival and proliferation. Akt is responsible for stimulating pro-survival signals through its kinase activity (15–17). One example is the phosphorylation of the pro-apoptotic Forkhead family of transcription factors. Following phosphorylation by Akt, a binding site is generated for the 14-3-3 protein, which in turn binds to and inactivates FOXO proteins by promoting their nuclear export (18–20). Another example of the pro-survival function of Akt is its ability to phosphorylate the pro-apoptotic protein Bad. Phosphorylation of Bad leads to 14-3-3 binding and results in loss of its ability to induce mitochondrial membrane permeability, thus preventing initiation of the intrinsic apoptotic pathway (21).
14-3-3 proteins are small acidic proteins that bind to phosphorylated serine/threonine residues as dimers. There are seven known isoforms of 14-3-3 in humans. They modify the function of their partner proteins by a multitude of mechanisms such as sequestration, alteration of protein-protein interactions, alteration of enzymatic activity, or by functioning as adaptor proteins (21–23). In several cases, protein phosphorylation by Akt generates a 14-3-3 binding site; both the Akt consensus sequence and the 14-3-3 binding sequence are similar (21–23).
Akt has been implicated in the down-regulation of p53 via MDM2. Studies have shown that MDM2 is phosphorylated by Akt at different sites, resulting in its stabilization (24–27). Other studies have shown nuclear translocation of MDM2 in response to phosphorylation by Akt. This results in decreased p53 transcriptional activity from MDM2 binding at the transactivation domain and increases the ubiquitylation of p53 (24, 25, 28–30). However, MDM2 stability, is not regulated solely through phosphorylation events. Previous studies have shown that MDMX stabilizes MDM2 after multimerization; in the absence of MDMX, MDM2 is less stable (8, 10, 11, 31). Importantly, MDMX degradation has been shown to be triggered by post-translational modifications such as phosphorylation in response to DNA damage or caspase cleavage to promote apoptosis (32–38).
Here we show that Akt is capable of phosphorylating MDMX on Ser367, generating a 14-3-3 binding site; this phosphorylation renders it more stable, thereby enhancing MDM2 stability. Moreover, this translates into p53 inactivation by preventing p53 transcriptional activity. Our work suggests that Akt is able to functionally inactivate p53 through its regulation of MDMX and thereby contributes to the oncogenic activity of Akt.
EXPERIMENTAL PROCEDURES
Preparation of Plasmids and Proteins—pcDNA3-MDMX(S367A) and pcDNA3-MDMX(P369R) mutants were generated by two-step site-directed PCR mutagenesis utilizing the following primers: Ser367 to Ala, 5′-TGTCGA-AGAACCATTGCGGCTCCTGTCGTT and 3′-TCTAAC-GACAGGAGCCGCAATGGTTCTTCG; and Pro369 to Arg, 5′-ATTTCGGCTCGAGTCGTTAGA and 3′-TCTAACGAC-TCGAGCCGAAAT. Plasmids containing constitutively active, myristoylated, hemagglutinin-tagged Akt with and without the pleckstrin homology domain and dominant-negative, hemagglutinin-tagged Akt(K179M) and Akt(T308A,S473A) were kind gifts of Drs. Ziv Radisavljevic and Brendan Manning (Harvard School of Public Health). Difopein and 14-3-3ε plasmids were kindly provided by Dr. Haian Fu (Emory University), and 14-3-3β, -γ, -ζ, and -σ plasmids were a kind gift of Dr. James DeCaprio (Dana-Farber Cancer Institute). GST2 protein production has been described previously (39).
Generation of Inducible Cell Lines—Full-length FLAG-MDMX, full-length FLAG-MDM2, FLAG-MDMX RFD, and FLAG-MDM2 RFD were cloned into pcDNA4 (Invitrogen). Plasmids were linearized and transfected into 293-TREx cells (Invitrogen). Cells were put under Zeocin (Invitrogen) selection, and positive clones that expressed the insert of interest under tetracycline were chosen.
Immunoaffinity Purification and Mass Spectrometry—Cells were induced with tetracycline, and cells were harvested 24 h post-induction. Cell lysates were prepared in 1% Triton X-100 lysis buffer and incubated with preconjugated agarose beads with anti-FLAG antibodies (Sigma). After boiling in Laemmli buffer, lysates were resolved by 12.5% SDS-PAGE. Gel was fixed in methanol and stained with Coomassie Blue. Gel bands were excised, washed in acetonitrile, and sent for digestion and tandem mass spectrometry (NIEHS Harvard School of Public Health Proteomics Facility, Boston, MA).
Cell Culture and Transfection—293T, 293-TREx, H1299, U2OS, OVCA420, LNCaP, and MCF7 cells (American Type Culture Collection) were maintained in minimal essential medium, Dulbecco's modified Eagle's medium, or RPMI 1640 medium supplemented with 10% fetal bovine serum. For serum starvation, cells were maintained in serum-free minimal essential medium for 15–18 h and then stimulated with human recombinant IGF-1 (Sigma) at 10–50 ng/ml. For experiments with the PI 3-kinase inhibitor, cells grown in serum-free medium were pretreated with 10 μm LY294002 (Sigma) for 30 min and then treated with IGF-1 as described above. Transfections were performed by calcium phosphate precipitation or the Lipofectamine method (Invitrogen).
Preparation of Whole Cell Extracts and Immunoprecipitation Analysis—Cells were transfected in 60-mm plates with 1–5 μg of DNA and harvested 24 h post-transfection. Cells were lysed as described previously (40). Protein concentrations were determined using Bio-Rad protein assay. After addition of 5×Laemmli buffer, the samples were incubated at 95 °C for 10 min and resolved by SDS-PAGE. For immunoprecipitation, cell lysates were prepared in 0.5% Triton X-100 lysis buffer and incubated with anti-FLAG M5 beads (Sigma) for an additional 2 h. Immune complexes and whole cell lysates were separated by SDS-PAGE. Proteins were transferred onto nitrocellulose membranes (Schleicher & Schuell) and probed with the following antibodies as indicated: anti-MDMX (BL1258, Bethyl Laboratories), anti-MDMX pS367 (a kind gift of Dr. Jiandong Chen) or BL1563 where noted, anti-MDM2 (Ab-1, Oncogene), anti-FLAG M2 (Sigma), anti-p53 (Ab-6, Oncogene), anti-Akt pS473 (Cell Signaling), anti-Akt (Cell Signaling), anti-pan-14-3-3β (Santa Cruz Biotechnology), anti-p21 (187, Santa Cruz Biotechnology), and anti-β-actin (AC-15, Sigma). Proteins were visualized with an enhanced chemiluminescence detection system (PerkinElmer Life Sciences).
In Vitro Kinase Assay—293T cells were transiently transfected with constitutively active or dominant-negative Akt. Another set of 293T cells was transiently transfected with FLAG-MDMX plasmids. MDMX immunoprecipitates were incubated with lysates expressing constitutively active or dominant-negative Akt in kinase assay buffer in the presence of 10 mm [γ-32P]ATP and incubated at 30 °C for 30 min. Samples were resolved on a 10% SDS-polyacrylamide gel. The gel was dried and analyzed by autoradiography.
RESULTS
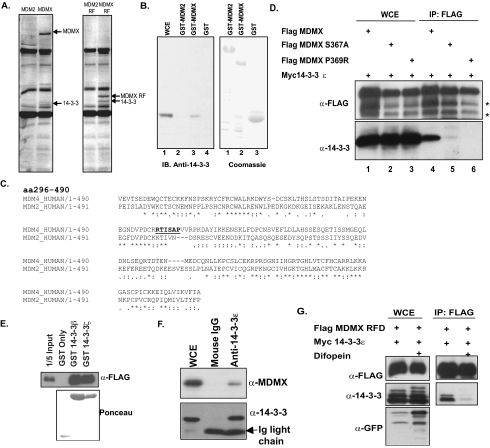
14-3-3 Is a Novel MDMX Binding Partner—To identify novel binding partners of MDMX, a proteomic screen was performed. An inducible cell line was utilized to express a full-length FLAG-tagged MDMX or MDM2 protein, as well as a FLAG-tagged mini-protein encoding the zinc RFDs of MDMX (amino acids 301–490) or MDM2 (amino acids 302–491). Tetracycline was used to induce the expression of these FLAG-tagged proteins in 293-TREx cells, and anti-FLAG antibody was used to purify the FLAG-tagged protein complexes. Immunoprecipitates were separated using SDS-PAGE, and unique bands were cut and sent for identification by mass spectrometry. As shown in Fig. 1A, a unique band associated with full-length MDMX and the zinc RING finger domain of MDMX but not with MDM2. Results from mass spectrometry analysis revealed that this band corresponded to the protein 14-3-3. The interaction between MDMX and 14-3-3 was verified in vitro using recombinant GST-MDMX fusion proteins (Fig. 1B).
FIGURE 1.
14-3-3 is an MDMX binding partner. A, 293-TREx cells were induced with tetracycline to express FLAG-tagged full-length MDM2 or MDMX or the MDM2 RFD or MDMX RFD. Immunoprecipitation was carried out followed by SDS-PAGE, and unique bands were sent for mass spectrometry. B, GST-MDM2 and GST-MDMX were incubated with lysate expressing 14-3-3ε to determine binding in vitro. WCE, whole cell extract; IB, immunoblotting. C, the sequence alignment of MDM2 and MDMX is shown. The underlined portion indicates the 14-3-3 binding motif. aa, amino acids. D, FLAG-tagged MDMX, MDMX(S367A), and MDMX(P369R) were coexpressed with 14-3-3 in 293T cells. Immunoprecipitation (IP) was carried out against FLAG-tagged proteins using M2 beads. Asterisks indicate degradation products often seen with MDMX overexpression. E, FLAG-tagged MDMX RFD was expressed in 293T cells, and lysates were incubated with GST beads, GST-14-3-3β, or GST-14-3-3ζ. F, Myc-14-3-3ε was coexpressed with FLAG-MDMX in 293T cells. Immunoprecipitation was carried out against Myc-14-3-3ε. G, FLAG-tagged MDMX RFD was coexpressed with 14-3-3ε and with or without difopein in 293T cells. FLAG immunoprecipitation was then carried out, followed by Western blot analysis with the indicated antibodies. GFP, green fluorescent protein.
Upon careful analysis of the MDMX sequence, we identified a 14-3-3 mode 1 binding motif (RXXSXP) in residues 364–369 (Fig. 1C, underlined). Additionally, the presence of a 14-3-3 binding motif in the MDMX sequence was confirmed using the Massachusetts Institute of Technology Scansite program (41). This sequence is specific to MDMX, as it is not found in the MDM2 sequence, further supporting our immunoprecipitation results (Fig. 1C, underlined).
14-3-3 proteins are small acidic proteins that typically interact with their ligands in a phosphorylation-dependent manner. To investigate whether 14-3-3 bound to MDMX in this manner, we subjected Ser367 to point-directed mutagenesis. Using site-directed PCR, we mutated the putative 14-3-3 binding site on MDMX from serine to alanine. As shown in Fig. 1D, the MDMX(S367A) point mutant was unable to bind 14-3-3 proteins. To rule out the possibility that alteration of the phosphorylation status of MDMX interferes with 14-3-3 binding, we mutated a conserved proline residue to an arginine, thereby eliminating the 14-3-3 binding motif. This MDMX(P369R) mutant was also unable to bind to 14-3-3, indicating that 14-3-3 is a bona fide binding partner of MDMX (Fig. 1D).
We next performed a GST pulldown assay using GST-14-3-3β and GST-14-3-3ζ to demonstrate direct binding of the C-terminal MDMX RFD (Fig. 1E). Immunoprecipitation of Myc-14-3-3 showed binding of full-length MDMX to 14-3-3 (Fig. 1F). In total, we observed binding of MDMX to the four 14-3-3 isoforms (β, ε, γ (data not shown), and ζ) we had available, but not to 14-3-3σ, which has been shown to be a p53 target gene (data not shown) (21).
Further evidence for specific 14-3-3 binding was shown using the 14-3-3 competitive inhibitor difopein. 14-3-3 binding to the MDMX RFD in the presence of difopein was not observed (Fig. 1G). Additionally, point mutations of MDMX or disruption of 14-3-3 binding did not interfere with binding to MDM2 (data not shown).
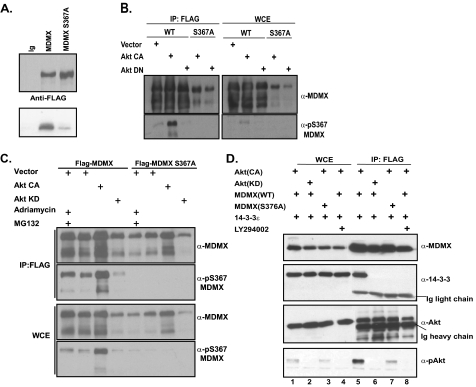
MDMX Is a Substrate of the Akt Kinase—14-3-3 proteins normally bind phosphorylated targets. Therefore, we investigated which kinase was responsible for phosphorylation of MDMX at Ser367. The sequence surrounding Ser367 appears to fit the consensus site RXRXXS of the serine/threonine protein kinase Akt. Although the sequence is not a perfect match, we tested whether the Akt pathway was mediating phosphorylation of MDMX. An in vitro kinase assay showed that Akt phosphorylated wild-type MDMX, but not the S367A mutant of MDMX (Fig. 2A). The residual band observed in the MDMX(S367A) lane suggests that Ser367 is the primary site for Akt-mediated phosphorylation; however, there may be other, yet uncharacterized Akt sites on MDMX. To further assess whether Akt was mediating phosphorylation of Ser367, FLAG immunoprecipitation was carried out on 293T cells cotransfected with FLAG-MDMX and either constitutively active Akt (CA-Akt) or dominant-negative Akt (DN-Akt). Western blot analysis utilizing a phospho-Ser367-specific antibody indicated that MDMX is indeed phosphorylated at this site in an Akt-dependent manner (Fig. 2B). Interestingly, when CA-Akt was coexpressed with MDMX, we also observed stabilization of MDMX, whereas the protein level of the S367A phosphorylation-deficient mutant was largely unchanged (Fig. 2B).
FIGURE 2.
Akt-mediated phosphorylation of MDMX. A, an in vitro kinase assay was carried out with immunopurified FLAG-tagged MDMX or MDMX(S367A) incubated with cell lysates expressing constitutively active Akt. B, 293T cells were cotransfected with CA-Akt or DN-Akt and FLAG-tagged MDMX or MDMX(S367A). FLAG immunoprecipitation (IP; left panel), whole cell extract analysis (WCE; right panel), and Western blot analysis were carried out using a phospho-specific antibody against Ser367. WT, wild-type. C, 293T cells were cotransfected and analyzed as in B with an additional sample treated with Adriamycin and the proteasome inhibitor MG132. Top panels, immunoprecipitation; bottom panels, whole cell extract. D, cotransfection of 293T cells and immunoprecipitation were carried out as in B with additional treatment with LY294002, and Western blot analysis was carried out with the indicated antibodies. Left, whole cell extract; right, immunoprecipitation.
Recent studies have shown that Ser367 of MDMX is phosphorylated in response to DNA damage. To test whether phosphorylation of Ser367 is specific to the DNA damage response, we compared the phosphorylation status of MDMX in response to Adriamycin treatment in cells transfected with CA-Akt or DN-Akt. Phosphorylation of Ser367 was observed in the presence of CA-Akt or vector only, as well as Adriamycin, but not KD-Akt (Fig. 2C). These results suggest, quite interestingly, that Ser367 phosphorylation is not a response specific to DNA damage. Of note, Akt-mediated phosphorylation was even stronger than Adriamycin-induced phosphorylation, implicating Akt as a physiologically relevant kinase for MDMX.
We next investigated the status of 14-3-3 binding with respect to Akt activity. Cotransfection of MDMX with CA-Akt led to 14-3-3 binding, whereas no binding was observed in the presence of DN-Akt (Fig. 2D). These results suggest that phosphorylation of Ser367 is required for 14-3-3 binding. Addition of the PI 3-kinase inhibitor LY294002 elicited the same effect on MDMX/14-3-3 binding as did DN-Akt (Fig. 2D).
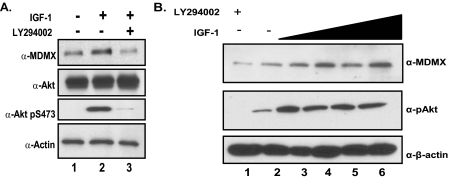
To determine whether endogenous Akt is capable of stabilizing MDMX, we treated MCF7 cells with IGF-1 to activate the PI 3-kinase/Akt pathway after serum starvation. As suggested by previous experiments, endogenous MDMX is stabilized by activation of the Akt pathway because MDMX protein levels increase in response to IGF-1 treatment after serum starvation; addition of the PI 3-kinase inhibitor LY294002 appears to diminish MDMX levels (Fig. 3A). These observations correlate with the phosphorylation of Akt at Ser473, which has been shown to be a good indicator of Akt activity. Additionally, the stabilization of MDMX in response to IGF-1 treatment was dose-dependent (Fig. 3B). These experiments suggest that endogenous MDMX protein levels are regulated by the PI 3-kinase/Akt pathway.
FIGURE 3.
Endogenous stabilization of MDMX mediated by Akt. A, MCF7 cells were serum-starved overnight and stimulated with IGF-1 for 4 h or pretreated with LY294002 for 30 min and then stimulated with IGF-1. B, MCF7 cells were serum-starved overnight and treated with increasing amounts of IGF-1 (1–50 ng/ml) for 4 h.
Stabilization of MDMX Results in Stabilization of MDM2 and Down-regulation of p53 Activity—Constitutively active Akt has been shown to function in a manner that promotes cell survival and proliferation, in part by stabilizing MDM2 and thus down-regulating p53 activity (24, 25, 29, 30). Our data support the hypothesis that Akt phosphorylates MDMX at Ser367, generates a 14-3-3 binding site, and leads to the binding of MDMX to 14-3-3, which results in its subsequent stabilization. Given the findings that MDMX is capable of stabilizing MDM2 and that the MDM2-MDMX heterocomplex is inherently more stable than the MDM2 homocomplex (10, 45, 46), we asked whether the effect of Akt on MDM2 was mediated by MDMX.
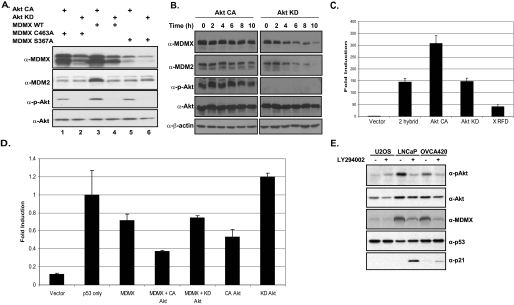
MDMX mutants were used to determine whether stabilization of MDM2 is dependent on MDMX in response to Akt activity. Overexpression of constitutively active Akt resulted in enhanced protein levels of wild-type MDMX, whereas overexpression of dominant-negative Akt did not (Fig. 4A). The levels of MDM2 are also increased in the presence of CA-Akt (Fig. 4A), as previous studies have shown. Interestingly, MDM2 levels appear to be unchanged in the presence of the MDMX(S367A) mutant (Fig. 4A). Moreover, coexpression with an MDMX RING mutant (C463A) that disrupts MDM2-MDMX binding failed to stabilize MDM2 levels, despite the fact that the RING mutant itself was stabilized by Akt. These data demonstrate that the stabilization of MDM2 by Akt is mediated by MDMX (Fig. 4A).
FIGURE 4.
Stabilization of MDMX mediated by Akt leads to MDM2 stabilization and p53 down-regulation. A, 293T cells were cotransfected with CA-Akt or KD-Akt and MDMX, MDMX(S367A), or MDMX(C463A). Western blot analysis was carried out with the indicated antibodies. WT, wild-type. B, 293T cells were cotransfected with MDMX, MDM2, and CA-Akt or KD-Akt. Cells were then treated with cycloheximide for the indicated time points. Western blot analysis was carried out with the indicated antibodies. C, a Gal4-Luc mammalian two-hybrid reporter system expressing Gal4-MDM2 and VP16-MDMX alone or with CA-Akt or DN-Akt or expressing MDMX RFD was used in 293T cells. D, H1299 cells were used for a luciferase assay to determine p53 transcriptional activity with the pG13 reporter and p53 alone or with MDMX, CA-Akt, KD-Akt, or a combination of MDMX and Akt. E, U2OS, LNCaP, and OVCA420 cells were treated with the PI 3-kinase inhibitor LY294002, and Western blot analysis was carried out with the indicated antibodies.
To test whether the half-lives of MDM2 and MDMX are extended by Akt activity, we cotransfected 293T cells with MDM2, MDMX, and either constitutively active Akt or kinase-dead Akt. Cells were then treated with cycloheximide, and protein levels were tracked for the indicated time points (Fig. 4B). The half-lives of both MDM2 and MDMX were dramatically increased in the presence of Akt activity, supporting previous result.
We next utilized a mammalian two-hybrid system to determine whether the presence of Akt increased the presence of an MDM2-MDMX complex. A luciferase reporter assay was used in which the reporter plasmid, pGal4-Luc, was coexpressed with a Gal4 DNA-binding domain fused to MDM2, and the VP16-activation domain was fused to MDMX to determine MDM2-MDMX binding (Fig. 4B). We have reported previously that expression of the MDMX RFD is capable of disrupting MDM2-MDMX binding (10); therefore, we used it as a negative control. In the presence of CA-Akt, we observed a 2-fold increase in signal compared with vector and KD-Akt, suggesting the ability of Akt to stabilize the MDM2-MDMX complex (Fig. 4B).
To determine the effects of stabilization of MDMX on p53 activity, we utilized another reporter assay. This time we chose to assess the level of p53 transcriptional activity. A luciferase assay was performed by cotransfecting H1299 cells, which are p53-deficient, with the p53-specific pG13-Luc reporter plasmid, Myc-p53, and wild-type MDMX, along with CA-Akt or KD-Akt. As shown in Fig. 4C, luciferase activity is increased when p53 is introduced into these cells and decreased in the presence of MDMX. When CA-Akt is coexpressed with MDMX, there is a reduction of p53 activity, compared with CA-Akt alone; KD-Akt had no effect on the ability of MDMX to suppress p53 activity. These results suggest that MDMX contributes to p53 suppression under Akt activity.
To further determine whether endogenous p53 and MDMX are affected by Akt, we utilized U2OS, LNCaP, and OVCA420 cells, the latter two of which have elevated Akt activity due to either phosphatase and tensin homolog (PTEN) deficiency (LNCaP) or Her-2 overexpression (OVCA420). We treated these cells with LY294002 to block Akt activity. In contrast with U2OS cells, in which Akt activity is relatively low, LNCaP and OVCA420 cells responded to the inhibitor with a significant decrease of Akt activity (Fig. 4D). LY294002 treatment also resulted in a decrease of MDMX steady-state levels, consistent with Akt-mediated stabilization of MDMX (Fig. 4D). Importantly, the effects of LY294002 correlated with increased p53 transcriptional activity (Fig. 4D). These data suggest that down-regulation of MDMX in response to suppression of Akt contributes to p53 activation.
Together, our data show that one mechanism by which Akt elicits its oncogenic function is through the stabilization of MDMX, potentially through 14-3-3 binding. MDMX stability is then translated into the stabilization of MDM2 and inactivation of p53.
DISCUSSION
A fundamental process in the development of cancer is the inactivation of the p53 tumor suppressor pathway. In some cases, this is achieved by mutation of the TP53 gene, resulting in a mutant protein unable to carry out its function as a transcription factor, although some tumors retain wild-type p53 (47). However, in those cases, p53 is functionally inactivated. One mechanism is through the up-regulation of the p53 negative regulator and E3 ligase MDM2. Amplifications of the MDM2 gene or stabilization of the MDM2 protein can result in the inactivation of p53 by increasing its degradation and inhibiting p53 transcriptional activity (25, 30, 48).
It has been reported that the pro-survival serine/threonine protein kinase Akt phosphorylates MDM2, resulting in its stabilization and nuclear translocation. This eventually leads to a functional inactivation of p53 (24, 25, 29, 30). Although these results strongly suggest that MDM2 mediates Akt-dependent inactivation of p53, the exact mechanism by which it does this remains unclear. Some studies have shown that Akt results in MDM2 nuclear translocation, whereas others do not; however, all studies indicate that MDM2 is stabilized by Akt. MDM2 possesses E3 ligase activity toward itself, and some reports suggest that MDM2 is inherently unstable; however, its homologue and binding partner, MDMX, which lacks the E3 ligase activity, is involved in its stabilization (45). MDMX itself has been shown to be an essential negative regulator of p53 (4, 42). We have shown recently that the MDM2-MDMX heterocomplex appears to be the predominant form in which both proteins exist in the cell and that disruption of this complex leads to MDM2 destabilization and p53 activation (10).
In pursuit of MDMX binding partners, we identified the 14-3-3 proteins. With the exception of 14-3-3σ, a p53 downstream target, all other 14-3-3 isoforms tested bound to MDMX. Upon further investigation, we determined that the 14-3-3 binding site on MDMX is generated by Akt. It is the specific phosphorylation of Ser367 mediated by Akt that stabilizes MDMX. Furthermore, we have shown that stabilization of MDMX by Akt results in the subsequent stabilization of MDM2; this appears to be dependent on its binding to MDMX, as a RING mutant unable to bind to MDM2 was incapable of stabilizing MDM2 in the presence of Akt activity. Our data support that Akt-induced MDM2 stabilization is mediated at least in part through its effect on MDMX.
Moreover, we have shown that the transcriptional activity of p53 is suppressed by Akt in an MDMX-dependent manner. We have also demonstrated that transcriptional activation of p53 correlates with decreased MDMX levels in response to a PI 3-kinase inhibitor that suppresses Akt activity.
Recent studies have shown that MDMX is phosphorylated in response to DNA-damaging agents (43). The Chk1 and Chk2 kinases have also been shown to phosphorylate Ser367, leading to 14-3-3 binding (34–36, 38, 44). In both cases, the outcome differed: in Chk1-mediated phosphorylation, MDMX was translocated to the cytoplasm; in Chk2-mediated phosphorylation, MDMX was degraded (34–36, 38, 44). It is possible that the damage response is mediated through additional phosphorylation sites other than Ser367 and that, depending on the type of damage, certain sites will be modified, leading to different outcomes. We have observed the phosphorylation of Ser367 under both growth-promoting and DNA-damaging conditions, suggesting that phosphorylation of other sites may be required for MDMX degradation. Indeed, reports also show that Ser403 and Ser342 are phosphorylated in response to DNA damage; it is possible that these sites are more important in the damage response (34), although more investigations will be necessary to address this issue. Furthermore, 14-3-3 proteins bind as dimers requiring two phosphorylation sites, and the function of 14-3-3 binding to MDMX is not yet clear. Although we have not observed an effect from 14-3-3 on altered or enhanced binding of MDMX to MDM2 (data not shown), we believe that Akt-mediated 14-3-3 binding leads to the stabilization of the MDM2-MDMX complex, most likely by preventing degradation of the complex. We also did not observe a change in translocation of MDMX when it was bound to 14-3-3 under growth-promoting conditions (data not shown). It is possible that under different cellular conditions, other 14-3-3 binding sites may be generated on MDMX, which may result in different outcomes. It is also possible that a 14-3-3 dimer binds MDMX and tethers it to another yet unknown protein under growth-promoting conditions.
Our data fit with the general hypothesis that the Akt pathway is a pro-survival/pro-growth pathway. We have shown that Akt generates a 14-3-3 binding site on MDMX, which leads to its stabilization and consequent stabilization of MDM2. The MDM2-MDMX heterocomplex is therefore stabilized and more efficient at suppressing p53 activity.
Acknowledgments
We thank Drs. Ziv Radisavljevic, Brendan Manning, Haian Fu, and James DeCaprio for reagents and the Harvard NIEHS Center for Environmental Health Proteomics Facility. We thank our summer intern Benigno Varela for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01CA85679–02 (to Z.-M. Y.) and T32ES07155 (to V. L.-P. and M. M. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GST, glutathione S-transferase; RFD, RING finger domain; CA, constitutively active; DN, dominant-negative; KD, kinase-dead; IGF-1, insulin-like growth factor 1; PI, phosphatidylinositol; E3, ubiquitin-protein isopeptide ligase.
References
- 1.Vogelstein, B., Lane, D., and Levine, A. J. (2000) Nature 408 307–310 [DOI] [PubMed] [Google Scholar]
- 2.Michael, D., and Oren, M. (2003) Semin. Cancer Biol. 13 49–58 [DOI] [PubMed] [Google Scholar]
- 3.Finch, R. A., Donoviel, D. B., Potter, D., Shi, M., Fan, A., Freed, D. D., Wang, C. Y., Zambrowicz, B. P., Ramirez-Solis, R., Sands, A. T., and Zhang, N. (2002) Cancer Res. 62 3221–3225 [PubMed] [Google Scholar]
- 4.Parant, J., Chavez-Reyes, A., Little, N. A., Yan, W., Reinke, V., Jochemsen, A. G., and Lozano, G. (2001) Nat. Genet. 29 92–95 [DOI] [PubMed] [Google Scholar]
- 5.Jackson, M. W., and Berberich, S. J. (2000) Mol. Cell. Biol. 20 1001–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shvarts, A., Steegenga, W. T., Riteco, N., van Laar, T., Dekker, P., Bazuine, M., van Ham, R. C., van der Houven van Oordt, W., Hateboer, G., van der Eb, A. J., and Jochemsen, A. G. (1996) EMBO J. 15 5349–5357 [PMC free article] [PubMed] [Google Scholar]
- 7.Gu, J., Kawai, H., Nie, L., Kitao, H., Wiederschain, D., Jochemsen, A. G., Parant, J., Lozano, G., and Yuan, Z.-M. (2002) J. Biol. Chem. 277 19251–19254 [DOI] [PubMed] [Google Scholar]
- 8.Badciong, J. C., and Haas, A. L. (2002) J. Biol. Chem. 277 49668–49675 [DOI] [PubMed] [Google Scholar]
- 9.de Graaf, P., Little, N. A., Ramos, Y. F., Meulmeester, E., Letteboer, S. J., and Jochemsen, A. G. (2003) J. Biol. Chem. 278 38315–38324 [DOI] [PubMed] [Google Scholar]
- 10.Kawai, H., Lopez-Pajares, V., Kim, M. M., Wiederschain, D., and Yuan, Z.-M. (2007) Cancer Res. 67 6026–6030 [DOI] [PubMed] [Google Scholar]
- 11.Linares, L. K., Hengstermann, A., Ciechanover, A., Muller, S., and Scheffner, M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 12009–12014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danovi, D., Meulmeester, E., Pasini, D., Migliorini, D., Capra, M., Frenk, R., de Graaf, P., Francoz, S., Gasparini, P., Gobbi, A., Helin, K., Pelicci, P. G., Jochemsen, A. G., and Marine, J. C. (2004) Mol. Cell. Biol. 24 5835–5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riemenschneider, M. J., Buschges, R., Wolter, M., Reifenberger, J., Bostrom, J., Kraus, J. A., Schlegel, U., and Reifenberger, G. (1999) Cancer Res. 59 6091–6096 [PubMed] [Google Scholar]
- 14.Ramos, Y. F., Stad, R., Attema, J., Peltenburg, L. T., van der Eb, A. J., and Jochemsen, A. G. (2001) Cancer Res. 61 1839–1842 [PubMed] [Google Scholar]
- 15.Downward, J. (2004) Semin. Cell Dev. Biol. 15 177–182 [DOI] [PubMed] [Google Scholar]
- 16.Brazil, D. P., Park, J., and Hemmings, B. A. (2002) Cell 111 293–303 [DOI] [PubMed] [Google Scholar]
- 17.Wendel, H. G., De Stanchina, E., Fridman, J. S., Malina, A., Ray, S., Kogan, S., Cordon-Cardo, C., Pelletier, J., and Lowe, S. W. (2004) Nature 428 332–337 [DOI] [PubMed] [Google Scholar]
- 18.Obsil, T., Ghirlando, R., Anderson, D. E., Hickman, A. B., and Dyda, F. (2003) Biochemistry 42 15264–15272 [DOI] [PubMed] [Google Scholar]
- 19.You, H., Jang, Y., You-Ten, A. I., Okada, H., Liepa, J., Wakeham, A., Zaugg, K., and Mak, T. W. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 14057–14062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basu, S. (2003) Cell Cycle 2 69–70 [PubMed] [Google Scholar]
- 21.Hermeking, H. (2003) Nat. Rev. Cancer 3 931–943 [DOI] [PubMed] [Google Scholar]
- 22.Dougherty, M. K., and Morrison, D. K. (2004) J. Cell Sci. 117 1875–1884 [DOI] [PubMed] [Google Scholar]
- 23.Yaffe, M. B. (2002) FEBS Lett. 513 53–57 [DOI] [PubMed] [Google Scholar]
- 24.Ashcroft, M., Ludwig, R. L., Woods, D. B., Copeland, T. D., Weber, H. O., MacRae, E. J., and Vousden, K. H. (2002) Oncogene 21 1955–1962 [DOI] [PubMed] [Google Scholar]
- 25.Feng, J., Tamaskovic, R., Yang, Z., Brazil, D. P., Merlo, A., Hess, D., and Hemmings, B. A. (2004) J. Biol. Chem. 279 35510–35517 [DOI] [PubMed] [Google Scholar]
- 26.Gottlieb, T. M., Leal, J. F., Seger, R., Taya, Y., and Oren, M. (2002) Oncogene 21 1299–1303 [DOI] [PubMed] [Google Scholar]
- 27.Milne, D., Kampanis, P., Nicol, S., Dias, S., Campbell, D. G., Fuller-Pace, F., and Meek, D. (2004) FEBS Lett. 577 270–276 [DOI] [PubMed] [Google Scholar]
- 28.Mayo, L. D., and Donner, D. B. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 11598–11603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogawara, Y., Kishishita, S., Obata, T., Isazawa, Y., Suzuki, T., Tanaka, K., Masuyama, N., and Gotoh, Y. (2002) J. Biol. Chem. 277 21843–21850 [DOI] [PubMed] [Google Scholar]
- 30.Zhou, B. P., Liao, Y., Xia, W., Zou, Y., Spohn, B., and Hung, M. C. (2001) Nat. Cell Biol. 3 973–982 [DOI] [PubMed] [Google Scholar]
- 31.Sharp, D. A., Kratowicz, S. A., Sank, M. J., and George, D. L. (1999) J. Biol. Chem. 274 38189–38196 [DOI] [PubMed] [Google Scholar]
- 32.Kawai, H., Wiederschain, D., Kitao, H., Stuart, J., Tsai, K. K., and Yuan, Z.-M. (2003) J. Biol. Chem. 278 45946–45953 [DOI] [PubMed] [Google Scholar]
- 33.Gentiletti, F., Mancini, F., D'Angelo, M., Sacchi, A., Pontecorvi, A., Jochemsen, A. G., and Moretti, F. (2002) Oncogene 21 867–877 [DOI] [PubMed] [Google Scholar]
- 34.Pereg, Y., Lam, S., Teunisse, A., Biton, S., Meulmeester, E., Mittelman, L., Buscemi, G., Okamoto, K., Taya, Y., Shiloh, Y., and Jochemsen, A. G. (2006) Mol. Cell. Biol. 26 6819–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lebron, C., Chen, L., Gilkes, D. M., and Chen, J. (2006) EMBO J. 25 1196–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin, Y., Dai, M. S., Lu, S. Z., Xu, Y., Luo, Z., Zhao, Y., and Lu, H. (2006) EMBO J. 25 1207–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereg, Y., Shkedy, D., de Graaf, P., Meulmeester, E., Edelson-Averbukh, M., Salek, M., Biton, S., Teunisse, A. F., Lehmann, W. D., Jochemsen, A. G., and Shiloh, Y. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 5056–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okamoto, K., Kashima, K., Pereg, Y., Ishida, M., Yamazaki, S., Nota, A., Teunisse, A., Migliorini, D., Kitabayashi, I., Marine, J. C., Prives, C., Shiloh, Y., Jochemsen, A. G., and Taya, Y. (2005) Mol. Cell. Biol. 25 9608–9620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu, J., Chen, D., Rosenblum, J., Rubin, R. M., and Yuan, Z.-M. (2000) Mol. Cell. Biol. 20 1243–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawai, H., Wiederschain, D., and Yuan, Z.-M. (2003) Mol. Cell. Biol. 23 4939–4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Obenauer, J. C., Cantley, L. C., and Yaffe, M. B. (2003) Nucleic Acids Res. 31 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Migliorini, D., Denchi, E. L., Danovi, D., Jochemsen, A., Capillo, M., Gobbi, A., Helin, K., Pelicci, P. G., and Marine, J. C. (2002) Mol. Cell. Biol. 22 5527–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meulmeester, E., Maurice, M. M., Boutell, C., Teunisse, A. F., Ovaa, H., Abraham, T. E., Dirks, R. W., and Jochemsen, A. G. (2005) Mol. Cell 18 565–576 [DOI] [PubMed] [Google Scholar]
- 44.Chen, L., Gilkes, D. M., Pan, Y., Lane, W. S., and Chen, J. (2005) EMBO J. 24 3411–3422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stad, R., Ramos, Y. F., Little, N., Grivell, S., Attema, J., van der Eb, A. J., and Jochemsen, A. G. (2000) J. Biol. Chem. 275 28039–28044 [DOI] [PubMed] [Google Scholar]
- 46.Singh, R. K., Iyappan, S., and Scheffner, M. (2007) J. Biol. Chem. 282 10901–10907 [DOI] [PubMed] [Google Scholar]
- 47.Chene, P. (2003) Nat. Rev. Cancer 3 102–109 [DOI] [PubMed] [Google Scholar]
- 48.Momand, J., Jung, D., Wilczynski, S., and Niland, J. (1998) Nucleic Acids Res. 26 3453–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]